Abstract
Free full text

MUC1: A Novel Metabolic Master Regulator
Abstract
MUC1, a type I transmembrane protein, is significantly overexpressed and aberrantly glycosylated in tumors of epithelial origin. By virtue of its aberrant signaling due to loss of apical-basal polarity in cancer, MUC1 regulates the metabolite flux at multiple levels. Serving as a transcriptional co-activator, MUC1 directly regulates expression of metabolic genes. By regulating receptor tyrosine kinase signaling, MUC1 facilitates production of biosynthetic intermediates required for cell growth. Also, via direct interactions, MUC1 modulates the activity/stability of enzymes and transcription factors that directly regulate metabolic functions. Additionally, by modulation of autophagy, levels of reactive oxygen species, and metabolite flux, MUC1 facilitates cancer cell survival under hypoxic and nutrient-deprived conditions. This article provides a comprehensive review of recent literature on novel metabolic functions of MUC1.
1. Introduction
MUC1 is a heterodimeric type I transmembrane protein that is normally expressed on the luminal surfaces of ductal epithelia [1, 2]. Due to its extensive O-glycosylation in the extracellular subunit, it provides protective and lubricative functions to the epithelial cells [3]. The transmembrane subunit has a 72 residue-long cytoplasmic tail, which is a highly active hub for multiple signaling interactions/functions [4]. MUC1 participates in outside-in signaling, sensing the extracellular milieu and reprogramming the transcriptional profiles of cells in response to the extracellular cues [5, 6]. MUC1 is significantly overexpressed in tumor tissues, and while the receptor-like functions of MUC1 provide motility and context-dependent adhesive/anti-adhesive functions to the cells, much of the oncogenic transformation induced by MUC1 is due to the aberrant signaling interactions of its cytoplasmic tail [7–9]. Under conditions of tissue injury and loss of apical-basal polarity in tumor cells, MUC1 loses its apical localization and interacts with otherwise basally located receptor tyrosine kinases that overhaul the signaling functions of MUC1 [4, 10, 11]. MUC1 directly interacts with a variety of transcription factors, and physically occupies multiple promoter elements [7, 12–14]. While MUC1 does not have a DNA binding domain, it acts as a transcriptional co-activator, and its presence in transcriptional complexes drastically alters recruitment of other co-factors and promoter specificity of transcription factors [14]. Transcriptional regulation and other oncogenic functions are significantly modulated by post-translational modifications of the cytoplasmic tail of MUC1 [4, 8, 9].
Multiple recent studies indicate that MUC1 causes transcriptional alterations that result in metabolic reprogramming in cancer cells. MUC1 interacts with both p53 and hypoxia-inducible factor-1 alpha (HIF-1α), two key transcription factors that directly regulate metabolic gene expression. Also, MUC1 regulates the expression of genes involved in multiple nutrient uptake and metabolic pathways [7, 12]. MUC1 expression leads to changes in metabolic flux during glycolysis, as well as in the pentose phosphate pathway (PPP), the tricarboxylic acid (TCA) cycle, and fatty acid biosynthesis pathways [12, 15]. PPP leads to the production of ribose, an essential building block for de novo DNA and RNA synthesis. As a consequence of MUC1 expression, the production of biosynthetic intermediates required for cell growth (i.e., biomass) is increased in cancer cells and cell proliferation is enhanced [12]. In addition to the transcriptional co-activator functions, MUC1 also directly modulates enzymatic functions of metabolic enzymes to regulate carbon flux [16]. Metabolic alterations are a hallmark feature of cancer and provide tumorigenic properties to cancer cells [17]. Thus, understanding the role of MUC1 in cancer metabolism has implications in devising novel therapeutic approaches for combating cancer.
