Abstract
Objective
To explore the relationship between biomarkers of pulmonary arterial hypertension (PAH), interferon (IFN)-regulated gene expression, and the alternative activation pathway in systemic sclerosis (SSc).Methods
Peripheral blood mononuclear cells (PBMCs) were purified from healthy controls, patients with idiopathic PAH, and SSc patients (classified as having diffuse cutaneous SSc, limited cutaneous SSc [lcSSc] without PAH, and lcSSc with PAH). IFN-regulated and "PAH biomarker" genes were compared after supervised hierarchical clustering. Messenger RNA levels of selected IFN-regulated genes (Siglec1 and MX1), biomarker genes (IL13RA1, CCR1, and JAK2), and the alternative activation marker gene (MRC1) were analyzed on PBMCs and on CD14- and CD14+ cell populations. Interleukin-13 (IL-13) and IL-4 concentrations were measured in plasma by immunoassay. CD14, MRC1, and IL13RA1 surface expression was analyzed by flow cytometry.Results
Increased PBMC expression of both IFN-regulated and biomarker genes distinguished SSc patients from healthy controls. Expression of genes in the biomarker cluster, but not in the IFN-regulated cluster, distinguished lcSSc with PAH from lcSSc without PAH. The genes CCR1 (P<0.001) and JAK2 (P<0.001) were expressed more highly in lcSSc patients with PAH compared with controls and mainly by CD14+ cells. MRC1 expression was increased exclusively in lcSSc patients with PAH (P<0.001) and correlated strongly with pulmonary artery pressure (r=0.52, P=0.03) and higher mortality (P=0.02). MRC1 expression was higher in CD14+ cells and was greatly increased by stimulation with IL-13. IL-13 concentrations in plasma were most highly increased in lcSSc patients with PAH (P<0.001).Conclusion
IFN-regulated and biomarker genes represent distinct, although related, clusters in lcSSc patients with PAH. MRC1, a marker for the effect of IL-13 on alternative monocyte/macrophage activation, is associated with this severe complication and is related to mortality.Free full text

Interferon and Alternative Activation of Monocyte/Macrophages in Systemic Sclerosis–Associated Pulmonary Arterial Hypertension
Abstract
Objective
To explore the relationship between biomarkers of pulmonary arterial hypertension (PAH), interferon (IFN)–regulated gene expression, and the alternative activation pathway in systemic sclerosis (SSc).
Methods
Peripheral blood mononuclear cells (PBMCs) were purified from healthy controls, patients with idiopathic PAH, and SSc patients (classified as having diffuse cutaneous SSc, limited cutaneous SSc [lcSSc] without PAH, and lcSSc with PAH). IFN-regulated and “PAH biomarker” genes were compared after supervised hierarchical clustering. Messenger RNA levels of selected IFN-regulated genes (Siglec1 and MX1), biomarker genes (IL13RA1, CCR1, and JAK2), and the alternative activation marker gene (MRC1) were analyzed on PBMCs and on CD14− and CD14+ cell populations. Interleukin-13 (IL-13) and IL-4 concentrations were measured in plasma by immunoassay. CD14, MRC1, and IL13RA1 surface expression was analyzed by flow cytometry.
Results
Increased PBMC expression of both IFN-regulated and biomarker genes distinguished SSc patients from healthy controls. Expression of genes in the biomarker cluster, but not in the IFN-regulated cluster, distinguished lcSSc with PAH from lcSSc without PAH. The genes CCR1 (P < 0.001) and JAK2 (P < 0.001) were expressed more highly in lcSSc patients with PAH compared with controls and mainly by CD14+ cells. MRC1 expression was increased exclusively in lcSSc patients with PAH (P < 0.001) and correlated strongly with pulmonary artery pressure (r = 0.52, P = 0.03) and higher mortality (P = 0.02). MRC1 expression was higher in CD14+ cells and was greatly increased by stimulation with IL-13. IL-13 concentrations in plasma were most highly increased in lcSSc patients with PAH (P < 0.001).
Conclusion
IFN-regulated and biomarker genes represent distinct, although related, clusters in lcSSc patients with PAH. MRC1, a marker for the effect of IL-13 on alternative monocyte/macrophage activation, is associated with this severe complication and is related to mortality.
Systemic sclerosis (SSc) is characterized by vascular injury, autoimmunity, and fibrosis in multiple organs, resulting in multifaceted clinical manifestations with high morbidity and mortality (1). Pulmonary arterial hypertension (PAH), one of the leading causes of mortality in SSc, is associated with elevated expression of markers of vascular damage as well as inflammation, suggesting that SSc-related PAH might be secondary to the inflammatory process (2). We recently showed that peripheral blood mononuclear cells (PBMCs) from patients with limited cutaneous SSc (lcSSc) with associated PAH had increased expression of a cluster of genes, including IL13RA1, ICAM1, and others (3), compared with PBMCs from lcSSc patients without PAH and compared with PBMCs from controls. Strikingly, a gradient of expression of these genes was apparent, showing higher gene expression in lcSSc patients without PAH than in controls and even higher expression in lcSSc patients with PAH than in lcSSc patients without PAH. In addition, several of the genes overexpressed by PBMCs have been previously associated with monocyte/macrophage cell types, suggesting that this cell type might be particularly important in mediating inflammation in lcSSc and lcSSc-related PAH (3).
Our group previously demonstrated that PBMCs from patients with diffuse cutaneous SSc (dcSSc) overexpress a cluster of interferon (IFN)-regulated genes, including Siglec1 (4), a surface protein found exclusively on macrophages and monocyte-derived dendritic cells. However, unlike altered gene expression associated with lcSSc-related PAH, increased Siglec1 expression by PBMCs in dcSSc does not correlate well with the Modified Rodnan Skin Thickness Score (MRSS) (4,5). In contrast, genes with elevated expression in lcSSc patients with PAH are not known to be regulated by IFN, suggesting that monocytes from lcSSc patients with PAH might be activated by another, non–type I IFN, cytokine.
