Abstract
Free full text

The carbohydrate-active enzymes database (CAZy) in 2013
Abstract
The Carbohydrate-Active Enzymes database (CAZy; http://www.cazy.org) provides online and continuously updated access to a sequence-based family classification linking the sequence to the specificity and 3D structure of the enzymes that assemble, modify and breakdown oligo- and polysaccharides. Functional and 3D structural information is added and curated on a regular basis based on the available literature. In addition to the use of the database by enzymologists seeking curated information on CAZymes, the dissemination of a stable nomenclature for these enzymes is probably a major contribution of CAZy. The past few years have seen the expansion of the CAZy classification scheme to new families, the development of subfamilies in several families and the power of CAZy for the analysis of genomes and metagenomes. This article outlines the changes that have occurred in CAZy during the past 5 years and presents our novel effort to display the resolution and the carbohydrate ligands in crystallographic complexes of CAZymes.
INTRODUCTION
Despite their similar chemical composition, carbohydrates can form an enormous number of combinations through the stereochemical variety of the hydroxyl groups that they carry, through the many possibilities to assemble monosaccharides one to another, and through the wealth of noncarbohydrate substituents that can decorate the resulting oligo- and polysaccharides. Complex carbohydrates are widely distributed in nature, where they mediate a multitude of biological functions, from carbon reserve, to structural molecules, or as the mediators of intra- and intercellular recognition within one organism or between organisms. The diversity of complex carbohydrates is controlled by a panel of enzymes involved in their assembly (glycosyltransferases) and their breakdown (glycoside hydrolases, polysaccharide lyases, carbohydrate esterases), collectively designated as Carbohydrate-Active enZymes (CAZymes). CAZymes have been classified in sequence-based families for >22 years (1–6) and this classification has become the standard of the field (7).
The first defining feature of CAZyme classification is that the families are defined based on significant amino acid sequence similarity with at least one biochemically characterized founding member (1). The consequence is that sequences that display too little similarity to ensure a significant alignment are not included, nor used to form putative families, as distant relatives of CAZymes may have other functions. Borderline cases are stored in the nonclassified section of each CAZyme category, awaiting biochemical characterization. A second defining feature is that our classification is made module by module. CAZymes are frequently modular proteins with a catalytic module harbouring a variable number of other discrete modules, which can be either catalytic or not. Thus a modular CAZyme can be assigned to several families if its constitutive modules belong to separate families. The third important feature is that we only analyse systematically protein sequences released in the daily releases of GenBank (ftp://ftp.ncbi.nih.gov/genbank/daily-nc), to avoid analysing unfinished sequences that may change accession number.
As early as 1991, it was noted that the sequence-based families of glycoside hydrolases grouped together enzymes of different substrate specificities (i.e. enzymes with ‘different’ EC numbers) (1) demonstrating that the acquisition of novel specificity has been commonplace during evolution. This feature was subsequently noted for the other classes of CAZymes (4,6). The processes by which a novel substrate specificity was acquired from a common ancestor leave detectable traces in the sequence of contemporary proteins. Thus, unexpectedly, the usual drawback of carbohydrates (their chemical resemblance) is at the origin of their success in the postgenomic era: CAZymes need to be specific to perform their biological functions. While the precise specificity of DNAses, RNAses, proteases and esterases is difficult or impossible to derive from their sequence alone, the CAZyme classification system allows in some cases the prediction of the broad category of carbohydrate substrate, based on the assignment to a family (8). This carries the potential to infer the glycobiological profile of an organism (or a community thereof) based on DNA sequence. However, the occurrence of enzymes that act on different substrates in the same family is a significant problem for the automated functional annotation of CAZyme-related genes. This can sometimes be overcome by the definition of subfamilies within families (9,10) (see below), but our current knowledge of the sequence-to-specificity relationships in CAZymes families is still largely insufficient and unevenly distributed for many families to allow unsupervised automated substrate prediction.
The Carbohydrate-Active Enzymes database (CAZy; http://www.cazy.org) was launched in 1999 to provide online and constantly updated access to the family classification of CAZymes. Coupled to the CAZypedia encyclopaedic resource (http://www.cazypedia.org), CAZy is the only comprehensive resource that correlates the sequence, structure and molecular mechanism of CAZymes. CAZy was presented in this journal in 2009 (11) and the present article outlines the changes that have been implemented in CAZy during the past 5 years.