2. Regulation of glucose metabolism
2.1. Direct transcriptional regulation of glycolytic gene expression
Multiple studies indicate an increase in glycolytic gene expression in primary tumors of multiple origins as well as in their respective metastatic sites [18–20]. Given the central role of aerobic glycolysis in managing tumor growth and metastasis, a number of studies have investigated the regulation of aerobic glycolysis both at the transcriptional and at the translational level. Thus far, studies have enlisted HIF-1α, p53, the mammalian target of rapamycin (mTOR), and c-myc as the prime regulators of aerobic glycolysis in tumors [21, 22]. MUC1, being a modulator of all these regulators, is also gaining similar recognition in the scientific community. For example, by studying the glucose uptake and consequent lactate production in MUC1-transformed rat fibroblasts, Kosugi et al. highlighted the role of MUC1 in the regulation of glycolysis [16]. The study recorded increased glycolysis via PI3K/Akt pathways in fibroblasts transformed by exogenously expressing MUC1 cytoplasmic tail. It is known that the phosphoinositide 3-kinase (PI3K)/Akt pathway induces expression of glucose transporters and stimulates the activities of hexokinase and phospho-fructokinase. Hence, Kosugi’s study correlates the increase observed in glycolysis with alterations in MUC1-induced gene expression. Similarly, a study by Chaika et al. lends support to the above findings, while demonstrating for the first time a direct regulation of metabolic gene expression by MUC1 [12]. Chaika et al. demonstrated that not only does MUC1 induce glucose uptake and glycolysis via HIF-1α modulation in pancreatic cancer cells, but the MUC1 cytoplasmic tail also directly occupies multiple promoters of glycolytic genes. Furthermore, MUC1 expression also increases the expression of some glycolytic genes, such as 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2), in a hypoxia/HIF-1α-independent manner. MUC1 directly occupies enolase 1 (ENO1) and phosphoglucomutase 2 (PGM2) glycolytic gene promoters and facilitates their expression even under normoxic conditions. Hypoxic conditions further enhance MUC1 occupancy on these glycolytic gene promoters. While ENO1 and PGM2 are not the rate-limiting enzymes of glycolysis, their marked overexpression drives glycolysis in the forward direction. Additionally, their importance is underscored by the fact that ENO is a biomarker of lung cancer and is also associated with tumor metastasis and resistance against chemotherapeutic interventions [23, 24]. Together, this evidence suggests a critical role of MUC1 in regulating glycolytic gene expression in tumors (Figure 1).
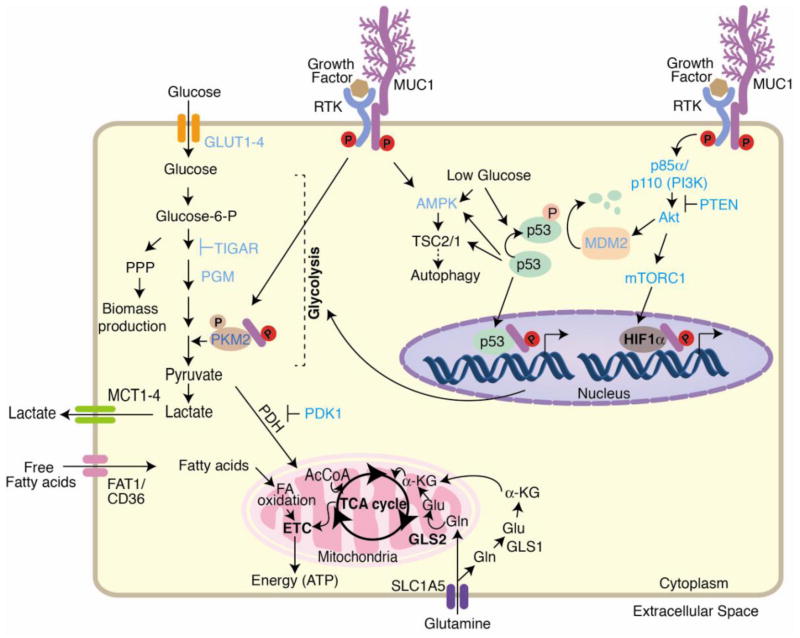
MUC1 regulates glycolytic flux by activating multiple signaling pathways, such as PI3K/Akt/mTOR, p53, and HIF-1α that regulate transcription of glycolytic genes. Under glucose-deprived conditions, MUC1 activates AMPK that facilitates survival by inducing autophagy. The interaction of MUC1 with RTKs increases RTK membrane stability. Thus, the complex facilitates signaling downstream of RTK, which can modulate PKM2 activity transcriptional activation of glycolytic genes. Reciprocally, RTKs phosphorylate MUC1 and facilitate signaling downstream of MUC1. MUC1 also directly interacts with glycolytic enzymes like PKM2 to modulate the carbon flux. As a net result of the changes in gene expression and enzymatic activities, carbon flux to glycolysis is enhanced, whereas carbon flux to TCA cycle is decreased. MUC1-regulated genes/enzymes are indicated in blue. Abbreviations in the figure include the following: glucose transporter (GLUT), pentose phosphate pathway (PPP), phosphoglucomutase (PGM), pyruvate kinase isozyme M2 (PKM2), acetyl coenzyme A (AcCoA), pyruvate dehydrogenase (PDH), pyruvate dehydrogenase kinase 1 (PDK1), monocarboxylate transporter 1 (MCT1), 5′-adenosine monophosphate-activated protein kinase (AMPK), tuberous sclerosis (TSC), α-ketoglutarate (α-KG), solute carrier family 1 member 5 (SLC1A5), insulin-like growth factor 1 (IGF-1), receptor tyrosine kinase (RTK), phosphoinositide 3 kinase (PI3K), phosphatase and tensin homolog (PTEN), tricarboxylic acid cycle (TCA cycle), fatty acid (FA), mammalian target of rapamycin complex 1 (mTORC1) and electron transport chain (ETC).