Monocytes can be activated through several pathways, the best defined of which are classical and alternative activation pathways (6-8). The classical activation pathway is mediated by IFNγ and/or lipopolysaccharide (LPS), enhancing microbicidal and tumoricidal capacity (7). The alternative activation pathway is mediated by IL-4 and/or IL-13. The main cellular effects of this pathway are activation of endocytosis, inhibition of nitric oxide production with consequent enhanced arginase activity, and induction of tissue remodeling and fibrosis (8). Although arginase is a well-known marker of alternative activation in mice, it is not regulated by IL-4/IL-13 in human monocytes, whereas expression of MRC1 (c-type mannose receptor 1) is highly induced by IL-4/IL-13 in human monocytes and is thus thought to represent a strong marker of alternative activation of monocyte/macrophages in humans (8).
To better understand monocyte/macrophage activation in the pathogenesis of lcSSc and lcSSc-related PAH, we explored the relationship between IFN-regulated genes, the recently described cluster of genes associated with lcSSc-related PAH, and the alternative activation pathway. We observed that IFN-regulated genes and biomarker genes represent distinct, although related, clusters. We found that the expression of markers of IFN activation, including Siglec1, expressed only by CD14+ cells, and MX1, expressed by both CD14+ and CD14− cells, was increased in all SSc patient subsets. Expression of MRC1 was found to be highly increased in lcSSc patients with PAH, was highly induced in vitro by IL-13, and correlated with pulmonary artery pressure and PAH-related mortality.
PATIENTS AND METHODS
Study participants
The Boston University Medical Center Institutional Review Board reviewed and approved the conduct of this study. Informed consent was obtained from all patients and healthy subjects. Subjects selected for this study included patients with dcSSc (n = 16) and lcSSc (n = 35) according to diagnostic (9) and subtype (10) criteria, as well as patients with idiopathic PAH (n = 8) and normal healthy controls (n = 10).
SSc disease duration was measured from the onset of the first non–Raynaud’s phenomenon symptom of SSc. Patients with lcSSc were stratified into those with and those without PAH based on echocardiography or right-heart catheterization; in all included patients with PAH, PAH was diagnosed by right-heart catheterization. Patients with pulmonary artery systolic pressures <35 mm Hg and a normal right ventricle on echocardiogram were considered not to have PAH (n = 22). Patients showing mean arterial systolic pressure >25 mm Hg with pulmonary capillary wedge pressure (PCWP) ≤15 mm Hg and pulmonary vascular resistance ≥3 Wood units were classified as having PAH (n = 13). Most patients with lcSSc had minimal or undetectable interstitial lung disease; however, 3 lcSSc patients (2 without PAH and 1 with PAH) with extensive lung fibrosis (as defined in ref. 11) were also included. Idiopathic PAH was diagnosed after exclusion of secondary causes of pulmonary hypertension and with the same criteria based on right-heart catheterization used to classify lcSSc patients as having PAH (see above). The mean ± SD arterial systolic pressure of patients with idiopathic PAH was 55.83 ± 11.30 mm Hg.
PBMCs and monocyte isolation
Blood was collected from healthy controls and patients either on the day of catheterization, or within 3 months of the date of echocardiography, in CPT tubes designed for one-step cell separation (Becton Dickinson). The sample was then immediately mixed and centrifuged at 1,800g at ambient temperature for 30 minutes. Positive selection was performed to isolate monocytes. After PBMC isolation, monocytes were magnetically labeled with CD14, and CD14+ and CD14− (flow-through) cell populations were isolated using a MACS MicroBeads Column (Miltenyi Biotec).
RNA isolation and microarray analysis hybridization
Total RNA from PBMCs, enriched monocytes retained on the column, and nonmonocyte cells (flow-through from the column) was extracted using RNeasy Mini kits (Qiagen). RNA was isolated using RNeasy minicolumns according to the protocol of the manufacturer (Qiagen). RNA samples were stored at −80°C.
Microarray data clustering
All microarray data from this study have been deposited in the GEO (Gene Expression Omnibus) database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE19617). Microarray data were clustered using Cluster 3.0 for Mac OSX (C Clustering Library 1.43), using hierarchical clustering. After selecting genes showing detectable expression in 80% of the samples, genes and then arrays were centered and normalized before clustering by gene.
Quantitative reverse transcription–polymerase chain reaction (RT-PCR)
Complementary DNA was synthesized from total RNA using reverse transcriptase (Gibco BRL) and random primers (Applied Biosystems) as described previously (12). RT-PCR was performed using an ABI StepOne Plus sequence detection system (Applied Biosystems) and TaqMan assay, according to an Applied Biosystems amplification protocol (User Bulletin 2: http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf). The comparative threshold method was used for data analysis, and 18S ribosomal RNA (rRNA) was used as control (the relative expression of the target RNA was calculated in relation to its values). Primers for 18S rRNA and messenger RNA (mRNA) of the following target genes were obtained from Applied Biosystems: IL13Ra1 (Hs00609817_m1), CCR1 (Hs00174298_m1), JAK2 (Hs00234567_m1), MX1 (Hs00895598_m1), Siglec1 (Hs00988063_m1), and MRC1 (Hs00267207_m1).
Stimulation of PBMCs from healthy controls and SSc patients
PBMCs were plated in 24- or 48-well plates at 37°C, at a concentration of 1–2 × 106 cells per well, in complete medium (RPMI 1640 supplemented with 10% fetal bovine serum, 20 mM L-glutamine, 100 IU/ml penicillin, and 100 gm/ml streptomycin). Stimulation consisted of coculture with one of the following reagents: lipoteichoic acid (1 μg/ml) as the Toll-like receptor 2 (TLR-2) agonist, poly(I-C) (25 μg/ml) as the TLR-3 agonist, ultrapure LPS derived from Escherichia coli O111:B4 (1 μg/ml) as the TLR-4 agonist, imiquimod R837 (5 μg/ml) as the TLR-7 agonist, and CpGA–oligodeoxy-nucleotide 2216 (1 μg/ml) as the TLR-9 agonist (all from InvivoGen). PBMCs were then incubated with or without (negative control) addition of recombinant human IL-13 (20 ng/ml; R&D Systems) for 18 hours and then either lysed in the Buffer RLT (Qiagen) for RNA preparation and analysis as described above or stained for flow cytometric analyses (see below).