WEBSITE DESIGN
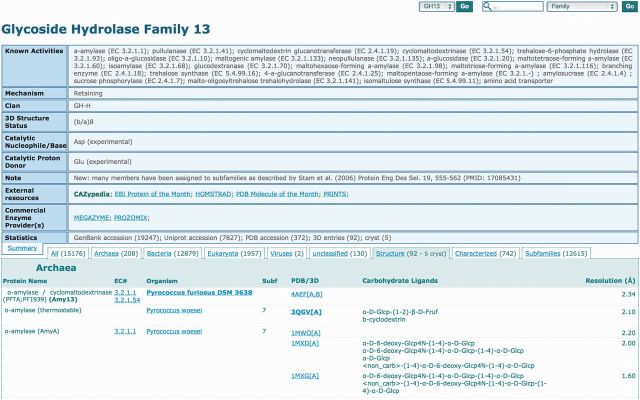
In March 2011, the website interface was deeply redesigned both in appearance (new layout, new colours and new logo) and in content. Thus new sections and new links have been added to commercial providers that list their products following the CAZy nomenclature. Other additions cover scientific meetings relevant to CAZymes, positions available and a ‘what’s new’ section that provides news on changes in the CAZy database. More interactivity in the display of information associated with each family was introduced (Figure 1). In particular, each family has now a specific tab, which lists those individual CAZymes that we believe have been experimentally characterized. Because the number of entries in several families had become impractical, the display was modified to just show the header for each family along with a series of tabs for access to subsets (All, Archea, Bacteria, Eukaryota, unclassified, Structure, Characterized). Each tab displays 1000 entries per page, except for the tab listing the characterized enzymes, where only 100 entries are shown per page. The search tool was also revisited and one can now search the entire site or specific fields such as CAZy family, taxonomic identifier, organism name, protein name, accessions in different databases (GenBank, Uniprot and Protein Data Bank (PDB)), known activities, EC number, mechanisms or clan.
NOVEL ENZYME CLASS
Because lignin is invariably found together with polysaccharides in the plant cell wall and because lignin fragments are likely to act in concert with polysaccharide lytic mono-oxygenases (LPMO), families of lignin degradation enzymes and of LPMOs have been used to define a new CAZy class that we have named ‘Auxiliary Activities’ to accommodate a broad range of enzyme mechanisms and substrates related to lignocellulose conversion (12).
DATABASE GROWTH
At the date of submission of this article, CAZy reports sequence information on almost 340 000 CAZymes, a staggering 225% increase compared with 5 years ago (Table 1). During the same period, the number of biochemically characterized CAZymes has grown by only 30% to 12 700 and the number of CAZymes with 3D structures has grown by ~78% (Table 1). Despite this growth, only ~1400 (0.4%) of the 340 000 CAZymes have a 3D structure solved to date. The past 5 years have seen the number of families covered by CAZy grow slowly to >330 at present. Five years ago the number of genome sequences analysed in CAZy was 750 (11). This number is now greater than 2800 (see below), representing a 3.8-fold increase. The continuously growing gap between the number of sequences and the number of biochemically or structurally characterized CAZymes is a direct consequence of the avalanche of genome sequences resulting from modern sequencing technologies combined with the much lower pace of experimental characterization of gene products. This gap would even be more considerable if one was to search and list CAZymes in nonfinished genomes.
Table 1.
Growth of the CAZy database during the past 5 years
| Protein class | Sequences Sept-2013 | Dec-2008 | Characterized Sept-2013 | Dec-2008 | With structure Sept-2013 | Dec-2008 |
|---|---|---|---|---|---|---|
| GH | 159 274 | 46 654 | 9221 | 6805 | 817 | 475 |
| GT | 119 910 | 40 863 | 1936 | 1846 | 139 | 83 |
| PL | 4043 | 1301 | 336 | 262 | 51 | 34 |
| CE | 15 856 | 5083 | 275 | 212 | 74 | 43 |
| CBM | 32 259 | 9210 | 663 | 570 | 280 | 166 |
| AA | 5801 |  464a 464a | 299 | 71a | 58 | 3a |
| Total | 337 143 | 103 111 | 12 730 | 9695 | 1419 | 801 |
aNumbers estimated from the literature: the AA category did not exist in December 2008.
DATABASE CONTENT: SUBFAMILIES
The occurrence of enzymes that act on different substrates in the same family prevents the straightforward functional annotation of CAZyme-related genes. The division of CAZyme families into subfamilies based on phylogenetic analysis has been explored as a possible approach to improve the relationship between sequence and specificity. Subfamily classification of GH5, GH13, GH30 and all of the PL families has shown that the majority of the defined subfamilies are monospecific, thus indicating that the correlation of substrate specificity with sequences is significantly better at the subfamily level than the family level (9,10,13). An additional benefit of the division into subfamilies is the identification of currently uncharacterized subfamilies that can subsequently be analysed experimentally to unveil potential new activities. Subfamilies are currently displayed for families GH5, GH13, GH30, AA1–AA5 and for all PL families. Many more families are currently evaluated for subfamily definitions. Care is taken that the subfamilies are defined in a robust manner to avoid confusion that would arise from constant redefinitions and resulting different naming conventions. We prefer to let the subfamilies ‘mature’ until we feel that the subfamily quality and stability is sufficient for public release.