2.2. Regulation of HIF stability and transcriptional activity
HIF-1α is an important transcription factor that plays a pivotal role in tumor cell survival under hostile microenvironments [25]. It not only regulates the cellular process under hypoxic conditions, but also upregulates drug efflux pumps to promote resistance against chemotherapies. Beyond its role in chemoresistance and angiogenesis, HIF-1α also orchestrates metabolic makeover for tumor cells [26, 27]. Accumulating evidence suggests that tumor-associated nutritional constraints and limited oxygen supply triggers HIF-1α-mediated glycolytic expression in genes. Interestingly, HIF-1α further diminishes the flux through mitochondrial TCA cycle by modulating the levels of pyruvate dehydrogenase kinase 1 (PDK1) [28]. PDK1 is an inhibitor of pyruvate dehydrogenase that converts pyruvate into acetyl-CoA. Thus, the overall effect of HIF-1α on glycolysis culminates in the reduction of oxidative phosphorylation and the consequent production of reactive oxygen species (ROS) in tumor cells. Because HIF-1α modulates multiple cellular processes in tumor cells, the stabilization and transactivation of HIF-1α is of key importance for the rapidly expanding tumor cells. Under normoxic conditions, HIF-1α is short lived due to its post-translational modifications, which include hydroxylation and ubiquitination by prolyl hydroxylases (PHDs) and von hippel-lindau (VHL) ubiquitin E3 ligase complex, respectively. However, under hypoxic conditions, as commonly present in tumors, HIF-1α is stabilized due to inactivity of PHD enzymes, which require oxygen as a substrate [29]. Beside oxygen levels, metabolic alterations such as oxoglutarate levels also modulate HIF-1α stability, with low oxoglutarate levels favoring reduced PHD activity and HIF-1α stability [30]. MUC1 manifests its metabolic functions due in part to its regulation of HIF-1α activity/stability. For example, a study by Yin et al. in 2007 presented a close relationship between HIF-1α and MUC1 [31]. In this study using an HCT116 cell line, Yin et al. reported that MUC1 mediates HIF-1α degradation under hypoxic conditions by increasing levels of PHD3, which in turn attenuates HIF-1α levels. Moreover, MUC1-associated ROS suppression was also proposed to underlie the reduced HIF-1α levels in the HCT116 cell line. Reciprocally, in a study by Aubert et al., HIF-1α directly occupied and regulated transcription from the MUC1 promoter in renal cell carcinoma [32]. Also, in a separate study of human lung adenocarcinoma cells, HIF-1α was found to modulate MUC1 expression under hypoxia [33]. A recent study demonstrated an interesting relationship between MUC1 and HIF-1α cross-regulation, with the latter being modulated by MUC1 in pancreatic cancer cells [12]. This study demonstrates that MUC1 overexpression maintains high HIF-1α protein levels due to reduced degradation of HIF-1α by PHDs. Furthermore, though levels of PHD2 and VHL ubiquitin E3 ligase are unchanged in MUC1-expressing cell lines, decreased levels of 2-oxoglutarate lead to reduced PHD activity, which underlies HIF-1α stability. As PHDs are iron (II)/2-oxoglutarate-dependent dioxygenases, decreased 2-oxoglutarate levels attenuate PHD activity, which in turn stabilizes HIF-1α. This was validated when 2-oxoglutarate treatments mitigated MUC1-induced HIF-1α stability. Similarly, a study by Kaira et al. demonstrated a positive correlation between MUC1 and HIF-1α expression in a series of histopathological samples [34]. This study demonstrated that depolarized expression of MUC1 significantly correlates with hypoxic conditions and HIF-1α and GLUT-1 levels, along with a poor outcome in the non small cell lung cancer patients. In addition to stabilizing HIF-1α, MUC1 promotes HIF-1α recruitment on the glycolytic gene promoters as well [12]. Furthermore, MUC1 directly interacts with p300, a HIF-1α co-activator, and recruits the latter onto glycolytic gene promoters in a hypoxia-dependent fashion. All these findings unanimously suggest an interplay between MUC1 and HIF-1α, which dictates the metabolic outcome by channelizing carbon flux in the hypoxic tumor microenvironments (Figure 2).