Flow cytometry
PBMCs were labeled with fluorescein isothiocyanate–conjugated mouse anti-human CD14 monoclonal antibody (BD PharMingen), allophycocyanin–Cy7–conjugated mouse anti-human CD206 (anti-MRC1) (BioLegend), and goat IgG anti-human IL-13 receptor antagonist type I (IL-13Ra1) antibody (R&D Systems) used with phycoerythrin-conjugated donkey anti-goat IgG (Jackson ImmunoResearch). Immunofluorescence was measured with a FACScan flow cytometer (BD Biosciences), and the data were analyzed with FlowJo software (Tree Star). The data are expressed as the percent of cells positive for each population.
IL-4 and IL-13 assay
Plasma was collected in EDTA blood collection tubes, aliquoted, and stored at −80°C. Plasma levels of human IL-13 and human IL-4 were analyzed by enzyme-linked immunosorbent assay (Thermo Scientific), including calibration curves using supplied assay diluents for human plasma with a range of 0–1,000 pg/ml.
Statistical analysis
Comparisons of RT-PCR expression and all clinical data were analyzed by one-way analysis of variance with Bonferroni adjustment for multiple comparisons. Two-group comparisons were analyzed by Student’s t-test. Correlations were tested by Pearson’s correlation coefficient.
RESULTS
Patient characteristics
Patient selection for microarray analyses was as described previously except that only patients with normal PCWP (≤15 mm Hg) were included in the studies reported here (3). Additional patients were selected for RT-PCR analysis, including lcSSc patients with or without PAH, dcSSc patients without PAH, and patients with idiopathic PAH. The mean ± SD age was slightly greater in lcSSc patients with PAH (69.86 ± 7.27 years) than in lcSSc patients without PAH (52.78 ± 10.44 years) or in dcSSc patients (49.56 ± 12.75 years). The mean ± SD disease duration was longer in lcSSc patients (11.51 ± 7.18 years in those without PAH and 9.53 ± 5.20 years in those with PAH) than in dcSSc patients (4.04 ± 7.51 years) (see additional clinical data in Supplemental Table I, available on the Arthritis & Rheumatism Web site at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN) 1529-0131). The mean ± SD MRSS in dcSSc patients was 22.75 ± 9.82. Patients with idiopathic PAH had a mean ± SD age of 54.17 ± 18.05 years and a mean ± SD disease duration of 26.83 ± 29.35 months.
IFN-regulated and PAH biomarker genes expressed by PBMCs in lcSSc patients cluster into distinct groups
Using our previously described microarray database (3), genes were clustered using average linkage, hierarchical, and supervised clustering, grouping healthy subjects, lcSSc patients without PAH, and lcSSc patients with PAH. This clustering showed increased expression of a cluster of IFN-regulated genes in selected lcSSc patients (Figure 1A). This cluster of genes is similar to that previously reported by our group in patients with dcSSc (4) and was found both in patients with PAH and in those without PAH. The IFN-regulated genes found in this cluster include many well-known IFN-regulated genes: IFN-induced with helicase C domain 1 I (IFIH1), myxovirus (influenza virus) resistance (MX1), IFN-induced protein with tetratrico-peptide repeat 1 (IFIT1 and IFIT2, IFN alpha-inducible proteins 2 and 3 (G1P2 and G1P3), IFI44, IFI44L, IFITM3, IFITM2, GBP1, IFN-inducible RNA-dependent protein kinase (PRKR), IFI27, IFN regulatory factor 7 (IRF7), and IFI35.
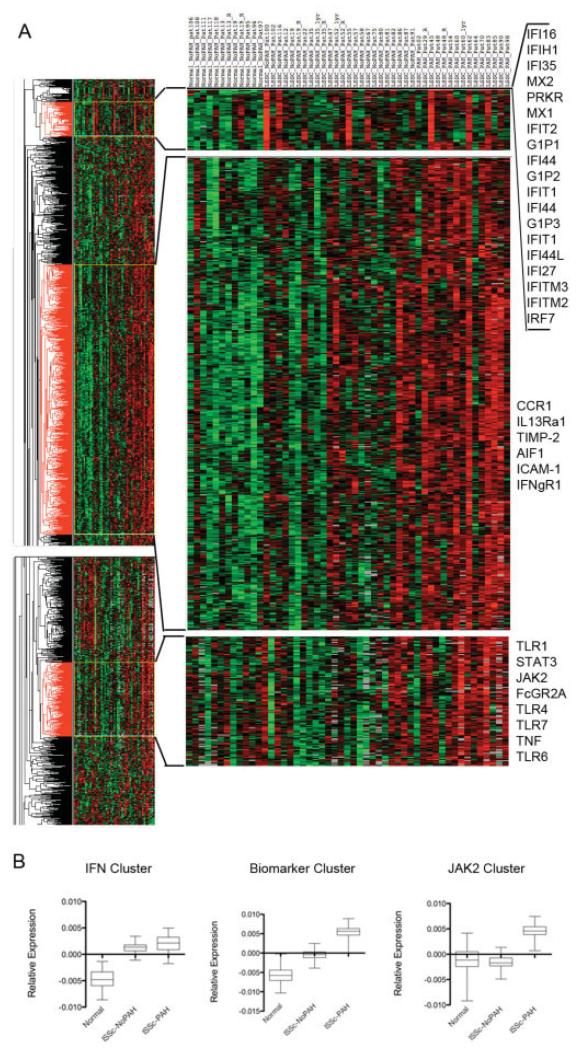
Biomarker and interferon (IFN)–regulated gene expression in limited cutaneous systemic sclerosis (lcSSc). A, Heat map showing the expression of genes clustered using average linkage, hierarchical, and supervised clustering. Individual subjects are listed at the top. Top right, Cluster of IFN-regulated genes; middle right, cluster of biomarker genes; bottom right, JAK2 cluster. Values above the mean expression level of each gene are indicated in red; those below the mean expression level are indicated in green. B, Relative expression of all genes in each cluster in A. Data are shown as box plots. Each box represents the 25th to 75th percentiles. Lines outside the boxes represent the minimum and maximum values. Lines inside the boxes represent the mean. lSSc-NoPAH = lcSSc without pulmonary arterial hypertension; lSSc-PAH = lcSSc with PAH.