DATABASE CONTENT: GENOMES
The collection of carbohydrate-active enzymes encoded by the genome of an organism (‘CAZome’) provides an insight into the nature and extent of the metabolism of complex carbohydrates of the species. The CAZomes of free-living organisms typically correspond to 1–5% of the predicted coding sequences. Extremely reduced CAZomes are characteristic of species with a strict intracellular parasitic lifestyle. Because of the massive chemical, structural and functional variability of carbohydrates, CAZome comparisons can highlight the adaptation of the CAZymes repertoire of species to their environment (14,15).
Since 2011, in addition to giving the family distribution, the new CAZy website displays the complete list of putative CAZymes (with accession numbers) of each genome that was analysed. At present, CAZy covers >2800 genomes in the following kingdoms: Bacteria (2351), Archea (158), Eukaryota (73), Viruses (240). The CAZomes listed in the CAZy website correspond to protein models of finished genomes, i.e. with proteins released in the daily releases of GenBank (ftp://ftp.ncbi.nih.gov/genbank/daily-nc). In a few cases, genomes with protein models not released as finished entries in GenBank but publically available, have been analysed and are presented in CAZy. However, for these few cases, the display only shows the number of proteins in each family, but does not feature the actual list of proteins.
Genomes are analysed using the CAZy pipeline, which combines Blast and HMM tools to compare protein models, respectively, with sequence and profile libraries created from the sequences of the catalytic and noncatalytic modules of the CAZy database. This is followed by a manual inspection by expert curators to resolve borderline cases (11). Our methodology provides coherent, expert and comparable sets of annotations. In this respect, one should note that the correspondence between CAZy families and those in PFAM (16)/INTERPRO (17) or DBCAN (18) is far from perfect. This is due to a variety of reasons that include different strategies, different thresholds, different goals, different methods, different training sets and different degrees of expert curation. An unfortunate consequence is that the CAZyme analysis of a genome performed with one method usually cannot be compared with that done with another.
There are two ways to get a genome analysed by CAZy: if the genome and encoded proteins are deposited as finished entries in GenBank (or EMBL or DDBJ) they will be analysed by our daily routines. Alternatively, if one wishes to perform a CAZy analysis before deposition to GenBank (or EMBL or DDBJ), one should approach us for collaboration. Metagenomic data are analysed exclusively in collaboration due to their usual large size.
DISPLAY OF STRUCTURAL INFORMATION
The CAZy database is not only used by those who wish to analyse genomes, but also by structural biologists who study the molecular details of substrate recognition by CAZymes. Until September 2013, the only information available in the structure pages of CAZy was the accession and macromolecule chain name(s) in the PDB (http://www.rcsb.org) (19). We have made a series of developments to provide additional information relevant to the 3D structure of CAZymes such as the resolution (for crystal structures) and a description of the carbohydrate ligands found in the CAZyme binding sites.
The resolution information is straightforward to generate, as it is present in the PDB files of structures solved by x-ray crystallography. When the resolution information is unavailable in the PDB file, the type of experimental method by which the structure was solved is given instead (powder diffraction or nuclear magnetic resonance).
On the other hand, the PDB does not provide any option to perform a comprehensive search for carbohydrate structures found in CAZyme binding sites and, unlike proteins or nucleic acids, the nomenclature for carbohydrate residues within PDB files is not standardized (20). In addition, the information on how the isolated carbohydrate residues are linked to each other is not described in PDB files. We thus extract the carbohydrate ligand information from PDB files using PDB-care (http://www.glycosciences.de/tools/pdb-care/) (21,22). The carbohydrate molecules covalently linked to an Asn or a Ser/Thr residue were discarded to eliminate N- and O-glycans to identify the carbohydrate ligands bound to CAZyme active sites. The latter are shown in the structure pages of CAZy following their IUPAC nomenclature.