MUC1 modulates the overall metabolic flux to decrease the cellular levels of 2-ketoglutarate (2-KG), while increasing succinate levels. 2-KG is a substrate for prolyl hydroxylases (PHDs), whereas succinate is an inhibitor. PHDs cause proline hydroxylation of HIF-1α and mark it for proteasomal degradation. Hence, the net result of MUC1 expression is an increased stability of HIF-1α. MUC1 also facilitates transcriptional activity of HIF-1α by recruiting p300, a co-activator for HIF-1α, to the transcriptional complexes at glycolytic gene promoters in a hypoxia-dependent fashion. Abbreviations include the following: prolyl hydroxylase domain-containing protein (PHD), von Hippel-Lindau tumor suppressor (VHL), hypoxia inducible factor 1, alpha subunit (HIF-1α), Ubiquitin (Ub), glucose transporter1 (GLUT1), pentose phosphate pathway (PPP), and monocarboxylate transporter (MCT).
2.3. Regulation of pyruvate kinase isoform 2 (PKM2) activity
Pyruvate kinase is a critical enzyme in the glycolysis pathway because it catalyzes the last irreversible step of the cycle, i.e. generation of ATP, by transferring the high-energy phosphate group from phosphoenolpyruvate (PEP) to ADP. The end product of the above reaction is either fed into the TCA cycle or converted to lactate via lactate dehydrogenase. The pyruvate kinase isoform 2 (PKM2) is predominantly present in proliferating cells like cancer cells. Here, as a result of its slow activity, PKM2 dictates the course of glycolysis by slowing down downstream flux to pyruvate while increasing the flux through PPP, resulting in increased nucleotide biosynthesis [35]. MUC1 regulates PKM2 in both positive and negative ways. The MUC1 cytoplasmic domain directly associates with the PKM2 C-domain and promotes its activity. In contrast, epidermal growth factor receptor (EGFR) phosphorylates the cytoplasmic tail of MUC1 (pYEKV motif), which suppresses PKM2 activity by binding to its Lys-433 residue [16]. Thus, MUC1 can promote PKM2 activity to generate ATP under conditions of limited growth stimuli, whereas MUC1 can suppress PKM2 to drive the glycolytic intermediates for amino acid and lipid biosynthesis when growth stimuli are present. Overall, this suggests a MUC1 precisely controls PKM2; in turn, this cross-talk is exploited by tumor cells to influence metabolic flux, specifically aerobic glycolysis, in order to meet the energy/biosynthetic requirements for growth and survival of the tumor cells.
2.4. Regulation of Mitochondrial activity
Mitochondrion, the powerhouse of cells, is the most-important metabolic organelle due to its role in ATP production and other metabolic processes in the cell [36]. Additionally, mitochondria replenish ROS and Ca2+ for the proper functioning of anabolic and catabolic processes in cellular systems [37]. Furthermore, mitochondria regulate cell fate because a number of pro-apoptotic proteins reside inside the mitochondria, including cytochrome c, apoptosis-inducing factor (AIF), and endonuclease G [38]. Of note, tumor cells have decreased carbon flux toward oxidative phosphorylation [39]. Given that the intrinsic pathway of apoptosis requires outer membrane permeabilization of mitochondria, release of cytochrome c into the cytosol, the reduced mitochondrial activity per molecule of glucose consumed enables mitochondria to acquire resistance to apoptosis. Thus, glycolytic makeover prevents the activation of the mitochondrial apoptotic pathway in the tumor cells. To date, various studies have evidenced that HIF-1α and p53 regulate mitochondrial activity in tumors. HIF-1α regulates oxidative phosphorylation by activating PDK1, which in turn inactivates pyruvate dehydrogenase and favors glycolysis [40, 41]. p53, on other hand, regulates mitochondrial activity though cytochrome c oxidase 2 (SCO2) and SCO1 proteins [42, 43]. Additionally, p53 regulates mitochondrial permeabilization through PUMA and Noxa [44, 45]. Interestingly, MUC1 regulates activities of both HIF-1α and p53. Furthermore, localization of MUC1 in the outer mitochondrial membrane indicates a direct regulation of mitochondrial activity by MUC1 [46, 47]. Furthermore, MUC1 attenuates the release of mitochondrial apoptogenic factors in response to DNA damage that is associated with most of tumors. MUC1-mediated regulation of ROS, discussed later in this article, is another way MUC1 regulates mitochondrial activity. By routing the flux of glucose away from the TCA cycle, MUC1 controls ROS levels. Consequently, inhibition of MUC1 increases mitochondrial ROS production [48]. Moreover, inhibition of MUC1 also leads to an increase in mitochondrial ATP synthase (β-F1-ATPase) expression and a reduction in ATP levels [49]. Thus, these observations strongly suggest a crucial role of MUC1 in maintaining mitochondrial function and integrity.