Furthermore, we observed a distinct cluster of genes, including genes that we recently reported as biomarkers that distinguish lcSSc patients from healthy controls and lcSSc patients with PAH from those without PAH (Figure 1A). Most of the biomarker genes clustered together, including interleukin-13 receptor, alpha 1 (IL13RA1), interferon gamma receptor 1 (IFNGR1), chemokine (C-C motif) receptor 1 (CCR1), allograft inflammatory factor 1 (AIF1), tissue inhibitor of metalloproteinases 2 (TIMP2), and intercellular adhesion molecule 1 (ICAM1). Notably, JAK2, one of the genes previously identified as a biomarker gene for lcSSc with PAH, clustered separately, although it showed a similar pattern of increased expression in lcSSc with PAH. This cluster was notable for also including the JAK2 signaling partner STAT3 and multiple TLR genes (TLR1, TLR4, TLR6, and TLR7) (for a complete list of genes in each of these clusters, see Supplemental Table II, available on the Arthritis & Rheumatism Web site at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN) 1529-0131).
Thus, IFN-regulated genes increased in a subset of lcSSc patients, and biomarker genes that distinguish healthy controls, lcSSc patients without PAH, and lcSSc patients with PAH clustered in groups. Although some patients, such as patients 45, 62, and 83, had up-regulated genes in both clusters, other patients had up-regulated genes in only one of these clusters, for example, patients 100 and 55 (see Figure 1A).
The average normalized gene expression in these 3 clusters was strikingly different. Whereas the biomarker cluster showed a graded increase in gene expression from healthy controls to lcSSc patients without PAH to lcSSc patients with PAH, the expression of IFN cluster genes was increased in lcSSc regardless of the presence of PAH. Notably, the JAK2 cluster of genes showed on average increased expression in lcSSc patients with PAH, but not in lcSSc patients without PAH or controls (Figure 1B).
Increased expression of IFN-regulated genes in monocytes and nonmonocyte cells from both lcSSc patients and dcSSc patients
Siglec1 is an IFN-regulated monocyte/macrophage marker that we previously showed to be highly expressed by the monocytes and tissue macrophages of dcSSc patients, and its expression in skin correlates with the MRSS (4,13). Increased expression of Siglec1 on PBMCs was observed in all SSc subtypes compared with healthy controls (P = 0.02) (Figure 2a). Expression of MX1, another gene highly induced by IFN but also up-regulated in nonmonocyte cell types (14), was also found to be higher in SSc patients than in healthy controls (P = 0.03), regardless of the SSc subtype (Figure 2c). MX1 and Siglec1 expression correlated positively (r = 0.35, P = 0.02).
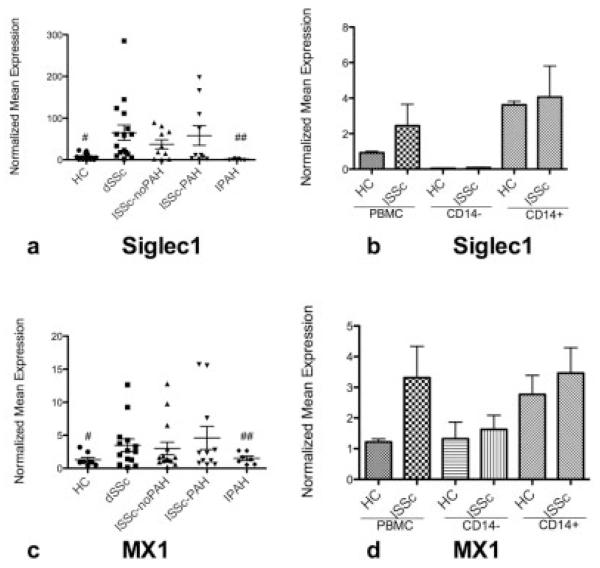
IFN-regulated gene expression in lcSSc and diffuse cutaneous SSc (dcSSc, or dSSc). a and c, Expression of Siglec1 (a) and MX1 (c) in peripheral blood mononuclear cells (PBMCs) from healthy controls (HC), dcSSc patients, lcSSc patients without PAH, lcSSc patients with PAH, and patients with idiopathic PAH (IPAH). b and d, Expression of Siglec1 (b) and MX1 (d) in PBMC, CD14−, and CD14+ populations in lcSSc patients without PAH (n = 4), lcSSc patients with PAH (n = 5), and healthy controls (n = 3) (lcSSc patients without PAH and lcSSc patients with PAH are combined). Data are expressed as the fold change normalized to mRNA expression in a single sample from a healthy control subject. Bars show the mean ± SEM. # and ## = P < 0.05 versus all SSc patients. See Figure 1 for other definitions.
For both Siglec1 and MX1, increased expression was seen in dcSSc patients, lcSSc patients without PAH, and lcSSc patients with PAH, without any significant difference between these 3 patient groups. In contrast, MX1 and Siglec1 were not highly expressed on PBMCs from patients with idiopathic PAH but had expression levels similar to those in the healthy control group (P > 0.05 for both) (Figures 2a and c). In order to better understand the importance of monocytes in the IFN cluster, we isolated CD14+ cells from the PBMCs of 9 lcSSc patients (4 without PAH and 5 with PAH) and 3 healthy controls. Siglec1 was expressed almost exclusively by CD14+ monocytes, whereas MX1 was found on both CD14+ and CD14− cell populations (Figures 2b and d), indicating that this signature is not limited to monocytes. The expression of Siglec1 and MX1 mRNA by PBMCs did not correlate with the MRSS or pulmonary artery pressure (data not shown).
