Abstract
Free full text

Placing the HIRA histone chaperone complex in the chromatin landscape
Summary
The HIRA chaperone complex, comprised of HIRA, UBN1 and CABIN1, collaborates with histone-binding protein ASF1a to incorporate histone variant H3.3 into chromatin in a DNA replication-independent manner. To better understand HIRA’s function and mechanism, we integrated HIRA, UBN1, ASF1a and histone H3.3 ChIP-seq and gene expression analyses. Most HIRA-binding sites co-localize with UBN1, ASF1a and H3.3 at active promoters and active and weak/poised enhancers. At promoters, binding of HIRA/UBN1/ASF1a correlates with the level of gene expression. HIRA is required for deposition of histone H3.3 at its binding sites. There are marked differences in nucleosome and co-regulator composition at different classes of HIRA-bound regulatory site. Underscoring this, we report novel physical interactions between the HIRA complex and transcription factors, a chromatin insulator and an ATP-dependent chromatin-remodelling complex. Our results map the distribution of the HIRA chaperone across the chromatin landscape and point to different interacting partners at functionally distinct regulatory sites.
Introduction
The HIRA chaperone complex, comprised of HIRA/UBN1/CABIN1, collaborates with histone binding protein ASF1a to incorporate the histone variant H3.3 into chromatin in a DNA replication-independent manner (Loppin et al., 2005; Ray-Gallet et al., 2002; Tagami et al., 2004). HIRA is required for early embryo development (Roberts et al., 2002; Szenker et al., 2012), and histone H3.3 is mutated in human cancer (Schwartzentruber et al., 2012; Wu et al., 2012).
Histone H3.3 is enriched at nucleosomes at transcription start sites (TSS) of genes, at enhancers and gene bodies of actively transcribed genes (Ahmad and Henikoff, 2002; Goldberg et al., 2010; Jin et al., 2009). Histone H3.3 contributes to nucleosome destabilization (Jin and Felsenfeld, 2007), and so is thought to facilitate nucleosome dynamics associated with transcription activation and ongoing transcription. The HIRA protein is required for deposition of histone H3.3 at many of these regions (Goldberg et al., 2010; Ray-Gallet et al., 2011). Consistent with this, HIRA is required for gene activation in some contexts (Dutta et al., 2010; Placek et al., 2009; Yang et al., 2011). Interestingly, the HIRA complex and its orthologs, together with histone H3.3, are also involved in chromatin silencing (Sherwood et al., 1993; van der Heijden et al., 2007).
The distribution of the HIRA chaperone complex across the epigenome is not known and there is a paucity of partner proteins known to participate in its diverse functions. To overcome this, we performed integrated ChIP-seq and gene expression analyses, and used this analysis to identify proteins that physically interact with the HIRA complex in chromatin regulation.
Results
Analysis of genome-wide distribution of HIRA, UBN1, ASF1a and histone H3.3
To gain insight into the function and regulation of the HIRA chaperone at distinct genomic sites, we performed ChIP-seq of endogenous HIRA, UBN1 and ASF1a in human HeLa cells. Analysis of the aligned reads yielded 8,296 HIRA peaks, 62,712 UBN1 peaks and 64,550 ASF1a peaks, compared to input DNA. 74% of HIRA peaks (6,147 out of 8,296) were co-occupied by both UBN1 and ASF1a (Fig. 1A and Table S1). To confirm these results, we performed anti-HIRA ChIP followed by quantitative PCR (ChIP-qPCR) at a single co-occupied HIRA/UBN1/ASF1a peak and flanking regions. This analysis confirmed enrichment of HIRA at the peak, relative to the flanking regions (Fig. 1B). Specific enrichment at this site was also observed with antibodies to UBN1 and ASF1a (Fig. 1C) as well as with 4 individual monoclonal antibodies to HIRA (Fig. S1A) by ChIP-qPCR. Indeed, across the whole genome, HIRA binding regions were coincident with UBN1 and ASF1a binding regions (Fig. 1D). We also performed ChIP of HIRA followed by semi-quantitative PCR at 9 distinct locations selected at random from the list of 6,147 HIRA/UBN1/ASF1a peaks; all 9 regions demonstrated enrichment in HIRA ChIP, compared to non-specific antibody (anti-GFP) (Fig. 1E, Table S2). Taken together, these data show that the core HIRA complex and ASF1a co-occupies at least several thousand discrete sites across the genome of proliferating human cells.