Not all carbohydrate structures are susceptible to automated description by PDB-care. In a number of cases, we have manually curated and provided IUPAC descriptions for structures that are unsuitable to PDB-care: (i) nonreducing glycans (cyclodextrins, sucrose and sucrose derivatives, trehalose, kestose, raffinose, nystose, etc.), (ii) ligands that contain both carbohydrate and noncarbohydrate moieties such as acarbose and acarbose derivatives, (iii) sulfur-containing oligosaccharides, (iv) fluorine-containing carbohydrates and (v) oligosaccharides containing 3,6-anhydro bridges. Table 2 displays examples of the manually handled cases. In addition, automated scripts have been devised to handle ~180 carbohydrate analogues that we denote <carb_like_ligandref> where ligandref corresponds to the three-letter ligand name given by the PDB. For instance, the carbohydrate-like inhibitor 1-deoxynojirimycin appears as <carb_like_NOJ>. The structural biology community is invited to contact us to report the possible errors that might have slipped through our curation process.
Table 2.
Examples of carbohydrate ligands treated manually
| Category | Common name | Display in CAZy structure pages | Example of PDB file |
|---|---|---|---|
| Nonreducing oligosaccharides | α-cyclodextrin | α-cyclodextrin | 3EDF |
| β-cyclodextrin | β-cyclodextrin | 3CGT | |
| Sucrose | α-D-Glcp-(1-2)-β-D-Fruf | 4FFH | |
| Raffinose | α-D-Galp-(1-6)-α-D-Glcp-(1-2)-β-D-Fruf | 1W2T | |
| Kestose | α-D-Glcp-(1-2)-β-D-Fruf-(1-2)-β-D-Fruf | 3LDR | |
| Nystose | α-D-Glcp-(1-2)-β-D-Fruf-(1-2)-β-D-Fruf-(1-2)-β-D-Fruf | 3LEM | |
| Thio-oligosaccharides | Thio-cellobiose | β-D-Glcp-(1-4)-β-D-Glcp4S | 4IPM |
| Thio-laminaribiose | β-D-Glcp-(1-3)-β-D-Glcp3S | 1J8V | |
| Thio-xylopentaose | β-D-Xylp-(1-4)-β-D-Xylp4S-(1-4)-β-D-Xylp4S-(1-4)-β-D-Xylp4S-(1-4)- β-D-Xylp4S | 3CUJ | |
| α-methyl-thio-cellopentaoside | β-D-Glcp-(1-4)-β-D-Glcp4S-(1-4)-β-D-Glcp4S-(1-4)-β-D-Glcp4S-(1-4)- α-D-Glcp4S-(1-1)-methyl | 1H5V | |
| Fluoro-oligosaccharides | 5-fluoro-β-D-glucose | β-D-Glcp5F | 4AMX |
| 2-deoxy-2-fluoro-α-D-glucose | α-D-Glcp2F | 1UYQ | |
| 5-fluoro-β-D-xylose | β-D-Xylp5F | 2XVK | |
| 3,6-anhydro oligosaccharides | Neoagarohexaose | α-L-3,6-anhydro-Galp-(1-3)-β-D-Galp-(1-4)-α-L-3,6-anhydro-Galp-(1-3)- β-D-Galp-(1-4)-α-L-3,6-anhydro-Galp-(1-3)-β-D-Galp | 2CDO |
| Porphyran/agarose hexasaccharide | α-L-Galp6SO3-(1-3)-α-D-Galp-(1-4)-α-L-3,6-anhydro-Galp-(1-3)- β-D-Galp-(1-4)-α-L-Galp6SO3-(1-3)-α-D-Galp | 4AW7 | |
| Agarooctaose | α-L-3,6-anhydro-Galp-(1-3)-β-D-Galp-(1-4)-α-L-3,6-anhydro-Galp-(1-3)- β-D-Galp-(1-4)-α-L-3,6-anhydro-Galp-(1-3)-β-D-Galp-(1-4)- α-L-3,6-anhydro-Galp-(1-3)-β-D-Galp | 4ATF | |
| Acarbose and its derivatives | Acarbose | <non_carb>-(1-4)-α-D-6-deoxy-Glcp4N-(1-4)-α-D-Glcp-(1-4)-β-D-Glcp | 3ZOA |
| Acarbose-derived trisaccharide | <non_carb>-(1-4)-α-D-6-deoxy-Glcp4N-(1-4)-α-D-Glcp | 1XCW | |
| Acarbose-derived pentasaccharide | α-D-6-deoxy-Glcp4N-(1-4)-α-D-Glcp-(1-4)-<non_carb>-(0-4)- α-D-6-deoxy-Glcp4N-(1-4)-α-D-Glcp-(1-4)-β-D-Glcp | 1PIG |
As of September 2013, >1400 CAZymes and modules thereof have a known 3D structure, corresponding to almost 6000 PDB entries out of which ~1500 carbohydrate (or carbohydrate analogue) ligands are now identified and presented in the structure tab of each CAZy family.