Increased expression of biomarker genes for lcSSc with PAH by PBMCs from SSc patients
Three highly overexpressed genes from the lcSSc with PAH biomarker clustering were selected for further investigation using RT-PCR on PBMCs from dcSSc patients, lcSSc patients without PAH, lcSSc patients with PAH, patients with idiopathic PAH, and healthy controls. These genes included IL13RA1, a receptor required for most actions of the Th2 cytokine IL-13 (15); CCR1, a target chemokine receptor of Th2 cytokines such as CCL6, CCL13, and CCL18 released by macrophages upon IL-13 or IL-4 stimulation and essential in mediating IL-13-induced inflammation (15,16); and JAK2, an intracellular signaling protein activated by IL-13 (17) that clustered in a group different from other biomarker genes (Figure 1A).
As we reported recently, lcSSc patients with PAH had on average higher mRNA expression of these 3 genes compared with lcSSc patients without PAH (Figures 3a, c, and e). PBMCs from lcSSc patients with PAH showed higher expression of CCR1 and JAK2 compared with PBMCs from dcSSc patients (P < 0.05 and P < 0.005, respectively) and compared with PBMCs from lcSSc patients without PAH (P < 0.05 for both). In contrast, IL13RA1 was found to be more highly expressed in all 3 subtypes than in healthy controls, but showed higher expression in dcSSc patients and lcSSc patients with PAH compared with controls (P < 0.05 for both). In contrast, PBMCs from patients with idiopathic PAH showed expression of these 3 genes similar to that in PBMCs from healthy controls (P > 0.05 for all) (Figures 3a, c, and e). Notably, although these genes fall into a cluster that is distinct from the IFN cluster, IFN-regulated Siglec1 expression correlated positively and significantly with IL13RA1 expression (r = 0.44, P < 0.005) and JAK2 expression (r = 0.35, P = 0.03). Similarly, IFN-regulated MX1 expression also correlated positively with IL13RA1 expression (r = 0.32, P = 0.02) but not with JAK2 expression (r = 0.39, P = 0.07). CCR1 expression correlated with both IL13RA1 expression (r = 0.63, P < 0.001) and JAK2 expression (r = 0.83, P < 0.001).
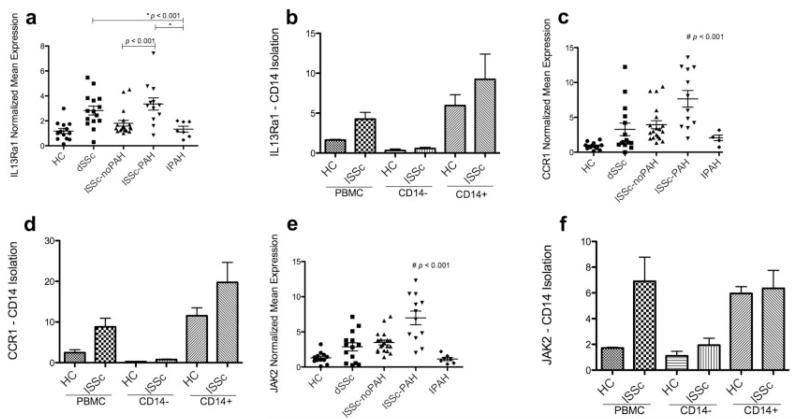
PAH biomarker gene expression in diffuse cutaneous SSc (dcSSc, or dSSc) and lcSSc. a, c, and e, Expression of IL13RA1 (a), CCR1 (c), and JAK2 (e) in peripheral blood mononuclear cells (PBMCs) from healthy controls (HC), dcSSc patients, lcSSc patients without PAH, lcSSc patients with PAH, and patients with idiopathic PAH (IPAH). b, d, and f, Expression of IL13RA1 (b), CCR1 (d), and JAK2 (f) in PBMC, CD14−, and CD14+ populations in lcSSc patients without PAH (n = 4), lcSSc patients with PAH (n = 5), and healthy controls (n = 3) (lcSSc patients without PAH and lcSSc patients with PAH are combined). Data are expressed as the fold change normalized to mRNA expression in a single sample from a healthy control. Bars show the mean ± SEM. # = P < 0.001, by analysis of variance. See Figure 1 for other definitions.
As was the case for Siglec1 (Figure 2b), IL13RA1 and CCR1 were expressed almost exclusively by CD14+ monocytes both in lcSSc patients (4 without PAH and 5 with PAH) and in healthy controls (Figures 3b and d), while JAK2 gene expression was found in both CD14+ and CD14− cell populations (Figure 3f). IL13RA1, CCR1, and JAK2 mRNA expression did not correlate with the MRSS or with PAP (data not shown).
Increased expression of MRC1, a marker of macrophage alternative activation, in lcSSc with PAH
Elevated IL13RA1 expression, mainly in the CD14+ cell population, suggested that alternative activation might contribute to monocyte activation in SSc. We therefore analyzed MRC1 expression in SSc PBMCs as a marker of alternative activation. MRC1 expression was highly and exclusively up-regulated in lcSSc patients with PAH compared with dcSSc patients, lcSSc patients without PAH, patients with idiopathic PAH, or healthy controls (overall P < 0.001) (Figure 4a). Additionally, MRC1 was the only biomarker studied whose expression correlated with PAP on catheterization of lcSSc patients (r = 0.52, P = 0.03) (Figure 4b). This positive correlation persisted if only lcSSc patients with PAH were analyzed (r = 0.61, P = 0.04). Those lcSSc patients with PAH who had higher PBMC MRC1 expression also had higher mortality during the 2-year followup period than did lcSSc patients without PAH (P = 0.02).