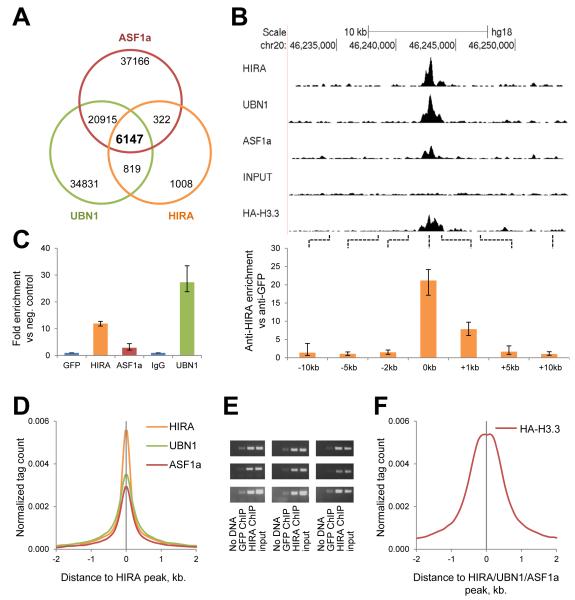
A) Venn diagram of overlap between HIRA, UBN1 and ASF1a peaks reveals a subset of 6,147 co-occupied regions. See also Table S1.
B) Representative HIRA, UBN1, ASF1a and HA-H3.3 ChIP-seq tracks and ChIP-qPCR validation of HIRA enrichment at peak compared to flanking regions. Error-bars show standard errors. See also Table S2.
C) ChIP-qPCR validation of the complex-binding region shown on panel (B) using a cocktail of monoclonal antibodies to HIRA or ASF1a or a rabbit polyclonal antibody to UBN1. Error-bars show standard errors. See also Fig. S1A and Table S2.
D) Normalized density of ChIP-seq tags of HIRA, UBN1 and ASF1a in a 4 kb window centered on a composite of all HIRA peaks.
E) ChIP-PCR validation of 9 different HIRA-binding regions identified by ChIP-seq. See also Table S2.
F) Normalized density of ChIP-seq tags of HA-H3.3 in a 4 kb window centered on composite of all HIRA/UBN1/ASF1a peaks.
Although HIRA is required for accumulation of histone H3.3 at many sites throughout the genome (Dutta et al., 2010; Goldberg et al., 2010; Placek et al., 2009; Yang et al., 2011), the genomic distribution of newly deposited histone H3.3 and the HIRA complex (nor ASF1a) have not been directly compared. Therefore, we assessed the genome-wide distribution of newly incorporated histone H3.3 by anti-HA ChIP-seq on HeLa cells after a short pulse of HA-histone H3.3 expression (less than 13 hours). By this approach, we identified 110,213 sites of HA-H3.3 deposition across the genome. Strikingly, 86% of all HIRA binding sites and 95% of co-occupied HIRA/UBN1/ASF1a sites were also enriched for HA-H3.3. Moreover, across the whole genome, co-occupied HIRA/UBN1/ASF1a sites were generally coincident with peaks of HA-H3.3 deposition (Fig. 1B, F). Furthermore, peaks of histone H3.3 were more pronounced at peaks of HIRA/UBN1/ASF1a than elsewhere in the genome (Fig. S1B). These results support the view that the HIRA complex and ASF1a collaborate to deposit histone H3.3 at their specific co-localization sites.
HIRA binds to chromatin at four distinct classes of genomic loci
To further characterize chaperone function we determined the distribution of regions co-occupied by HIRA, UBN1 and ASF1a between promoter, genic and intergenic regions: 22% of the 6,147 HIRA/UBN1/ASF1a peaks mapped to gene promoters and the rest to either gene body (38%) or intergenic regions (40%) (Fig. S1C, D).