FUTURE DIRECTIONS
CAZy is a knowledge-based resource that aims to link the sequence, the specificity and the 3D structural features of CAZymes. How these enzymes achieve selective recognition of target substrates that display only subtle stereochemical differences is key to prediction of substrate specificity. While this is already achievable for a few subfamilies, we are still a long way from a reliable automated substrate (and/or product) prediction for all CAZymes encoded by a genome. We believe that subfamily-based target selection for experimental investigation of CAZymes will progressively fill the knowledge gap that will allow reliability in future functional predictions.
FUNDING
Agence Nationale de la Recherche, grant BIP:BIP [ANR-10-BINF-03-04]. Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
We thank Thomas Lütteke (Justus-Liebig University Gießen, Institute of Veterinary Physiology and Biochemistry, Germany) for providing us the PDB-care program, and Kirk M. Schnorr (Novozymes, Bagsvaerd, Denmark) for his help with proof reading this manuscript. Bernard Henrissat is a Honorary Professor of Glycomics at the Faculty of Health and Medical Sciences, University of Copenhagen, Denmark.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkt1178
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/42/D1/D490/3617205/gkt1178.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Unique Fn3-like biosensor in σ<sup>I</sup>/anti-σ<sup>I</sup> factors for regulatory expression of major cellulosomal scaffoldins in Pseudobacteroides cellulosolvens.
Protein Sci, 33(11):e5193, 01 Nov 2024
Cited by: 1 article | PMID: 39470320
Genome-wide analysis of the Amorphophallus konjac AkCSLA gene family and its functional characterization in drought tolerance of transgenic arabidopsis.
BMC Plant Biol, 24(1):1033, 31 Oct 2024
Cited by: 0 articles | PMID: 39478464 | PMCID: PMC11526714
Environmental Driving of Adaptation Mechanism on Rumen Microorganisms of Sheep Based on Metagenomics and Metabolomics Data Analysis.
Int J Mol Sci, 25(20):10957, 11 Oct 2024
Cited by: 0 articles | PMID: 39456741 | PMCID: PMC11508146
Comparative genomic analysis of pathogenic factors of Listeria spp. using whole-genome sequencing.
BMC Genomics, 25(1):935, 07 Oct 2024
Cited by: 0 articles | PMID: 39375592 | PMCID: PMC11457443
Elucidation of the noncovalent interactions driving enzyme activity guides branching enzyme engineering for α-glucan modification.
Nat Commun, 15(1):8760, 09 Oct 2024
Cited by: 0 articles | PMID: 39384762 | PMCID: PMC11464733
Go to all (3,036) article citations
Other citations
Wikipedia (Showing 10 of 78)
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_76
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_78
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_79
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_8
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_80
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_81
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_85
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_88
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_89
- https://en.wikipedia.org/wiki/Glycoside_hydrolase_family_9
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (Showing 16 of 16)
-
(1 citation)
PDBe - 1J8VView structure
-
(1 citation)
PDBe - 4AMXView structure
-
(1 citation)
PDBe - 3ZOAView structure
-
(1 citation)
PDBe - 2XVKView structure
-
(1 citation)
PDBe - 3CGTView structure
-
(1 citation)
PDBe - 4ATFView structure
-
(1 citation)
PDBe - 4IPMView structure
-
(1 citation)
PDBe - 1PIGView structure
-
(1 citation)
PDBe - 1UYQView structure
-
(1 citation)
PDBe - 2CDOView structure
-
(1 citation)
PDBe - 3EDFView structure
-
(1 citation)
PDBe - 4FFHView structure
-
(1 citation)
PDBe - 1XCWView structure
-
(1 citation)
PDBe - 1H5VView structure
-
(1 citation)
PDBe - 4AW7View structure
-
(1 citation)
PDBe - 1W2TView structure
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database.
Glycobiology, 20(12):1574-1584, 09 Aug 2010
Cited by: 200 articles | PMID: 20696711
The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics.
Nucleic Acids Res, 37(database issue):D233-8, 05 Oct 2008
Cited by: 2963 articles | PMID: 18838391 | PMCID: PMC2686590
The carbohydrate-active enzyme database: functions and literature.
Nucleic Acids Res, 50(d1):D571-D577, 01 Jan 2022
Cited by: 589 articles | PMID: 34850161 | PMCID: PMC8728194
Carbohydrate binding modules: Compact yet potent accessories in the specific substrate binding and performance evolution of carbohydrate-active enzymes.
Biotechnol Adv, 73:108365, 26 Apr 2024
Cited by: 0 articles | PMID: 38677391
Review