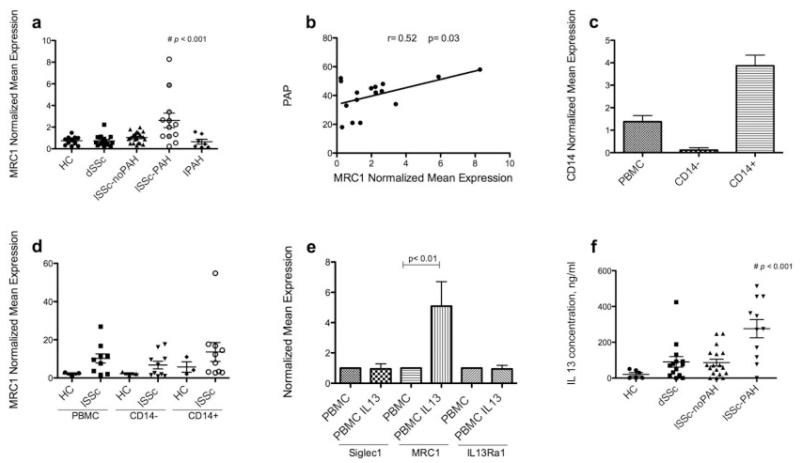
MRC1 expression in SSc patient populations and MRC1 expression by monocytes. a, Expression of MCR1 in peripheral blood mononuclear cells (PBMCs) from healthy controls (HC), patients with diffuse cutaneous SSc (dcSSc, or dSSc), lcSSc patients without PAH, lcSSc patients with PAH, and patients with idiopathic PAH (IPAH). Shaded circles represent lcSSc patients with PAH who died during the 2 years of followup in this study. b, Linear regression analysis of the relationship between the mean pulmonary artery pressure (PAP) by right-heart catheterization and the expression of MRC1 in PBMCs from lcSSc patients with PAH. c, Expression of CD14 on PBMC, CD14−, and CD14+ populations from SSc patients and controls. d, Expression of MRC1 on PBMC, CD14−, and CD14+ populations in lcSSc patients without PAH (n = 4), lcSSc patients with PAH (n = 5), and healthy controls (n = 3) (lcSSc patients without PAH and lcSSc patients with PAH are combined). e, Expression of Siglec1, MRC1, and IL13RA1 on PBMCs from healthy controls (n = 7) after 18 hours of stimulation with 20 ng/ml interleukin-13 (IL-13) or without IL-13 stimulation (control). f, Plasma IL-13 concentration in healthy controls, dcSSc patients, lcSSc patients without PAH, and lcSSc patients with PAH. Data in a and c-e are expressed as the fold change normalized to mRNA expression in a single sample from a healthy control. Bars show the mean ± SEM. # = P < 0.001, by analysis of variance. ELISA = enzyme-linked immunosorbent assay (see Figure 1 for other definitions).
MRC1 is more highly expressed in CD14+ cells and is induced by IL-13
In order to clarify the cell type responsible for increased MRC1 expression, we analyzed MRC1 mRNA expression by CD14+ monocytes from lcSSc patients and controls. CD14+ cell isolation was effective, as shown by CD14 expression in the fractionated cell populations (Figure 4c), and MRC1 expression in these samples was higher in CD14+ cells (Figure 4d), although MRC1 was also expressed at higher levels in CD14− cells from lcSSc patients than in those from healthy controls. To confirm previously reported findings, we tested the effect of IL-13 on PBMCs from 7 healthy controls and found dramatic up-regulation of MRC1 expression after 18 hours of IL-13 stimulation, with no effect on Siglec1 or IL13RA1 expression (Figure 4e).
IL-13 in lcSSc patients with PAH
IL-13 was analyzed in the plasma of lcSSc patients and healthy controls. The highest concentration of IL-13 was found in lcSSc patients with PAH (P < 0.001) (Figure 4f). Plasma IL-4 levels were undetectable in samples from lcSSc patients or healthy controls (data not shown).
IL-13 induces surface expression of MRC1 but not IL13RA1 by CD14+ monocytes
In order to further investigate whether higher mRNA levels of MRC1 are more likely related to IL-13 induction or are instead a reflection of increased IL13RA1 expression, PBMCs from an lcSSc patient with PAH were isolated and stimulated with IL-13. CD14, MRC1 (CD206), and IL13RA1 were analyzed by flow cytometry. MRC1 was highly induced in CD14+ cells after IL-13 treatment (by 4.81% before treatment and by 12.8% after treatment) (Figure 5a). As shown in Figure 5b, despite detectable baseline expression of IL13RA1 on the majority of PBMCs (51.5%), no significant induction of IL13RA1 was observed after IL-13 stimulation (53.5%), in contrast to increased PBMC expression of MRC1 after IL-13 stimulation. The high MRC1 induction and the contrasting absence of IL13RA1 expression on PBMCs after IL-13 stimulation were confirmed by RT-PCR (Figure 5c).
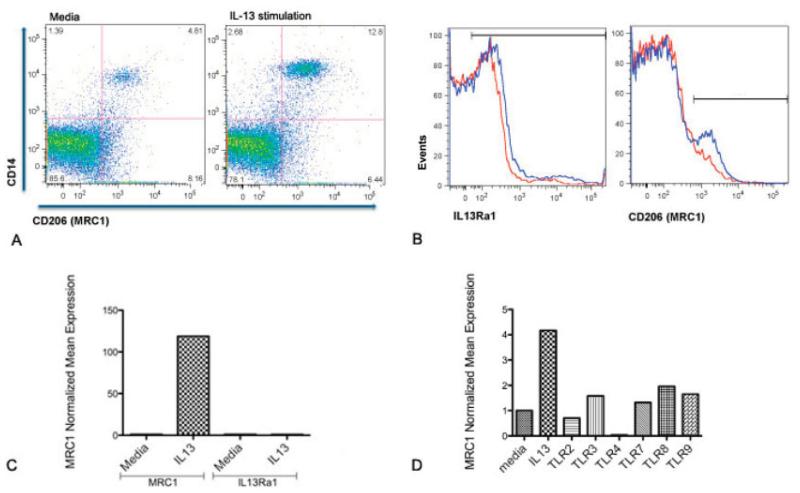
MRC1 (CD206) surface expression on peripheral blood mononuclear cells (PBMCs) from lcSSc patients with PAH. A, Double staining (CD14+CD206+) of PBMCs before (4.81%) and after (12.8%) stimulation with interleukin-13 (IL-13). B, Induction of IL13RA1 and CD206 surface expression on all PBMCs before (red lines) and after (blue lines) 3 hours of stimulation with 20 ng/ml IL-13. After IL-13 stimulation, positive cell expression of IL13RA1 changed from 51.5% (red) to 53.5% (blue), and positive cell expression of CD206 changed from 8.45% (red) to 15.3% (blue). Horizontal bars represent stained cells. C, Expression of IL13RA1 MRC1 and IL13RA1 mRNA before and after IL-13 stimulation. D, MRC1 mRNA expression on healthy PBMCs stimulated for 24 hours with IL-13 or with several Toll-like receptor (TLR) ligands. Data are expressed as the fold change normalized to mRNA expression in a single sample from a healthy control. See Figure 1 for other definitions.