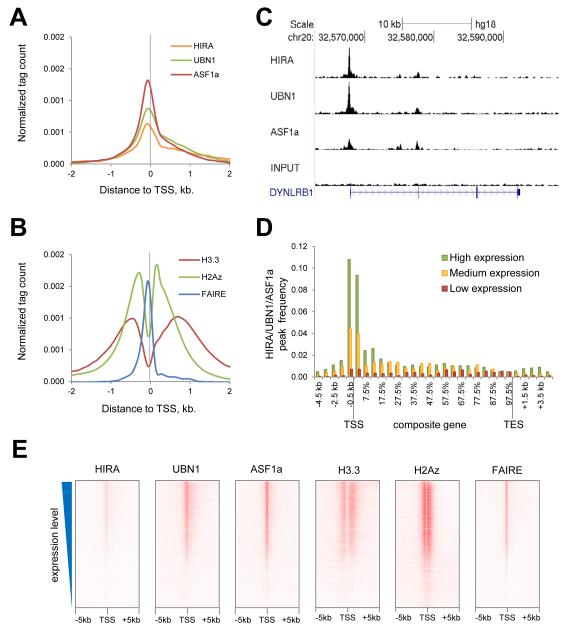
To better characterize HIRA’s binding across the genome, we performed unsupervised clustering of all 8,296 HIRA-binding sites according to their overlap with UBN1, ASF1a, HA-H3.3 and other chromatin proteins and genomic features annotated in HeLa cells in publicly available databases. In addition, we performed formaldehyde-assisted isolation of regulatory elements (FAIRE) to identify accessible regions of DNA in the same cells, and incorporated this into the analysis (Table S1). This analysis separated HIRA-binding sites into four distinct clusters, 1, 2, 3 and 4 (Fig. 2A). Clusters 1-3 were comprised largely of HIRA peaks co-occupied by UBN1 and ASF1a, while cluster 4 was predominantly comprised of HIRA-only peaks, lacking UBN1 and ASF1a.

A) Unsupervised clustering identifies 4 distinct clusters of HIRA peaks.
B) Heatmaps of normalized density of ChIP-seq tags of HA-H3.3, H2Az and FAIRE in a 10 kb window centered on HIRA peak. HIRA peaks are arranged in the same ordered as in panel (A) and grouped in four clusters. See also Fig. S1.
C) siRNA-mediated knock down of HIRA impairs HA-H3.3 incorporation at representative regions of clusters 1, 2 and 3 as measured by HA ChIP-qPCR. Error-bars show standard errors. See also Fig. S2A for confirmation of efficient protein knockdown and Table S2 for regions location.
Cluster 1 is enriched in H3K4me1/3, H3K27ac, H2Az, p300 and transcription factor c-Myc (Fig. 2A). Almost all regions in this cluster are also FAIRE positive and DNase hypersensitive (DNaseHS), and exhibit very low overlap with RNA polymerase II, CpG islands or promoters of annotated genes. Such a binding pattern is best associated with active enhancers (Ernst et al., 2011; Heintzman et al., 2009; Rada-Iglesias et al., 2011), suggesting that cluster 1 represents HIRA/UBN1/ASF1a binding at these regulatory elements.
Cluster 2 shows a strong overlap with gene promoters, RNA polymerase II, CpG islands, H2Az, H3K4me3 (but not H3K4me1), H3K27ac and transcription factor c-Myc (Fig. 2A). Almost all regions in this cluster are FAIRE positive and DNaseHS. This signature is most consistent with promoters of actively transcribed genes (Ernst et al., 2011; Heintzman et al., 2009; Rada-Iglesias et al., 2011).
Cluster 3 is enriched in H3K4me1, but, compared to cluster 1, shows less overlap with p300, H3K27ac and c-Myc and less DNaseHS and FAIRE (Fig. 2A). Moreover, cluster 3 shows minimal overlap with promoters, CpG islands and RNA polymerase II. Consequently, cluster 3 is most consistent with weak or poised enhancers (Ernst et al., 2011; Heintzman et al., 2009; Rada-Iglesias et al., 2011).
Cluster 4 is comprised largely of the 1008 genomic sites that bind HIRA, but neither UBN1 nor ASF1a (HIRA-only peaks) (Fig. 1A and and2A).2A). Relaxing the stringency of the algorithm for calling ASF1a and UBN1 peaks failed to generate overlap between UBN1 and ASF1a peaks and all the HIRA peaks (Fig. S1E). Also, independent re-analysis of read numbers confirmed that those HIRA peaks which scored negative for both ASF1a and UBN1 showed only very few reads in UBN1 and ASF1a ChIPs at these regions (Fig. S1F). These analyses support the notion that these apparent HIRA-only peaks genuinely lack enrichment of UBN1 and ASF1a, and so are qualitatively distinct from the co-occupied HIRA/UBN1/ASF1a peaks. Consistent with this idea, unlike the HIRA/UBN1/ASF1a-bound regions of the genome, HIRA-only peaks were largely FAIRE and DNase HS-negative, lacked active histone marks (H3K4me3, H3K27ac and H3K9/14ac) (Fig. 2A and Fig. S1G) and overlapped poorly with many chromatin regulatory proteins analyzed as part of the ENCODE project (Fig. S1H and Table S3). Based on the analysis in Fig. 2A, a large proportion of cluster 4 HIRA-binding sites contains detectable H2Az, but a more quantitative analysis, evaluating the number of reads and not simply the presence or absence of binding, showed these sites to be very poor binders of H2Az (Fig. 2B). Most surprisingly, in striking contrast to HIRA/UBN1/ASF1a peaks, the HIRA-only sites were also largely depleted of histone H3.3 (Fig. 2A and Fig. S1G, I). Taken together, these data suggest that HIRA binds to some sites in the genome in the absence of UBN1 and ASF1a and without steady state enrichment of histone H3.3. Together, this indicates a very different, but currently unknown, function for HIRA at these sites.
To gain further insight into HIRA clusters 1-4, we also plotted quantitative heat maps of the abundance of newly synthesized HA-histone H3.3, H2Az and FAIRE accessibility, 5kb either side of the centered HIRA peak, and with the HIRA-binding loci vertically ordered as in Fig. 2A (Fig. 2B). These plots underscored the difference between clusters 1-4. At cluster 2, as expected for promoters, HA-H3.3 and H2Az both showed a bimodal distribution, indicative of H3.3/H2Az-containing nucleosomes either side of the nucleosome-free, FAIRE-positive TSS (Jin et al., 2009). Interestingly, the nucleosome free region was positioned very close to the centered HIRA peak (Fig. 2B). Thus, at promoters, HIRA/UBN1/ASF1a is essentially localized to the nucleosome-free TSS. While the active enhancers in cluster 1 were also characterized by coincident HIRA/UBN1/ASF1a and FAIRE peaks, the distribution of HA-H3.3 and H2Az was quite different to the promoters in cluster 2 (Fig. 2B). Cluster 1 HIRA/UBN1/ASF1a peaks were less rich in H2Az, and the bimodal distribution of HA-H3.3 and H2Az was less apparent. The weak/poised enhancers of cluster 3 were also characterized by coincident HIRA/UBN1/ASF1a, FAIRE and HA-H3.3 peaks (Fig. 2B). As at cluster 1, the HA-H3.3 was localized to a monomodal, not bimodal, peak. There was relatively little H2Az at cluster 3.
Knock down of HIRA decreased total incorporation of histone H3.3 into chromatin, as judged by total chromatin-bound (insoluble in Triton X-100) histone H3.3 and total DNA co-precipitated in anti-HA-H3.3 ChIP (Fig. S2A, B). At specific regions, knock down of HIRA specifically blocked binding of ectopically expressed epitope-tagged H3.3 at TSS and promoters (cluster 2) (Fig. 2C and Fig. S2B), with a much lesser effect on total endogenous H3 (H3.1, H3.2 and H3.3) at the same sites (Fig. S2B). Similarly, loss of HIRA resulted in greatly reduced incorporation of histone H3.3 at cluster 1 and 3 enhancer regions (Fig. 2C).
These results indicate that the HIRA complex and ASF1a co-localize with histone H3.3 at diverse regulatory elements throughout the genome (active promoters, strong enhancers and weak/poised enhancers) and is required for deposition of H3.3 at these sites. Significantly, the localization of histone H3.3 and H2Az differs quantitatively and qualitatively between these different classes of HIRA/UBN1/ASF1a-bound regulatory element.
Binding of HIRA/UBN1/ASF1a at TSS correlates with gene expression
The previous comparison of HIRA-binding to nucleosome composition and positioning (Fig. 2B) suggested that cluster 2 is comprised of HIRA/UBN1/ASF1a bound to gene promoter TSS. Indeed, when HIRA, UBN1 and ASF1a binding was analyzed at a composite of all genes, the three components were found to bind just upstream of the TSS, coincident with the FAIRE-positive nucleosome-free region between the H3.3/H2Az nucleosomes (Fig. 3A-B). The composite plot in Fig. 3A reflects the average distribution at individual genes (Fig. 3C). Significantly, the complex was markedly enriched at the promoters of highly expressed genes, but almost absent from the promoters of repressed genes (Fig. 3D), demonstrating a positive correlation between HIRA/UBN1/ASF1a-binding and gene expression level. In this respect, HIRA/UBN1/ASF1a-binding was very similar to H3.3, H2Az and FAIRE accessibility (Fig. 3E). Taken together, these results show that a proportion of the HIRA/UBN1/ASF1a complex is localized to TSS of highly expressed genes.
A) Normalized density of ChIP-seq tags of HIRA, UBN1, ASF1a in a 4 kb window of a composite promoter centered on TSS of all genes.
B) Normalized density of ChIP-seq tags of H3.3, H2Az, FAIRE in a 4 kb window of a composite promoter centered on TSS of all genes.
C) Example of characteristic distribution of HIRA, UBN1 and ASF1a across the DYNLRB1 gene.