TLR ligands do not induce MRC1
Since our group has shown previously that TLR ligands through type I IFN induce Siglec1 on monocytes in SSc patients (4), we analyzed MRC1 expression after TLR ligand stimulation on healthy PBMCs. MRC1 expression was not significantly induced by any TLR ligand, although MRC1 expression was highly induced by IL-13 stimulation (Figure 5d); as expected, Siglec1 expression was strongly induced by several TLR ligands but was not stimulated by IL-13 (Figure 5d).
DISCUSSION
Our data suggest that circulating monocytes in lcSSc patients are activated by several circulating cytokines, including type I IFN and IL-13, and show that the IL-13– but not the IFN-regulated gene signal is associated with lethal PAH. Furthermore, our results show that PBMC gene expression biomarkers of lcSSc with PAH and IFN-regulated genes cluster into separate groups, reflecting different patterns of expression, with an increased number of IFN-regulated genes seen in PBMCs from all SSc patients, including dcSSc patients and lcSSc patients with or without PAH.
MRC1 is highly expressed in lcSSc patients with PAH, correlating strongly with PAP by right-heart catheterization and patient mortality. MRC1 is a distinctive marker of IL-4/IL-13–activated macrophages (18) and can mediate signals from endogenous glycoproteins, inhibiting the Th1-skewing cytokine IL-12 and stimulating production of antiinflammatory cytokines such as IL-10 (19). Notably, increased MRC1 expression was seen almost exclusively in our lcSSc patients with PAH, with no increased expression in PBMCs from dcSSc patients or lcSSc patients without PAH. These data suggest that MRC1 expression might play a significant role in regulating the immune response in lcSSc patients with PAH. Supporting the notion of a role for IL-13 in this process, IL-13, but not IL-4, was found at higher concentrations in the plasma of lcSSc patients with PAH.
Although MRC1 was not present on the microarray analyses previously carried out, several of the biomarker genes we identified from that analysis suggested a possible role of IL-13 in stimulating biomarker signature gene expression. We therefore analyzed 2 genes from our lcSSc with PAH biomarker cluster with roles in IL-13 signaling: IL13RA1 and JAK2. IL13RA1 is a subunit of the primary IL-13 receptor (IL-4Ra/IL-13Ra1) that mediates the important immunoregulatory role of IL-13 on monocytes and macrophages; it up-regulates vascular cell adhesion molecules in endothelial cells and increases type I collagen synthesis and chemokine expression in human dermal fibroblasts (15). Binding of IL-13 to its receptor provokes phosphorylation of the JAKs, and in human coronary artery endothelial cells, the inhibition of angiogenesis caused by IL-13 was completely abolished with JAK2 depletion (17). IL-13 also activates JAK2 in monocytes (20). The highest expression of JAK2 in lcSSc patients with PAH was found in CD14+ cells, but JAK2 expression in lcSSc patients with PAH was also found in CD14− cells, and increased IL13RA1 expression in both dcSSc and lcSSc was found exclusively on CD14+ cells (monocytes).
Consistent with the notion of IL-13 activation of macrophages in lcSSc patients with PAH, very high plasma concentrations of IL-13 were found almost exclusively in lcSSc patients with PAH. Using more strict criteria for PAH (right-heart catheterization), our data confirm data recently reported by Gourh et al (21). Taken together, our data support the notion that IL-13 stimulates monocyte/macrophages in lcSSc patients with PAH, provoking an alternative activation pathway reflected by high expression of MRC1 and possibly also contributing to defective angiogenesis through JAK2.
Although the association of PAH with inflammatory diseases such as SSc has increased interest in immune mediators affecting pulmonary vasculature, it remains uncertain how IL-13 might contribute to PAH. Several recent reports have emphasized potential roles of IL-13- and Th2-skewed responses in murine models (22,23). Daley et al have shown that arterial muscularization dependent on IL-13 can be stimulated through 2 different antigen challenge models (23). These mice did not develop pulmonary hypertension, and although pulmonary muscularization, a common feature of PAH, was dependent on IL-13, it could not be induced by IL-13 treatment alone, indicating that other factors are required, which is consistent with past reports showing that IL-13 induces only mild vascular changes (24). Another recent report on patients with idiopathic PAH indicated that increased IL-13 expression was found in inflammatory perivascular lung infiltrates (22). Surprisingly, rather than stimulating, IL-13 inhibited 2 key features of PAH, pulmonary artery smooth muscle cell proliferation and endothelin 1 expression in vitro (22). Thus, these in vitro results are difficult to reconcile with the in vivo effects of IL-13 in mice, which support a role for IL-13 in PAH, particularly in vascular smooth muscle hyperplasia.
In striking contrast to up-regulated expression of PAH biomarker and MRC1 genes in PBMCs from lcSSc patients with PAH, PBMCs from patients with idiopathic PAH showed no increased expression of these genes. This observation points to fundamental differences in the pathogenesis of lcSSc with PAH and idiopathic PAH, suggesting that lcSSc with PAH may be more driven by an inflammatory process than is idiopathic PAH.
Although our studies do not support a strong role for type I IFN in lcSSc with PAH, the association between the biomarker and IFN-regulated genes remains intriguing and suggests that the pathways stimulating these 2 cytokines (IL-13 and IFN) are connected in some manner. Several case reports over the years have highlighted a possible association of IFN therapy with PAH (25-28), and recently a weak association of PAH with serum IFNγ-inducible 10-kd protein (i.e., CXCL10, a protein highly regulated by IFN) has been reported (29). In the context of other cytokines, IFN may contribute to inducing PAH in at least some lcSSc patients.