D) Composite distribution of co-occupied HIRA/UBN1/ASF1a peaks across high, medium or low expressed genes. Probes on the Affymetrix expression array were rank ordered by average expression level in proliferating HeLa cells. Probes were mapped to Ensembl genes and the top (high), bottom (low) and middle (medium) expressed 2000 genes selected. HIRA/UBN1/ASF1a peak frequency across a composite gene assembled from each group of 2000 genes was plotted. The x-axis shows the position along the gene, where the distance between TSS and TES is in % of gene length and regions upstream of TSS and downstream of TES are in bp.
E) Heatmaps of normalized density of ChIP-seq tags of HIRA, UBN1, ASF1a, H3.3, H2Az and FAIRE in a 10 kb window centered on TSS. TSS are rank ordered according to the expression level of the corresponding transcript.
Binding partners of HIRA complex and ASF1a
Fig. 2A reveals a marked overlap between HIRA/UBN1/ASF1a binding sites and binding of transcription factors and transcription regulators. Strikingly, 76% of HIRA/UBN1/ASF1a peaks co-localize with at least one protein from four families: human SWI/SNF ATP-dependent chromatin remodeling complexes (BRG1, INI1, BAF155 and BAF170), AP-1 (c-Fos, c-Jun, JunD (clusters 1 and 3)), c-Myc/Max (clusters 1 and 2) and TFAP2 (TFAP2A and TFAP2C (clusters 1 and 2)) (Fig. 2A, Fig. 4A and Table S3). The most robust overlap was observed with various members of the SWI/SNF family of chromatin remodelers. Specifically, 57%, 41%, 41% and 36% of HIRA/UBN1/ASF1a peaks overlapped with BAF155, BAF170, INI1 and BRG1 respectively (Table S3). This overlap was particularly marked at active enhancers and promoters (clusters 1 and 2, respectively) (Fig. 2A). These transcription regulators represent candidate HIRA/UBN1/ASF1a-bound regulatory partners.

A) Overlap between HIRA/UBN1/ASF1a peaks and various proteins studied under the ENCODE project. See also Table S3.
B) Immunoprecipitation of endogenous HIRA from nuclear lysates co-precipitates other members of the chaperone complex (UBN1, ASF1a) as well as transcription factors (c-Jun, c-Myc, GTF2i), members of SWI/SNF chromatin remodelers (BRG1, BRM, INI1) and CTCF but not TCF4 or MCM2. See also Fig. S3.
C) Co-precipitation of ectopically expressed epitope tagged HA-HIRA and FLAG-BRG1. D) Co-precipitation of endogenous members of UBN1, ASF1a, CABIN1 and SWI/SNF complex (BRG1/INI1) from nuclear lysates.
E) HIRA/UBN1/ASF1a colocalizes with BRG1 and INI1 at the TSS of highly expressed genes. Distribution of genic BRG1, BRG1-positive
HIRA/UBN1/ASF1a, INI1 and INI1-positive HIRA/UBN1/ASF1a peaks plotted against the normalized gene coordinate (x-axis), with genes sorted according to their level of expression in HeLa cells. Grey windows show at least 2-fold enrichment of HIRA/UBN1/ASF1a relative to input.
To confirm whether these candidates for HIRA/UBN1/ASF1a binding partners identified by ChIP-seq are bona fide interacting proteins, we tested specific interactions by immunoprecipitation-western blot analysis. Transcription factors c-Myc, c-Jun, GTF2I (a multi-functional promoter-binding transcription factor (Roy, 2012)), chromatin remodelers of the SWI/SNF family (BRG1, BRM and INI1) and chromatin insulator CTCF (enriched in clusters 1 and 2 (Fig. 2A)) were all specifically co-precipitated with endogenous HIRA from HeLa lysates, while an abundant chromatin-binding protein MCM2 and transcription factor TCF4 were not (Fig. 4B). The interaction between HIRA and BRG1 was additionally confirmed using ectopically expressed epitope tagged proteins (Fig. 4C).
Antibodies to UBN1 and ASF1a also co-precipitated subunits of SWI/SNF, BRG1 and INI1 (Fig. 4D). Conversely, antibodies to BRG1 and INI1 co-precipitated HIRA, UBN1a, ASF1a and CABIN1 (Fig. 4D). Confirming appropriate specificity and sensitivity of these assays, only antibodies to ASF1a co-precipitated MCM2 (Fig. 4D). The DNA replication-independent chromatin regulators HIRA, UBN1, INI1 and BRG1 do not interact with the DNA replication helicase MCM2, while ASF1a does bind to MCM2 due to its HIRA/UBN1/CABIN1-independent role in DNA replication-coupled nucleosome assembly (Groth et al., 2007)). Co-precipitation of HIRA, BRG1 and INI1 was largely resistant to denaturation of DNA by ethidium bromide in the lysis buffer, and occurred even after digestion of chromatin to predominantly mono and dinucleosomes (Fig. S3A), suggesting that the co-precipitation does not reflect an indirect interaction mediated by long-range chromatin structure.
To verify close physical proximity between HIRA and the BRG1/INI1 complex, we used the proximity ligation assay (PLA), an epifluorescence-based method that scores physical close proximity of target proteins at the molecular level (Fredriksson et al., 2002). Using in situ PLA under stringent conditions designed to remove proteins not stably bound to chromatin, we demonstrated that HIRA is located in close proximity to BRG1 and INI1 (Fig. S3B, C). This assessment of proximity was specific, by reference to cells in which HIRA was knocked down by shRNA and antibodies to several proteins not known to interact with HIRA (DNMT1, MCM2, UACA, ATRX, XRN1, MBD2, LSH, EDC4; Fig. S3B, C). In sum, targeted immunoprecipitation-western blot analyses and PLA assays verified many of the physical interactions indicated by ChIP-seq. Of particular note, BRG1/INI1 appears to physically interact, directly or indirectly, with multiple members of the HIRA complex and ASF1a.
To further investigate the HIRA/UBN1/ASF1a and BRG1/INI1 interaction, we more closely compared the genome wide distribution of co-occupied HIRA/UBN1/ASF1a binding sites with previously published data describing the genome wide distribution of BRG1 and INI1, also in HeLa cells (Euskirchen et al., 2011). This analysis revealed co-incident HIRA/UBN1/ASF1a and BRG1 peaks, overlapping with H3.3-containing nucleosomes (e.g. Fig. S3D). Like HIRA/UBN1/ASF1a, BRG1/INI1 has been previously reported to be enriched at many gene TSS and unsupervised clustering revealed marked overlap of HIRA/UBN1/ASF1a and BRG1/INI1 at active gene promoters (cluster 2, Fig. 2A). Indeed, at genic regions co-localization between HIRA/UBN1/ASF1a and BRG1/INI1 was most prominent at the TSS of highly expressed genes (Fig. 4E).
Discussion
More than six thousand loci are co-occupied by HIRA, UBN1 and ASF1a across the genome. We also find a striking co-localization with histone H3.3, its preferred deposition substrate, at 95% (5,867) of these sites. HIRA contributes to total deposition of histone H3.3 in the genome and at all its specific binding sites tested. Co-occupied HIRA, UBN1 and ASF1a binding sites occur at three main regulatory elements; namely, promoters of active genes and active and weak/poised enhancers. At active promoters, histone H3.3 and H2Az both show a bimodal distribution reflecting H3.3/H2Az-containing nucleosomes either side of the TSS. However, active and weak/poised enhancers exhibit monomodal H3.3 and H2Az peaks. Active enhancers bind more H3.3 and H2Az than weak/poised enhancers. These results extend previous studies to further distinguish between different local nucleosome structures at distinct regulatory elements (Ernst et al., 2011; Heintzman et al., 2009; Rada-Iglesias et al., 2011).
At gene promoters, HIRA, UBN1 and ASF1a bind at the FAIRE-positive “nucleosome-free” region just upstream of the TSS, and binding of all 3 factors, as well as H2Az and H3.3 either side of the TSS, shows a striking correlation with gene expression. The nucleosome-free region is thought to dynamically cycle between the nucleosome-bound and unbound state (Jin et al., 2009). The chaperone complex likely contributes to these dynamics. Interestingly, while histone H3.3 accumulates at the 3′ end of gene bodies of actively transcribed genes (Goldberg et al., 2010), we did not observe enrichment of HIRA/UBN1/ASF1a at these regions (Fig. 3C, D). This suggests that HIRA/UBN1/ASF1a is more stably bound to TSS, where there is perhaps a more long term requirement in anticipation of transcription initiation, compared to gene bodies where it is only transiently required in conjunction with transcription-coupled nucleosome re-assembly.
Surprisingly, HIRA binds to at least 1000 sites across the genome, in the absence of UBN1 and ASF1a (HIRA-only binding sites). The chromatin landscape of these HIRA-only sites is very different from combined HIRA/UBN1/ASF1a binding sites. Most notably, HIRA-only binding sites are not enriched in H3.3, suggesting a quite different function for HIRA in the absence of UBN1 and ASF1a. To date, these 1000 HIRA-only binding sites have not revealed other features in common, so the nature of this function is currently unknown.