CCR1, a chemokine receptor expressed at high levels on blood monocytes, tissue macrophages, neutrophils, and eosinophils, and at low levels on T cells (16), was also increased in lcSSc patients with PAH, essentially only in the monocyte/macrophage population. Previous gene expression profiling of PBMCs has been shown to discriminate PAH patients from healthy subjects (30,31) and has identified several up-regulated genes both in patients with idiopathic PAH and in lcSSc patients with PAH, including chemokines such as CCL3 with CCR1 as its receptor, and CCL4 (31). These chemokines are chemoattractants for monocyte/macrophages, and inhibition of these genes can prevent pulmonary hypertension in the monocrotaline model (32), supporting the key role of monocyte/macrophages in PAH.
In summary, we confirmed our previously described IFN signature in dcSSc patients (4) and extended these observations to lcSSc. Several IFN-regulated genes were found to be highly expressed, including Siglec1, found exclusively on CD14+ cells, and MX1, found on both CD14+ and CD14− cells, a protein that has a well-characterized antiviral role and is strictly dependent on type I IFN (33). This high expression was observed in some but not all SSc patients and did not correlate with clinical disease activity measures such as the MRSS and pulmonary artery pressure. Despite the lack of correlation between IFN-regulated gene expression by PBMCs, we have shown that skin expression of several IFN-regulated genes correlates highly with the MRSS in dcSSc patients (13). In contrast to PBMC IFN-regulated genes, the biomarker cluster effectively distinguishes lcSSc patients without PAH from lcSSc patients with PAH, including the genes analyzed in more detail here (CCR1 and JAK2). Furthermore, MRC1, a marker of alternative activation of monocyte/macrophages, correlated with pulmonary artery pressure and mortality, with the highest expression in lcSSc patients with PAH. We suggest that IL-13–activated monocyte/macrophages may have a particularly important role in the development of PAH in lcSSc, with MRC1 as an important marker.
Supplementary Material
A&R Table 1
A&R supplement
Acknowledgments
ROLE OF THE STUDY SPONSOR
Actelion Pharmaceuticals was not involved in the study design, data collection, data analysis, or writing of the manuscript, and publication of the manuscript was not contingent upon their approval.
Supported by Actelion Pharmaceuticals (grant to Dr. Lafyatis) and by the NIH (National Institute of Arthritis and Musculoskeletal and Skin Diseases grant U01-AR-055063 to Dr. Lafyatis). Dr. Pendergrass’ work was supported by the NIH (National Institute of Arthritis and Musculoskeletal and Skin Diseases T32 training grant AR-007575-11). Ms Affandi’s work was supported by the Dutch Arthritis Association (grant NR-10-1-301). Dr. Whitfield’s work was supported by the Arthritis Foundation (Hulda Irene Duggan Arthritis Investigator Award). Dr. Farber’s work was supported by the Scleroderma Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Lafyatis had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Christmann, Hayes, Farber, Lafyatis.
Acquisition of data. Christmann, Hayes, Pendergrass, Padilla, Farina, Affandi, Farber, Lafyatis.
Analysis and interpretation of data. Christmann, Hayes, Farina, Affandi, Whitfield, Farber, Lafyatis.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1002/art.30318
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4030759?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Immune mechanisms in fibrotic interstitial lung disease.
Cell, 187(14):3506-3530, 01 Jul 2024
Cited by: 0 articles | PMID: 38996486
Review
Nintedanib downregulates the profibrotic M2 phenotype in cultured monocyte-derived macrophages obtained from systemic sclerosis patients affected by interstitial lung disease.
Arthritis Res Ther, 26(1):74, 20 Mar 2024
Cited by: 1 article | PMID: 38509595 | PMCID: PMC10953168
The Pathogenesis of Systemic Sclerosis: The Origin of Fibrosis and Interlink with Vasculopathy and Autoimmunity.
Int J Mol Sci, 24(18):14287, 19 Sep 2023
Cited by: 9 articles | PMID: 37762589 | PMCID: PMC10532389
Review Free full text in Europe PMC
The critical importance of epigenetics in autoimmune-related skin diseases.
Front Med, 17(1):43-57, 22 Feb 2023
Cited by: 6 articles | PMID: 36811762
Review
Current Trends in Vascular Biomarkers for Systemic Sclerosis: A Narrative Review.
Int J Mol Sci, 24(4):4097, 17 Feb 2023
Cited by: 4 articles | PMID: 36835506 | PMCID: PMC9965592
Review Free full text in Europe PMC
Go to all (88) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE19617
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Increased expression of endoplasmic reticulum stress and unfolded protein response genes in peripheral blood mononuclear cells from patients with limited cutaneous systemic sclerosis and pulmonary arterial hypertension.
Arthritis Rheum, 65(5):1357-1366, 01 May 2013
Cited by: 43 articles | PMID: 23400395 | PMCID: PMC3636187
A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists.
Arthritis Rheum, 56(3):1010-1020, 01 Mar 2007
Cited by: 214 articles | PMID: 17328080
The HLA-B*35 allele modulates ER stress, inflammation and proliferation in PBMCs from Limited Cutaneous Systemic Sclerosis patients.
Arthritis Res Ther, 17:363, 16 Dec 2015
Cited by: 8 articles | PMID: 26669670 | PMCID: PMC4704539
The heart and pulmonary arterial hypertension in systemic sclerosis.
Acta Clin Belg, 71(1):1-18, 05 Feb 2016
Cited by: 5 articles | PMID: 27075793
Review
Funding
Funders who supported this work.
NIAMS NIH HHS (6)
Grant ID: R03 AR062721
Grant ID: U01 AR055063
Grant ID: U01-AR-055063
Grant ID: P50 AR060780
Grant ID: R01 AR051089
Grant ID: T32 AR-007575-11