We identified proteins that bind directly or indirectly to ASF1a and/or the HIRA complex, namely c-Jun, c-Myc, GTF2I, CTCF and BRG1/INI1. Their interaction with the H3.3 chaperone is likely to direct and modulate histone chaperone activity. A physical interaction between the HIRA complex and ASF1a and BRG1 and INI1 is consistent with previous reports that have linked members of the HIRA complex and SWI/SNF ATP-dependent chromatin remodeling factors in model organisms (Dimova et al., 1999; Konev et al., 2007; Moshkin et al., 2002). Our ChIP-seq analysis indicates that the HIRA/UBN1/ASF1a interaction with BRG1/INI1 likely occurs preferentially at active promoters and enhancers, compared to weak/poised enhancers. This illustrates a general conclusion of our analysis that the nucleosome and co-regulator composition of the chaperone’s binding sites varies considerably between the different types of regulatory elements. Presumably, different networks of physical and functional interactions, involving HIRA/UBN1/ASF1a, dictate the distinct functional properties of active promoters and active and weak/poised enhancers.
Experimental procedures
See Suppl. information for more details and references.
HIRA, UBN1 and AFS1a ChIP
HeLa cells were cross-linked with 1.5mM EGS in PBS for 45 min at room temperature, followed by treatment with 1% formaldehyde for 15 minutes. After quenching with glycine, the cells were harvested and sonicated to produce soluble chromatin with DNA fragments in the range of 150-300 bp. For ChIP, this fragmented chromatin was incubated with antibodies to HIRA, UBN1 or ASF1a pre-immobilized on Dynabeads.
Histone ChIP
HeLa cells were cross-linked with 1% formaldehyde in DMEM for 15 minutes. After that, the procedure was similar to HIRA ChIP.
FAIRE
FAIRE DNA was purified from the same inputs that were used for HA-H3.3 ChIPs as described previously (Giresi et al., 2007). Briefly, input sample was extracted twice with phenol – chloroform – isoamyl alcohol mixture and FAIRE DNA was recovered from the aqueous phase using Qiagen PCR clean-up kit.
Massively parallel sequencing and data analysis
Libraries were prepared from 10-20 ng of ChIP or input DNA using Illumina ChIP-seq kit according to the manufacturer’s instructions and the resulting libraries were sequenced on GAIIx to yield about 30 million raw reads. ChIP-seq or input reads were mapped to the human genome (hg18) using the Bowtie alignment software. Only unique reads mapping to a single location were retained. Determination of Enriched Regions was performed using the USeq package and reads were visualized using the UCSC browser. Results presented are analyzed from a single ChIP-seq reaction of each of UBN1 and ASF1a, but results are representative of two independent ChIP-seq reactions for HIRA.
Acknowledgments
We thank Pawel Herzyk, Julie Galbraith and William Clark for DNA sequencing; David Strachan and Lynn McGarry for image analysis; and Sarah Kinkley, Adam Woolfe, David Vetrie and Koorosh Koorfi for critical discussions. We thank members of the Adams lab and NIA program project members for critical discussions. Work in GA’s lab was supported by la Ligue Nationale contre le Cancer (Equipe labellisée Ligue). Work in the lab of PDA was funded by CR-UK program project, C10652/A10250, and program project NIA AG031862-02.
Footnotes
GEO accession numbers
HIRA ChIP-seq - GSM1095934
UBN1 ChIP-seq - GSM1095936
ASF1a ChIP-seq - GSM1095933
Corresponding input DNA - GSM1095935
Newly synthesized HA-H3.3 ChIP-seq - GSM1095930
Corresponding FAIRE-seq - GSM1095931
Corresponding input DNA - GSM1095932
Expression microarrays of siHIRA-treated HeLa (GSM1095926, GSM1095927, GSM1095928, GSM1095929) and scrambled siRNA-treated HeLa (GSM1095922, GSM1095923, GSM1095924, GSM1095925).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad K, Henikoff S. The histone variant h3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. [Abstract] [Google Scholar]
- Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. [Abstract] [Google Scholar]
- Dutta D, Ray S, Home P, Saha B, Wang S, Sheibani N, Tawfik O, Cheng N, Paul S. Regulation of angiogenesis by histone chaperone HIRA-mediated Incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J Biol Chem. 2010;285:41567–41577. [Europe PMC free article] [Abstract] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. [Europe PMC free article] [Abstract] [Google Scholar]
- Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, Rozowsky J, Bhardwaj N, Gerstein MB, Snyder M. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011;7:e1002008. [Europe PMC free article] [Abstract] [Google Scholar]
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–477. [Abstract] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. [Europe PMC free article] [Abstract] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. [Europe PMC free article] [Abstract] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. [Abstract] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. [Europe PMC free article] [Abstract] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. [Europe PMC free article] [Abstract] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. [Europe PMC free article] [Abstract] [Google Scholar]
- Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. [Europe PMC free article] [Abstract] [Google Scholar]
- Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. [Abstract] [Google Scholar]
- Moshkin YM, Armstrong JA, Maeda RK, Tamkun JW, Verrijzer P, Kennison JA, Karch F. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 2002;16:2621–2626. [Europe PMC free article] [Abstract] [Google Scholar]
- Placek BJ, Huang J, Kent JR, Dorsey J, Rice L, Fraser NW, Berger SL. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J Virol. 2009;83:1416–1421. [Europe PMC free article] [Abstract] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. [Europe PMC free article] [Abstract] [Google Scholar]
- Ray-Gallet D, Quivy J-P, Scamps C, Martini EM-D, Lipinski M, G A. HIRA Is Critical for a Nucleosome Assembly Pathway Independent of DNA Synthesis. Mol Cell. 2002;9:1091–1100. [Abstract] [Google Scholar]
- Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol Cell Biol. 2002;22:2318–2328. [Europe PMC free article] [Abstract] [Google Scholar]
- Roy AL. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I: 10 years later. Gene. 2012;492:32–41. [Europe PMC free article] [Abstract] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. [Abstract] [Google Scholar]
- Sherwood PW, Tsang SV, Osley MA. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:28–38. [Europe PMC free article] [Abstract] [Google Scholar]
- Szenker E, Lacoste N, Almouzni G. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep. 2012;1:730–740. [Abstract] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. [Abstract] [Google Scholar]
- van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet. 2007;39:251–258. [Abstract] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang JH, Song Y, Seol JH, Park JY, Yang YJ, Han JW, Youn HD, Cho EJ. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc Natl Acad Sci U S A. 2011;108:85–90. [Europe PMC free article] [Abstract] [Google Scholar]
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.celrep.2013.03.026
Article citations
Differential modulation of polycomb-associated histone marks by cBAF, pBAF, and gBAF complexes.
Life Sci Alliance, 7(11):e202402715, 29 Aug 2024
Cited by: 1 article | PMID: 39209535 | PMCID: PMC11361369
Multifunctional histone variants in genome function.
Nat Rev Genet, 13 Aug 2024
Cited by: 0 articles | PMID: 39138293
Review
Histone H3.3 chaperone HIRA renders stress-responsive genes poised for prospective lethal stresses in acquired tolerance.
Genes Cells, 29(9):722-734, 08 Jul 2024
Cited by: 1 article | PMID: 38977420 | PMCID: PMC11447927
Comparative Review on Cancer Pathology from Aberrant Histone Chaperone Activity.
Int J Mol Sci, 25(12):6403, 10 Jun 2024
Cited by: 0 articles | PMID: 38928110
Review
Replisome Proximal Protein Associations and Dynamic Proteomic Changes at Stalled Replication Forks.
Mol Cell Proteomics, 23(5):100767, 13 Apr 2024
Cited by: 0 articles | PMID: 38615877
Go to all (94) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex.
Mol Cell Biol, 31(19):4107-4118, 01 Aug 2011
Cited by: 63 articles | PMID: 21807893 | PMCID: PMC3187368
UBN1/2 of HIRA complex is responsible for recognition and deposition of H3.3 at cis-regulatory elements of genes in mouse ES cells.
BMC Biol, 16(1):110, 03 Oct 2018
Cited by: 19 articles | PMID: 30285846 | PMCID: PMC6171237
Identification of an ubinuclein 1 region required for stability and function of the human HIRA/UBN1/CABIN1/ASF1a histone H3.3 chaperone complex.
Biochemistry, 51(12):2366-2377, 16 Mar 2012
Cited by: 24 articles | PMID: 22401310 | PMCID: PMC3320765
A Molecular Prospective for HIRA Complex Assembly and H3.3-Specific Histone Chaperone Function.
J Mol Biol, 429(13):1924-1933, 19 Nov 2016
Cited by: 22 articles | PMID: 27871933 | PMCID: PMC5438300
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Cancer Research UK (1)
Grant ID: C10652/A10250
NIA NIH HHS (2)
Grant ID: P01 AG031862
Grant ID: AG031862-02