3. Regulation of Carbon flux by MUC1
3.1. Modulation of flux though glycolysis
Multiple reports indicate a unique metabolic makeover caused by cancerous cells that results in their rapid proliferation [39, 50]. Irrespective of mitochondrial respiration, cancer cells switch to different metabolic pathways to harness large amounts of biomass without compromising the ATP production, thus obtaining an edge over the normal cells in terms of cell growth and division [39, 50]. One such universal pathway is the aerobic glycolysis pathway, which actively utilizes large amounts of glucose and compensates for mitochondrial dysfunction in tumor cells. Given the prime role of this pathway in proliferating cells, it is not surprising that a number of signaling molecules and networks regulate aerobic glycolysis. Interestingly, MUC1 is one such factor that directly and indirectly modulates this crucial pathway. Multiple studies provide direct evidence for the role of MUC1 in modulating aerobic glycolysis, where MUC1 expression facilitates glucose uptake and lactate production in a variety of cell types and tumor models [12, 16, 49]. MUC1 stimulates aerobic glycolysis, in part, through its direct association with PKM2 at the cys474 residue. In addition to PKM2, MUC1 also interacts with the p85 subunit of phosphoinositide 3-kinase (PI3K) and activates PI3K/Akt/mTOR pathway, which regulates glycolysis [51]. Notably, Akt is an important signaling molecule that promotes a glycolytic shift by increasing the cell-surface localization of glucose transporters; Akt also maintains the function of hexokinase in tumor cells. Hence, MUC1 also controls glucose flux in glycolysis through the precise regulation of the P13K/Akt pathway.
Another important link between MUC1 and glycolysis is p53, an extensively studied tumor suppressor gene. Specifically, p53 regulates the metabolic fate of tumor cells by controlling transcription of multiple genes during glycolysis. By decreasing the expression of GLUT1 and GLUT4, along with ubiquitination and degradation of PGM, p53 counterchecks aerobic glycolysis and promotes oxidative phosphorylation in tumors [52]. Furthermore, p53 regulates the expression of the TP53-induced glycolysis and apoptosis regulator (TIGAR), which counteracts the key regulatory enzyme phosphofructokinase through dephosphorylation of fructose-2,6-bisphosphate to fructose-6-phosphate [53]. Interestingly, p53 can also control glycolysis through the negative regulation of PI3K/Akt and mTOR signaling pathways. Importantly, a reciprocal interrelationship exists between MUC1 and p53 with regard to their roles in transcriptional regulation. In particular, MUC1 directly binds to the regulatory domain of p53 and regulates p53-responsive genes such as p21 and Bax [7]. Furthermore, MUC1 can regulate promoter occupancies by p53. Though it is unclear if MUC1 also exploits other pools of p53-regulated genes, but MUC1-p53 association does indicate a potential mechanism that most tumors utilize to potentiate glucose flux under restrictive environmental conditions.
In addition to PI3K/mTOR and p53, HIF-1α presents another critical link between MUC1 and aerobic glycolysis. Compelling evidence indicates that HIF-1α drives glycolytic flux in fast-growing tumors through direct activation of GLUT1, PDK1, and pyruvate kinase type M2 (PKM2) [22, 28]. Furthermore, HIF-1α modulates the expression and activity of key glycolytic enzymes such as HKI, HKII, PFKFB3, ALDO-A, ALDO-C, ENO-alpha, PFK-L, PGK1, PKM2, PDK1, and LDHA, thus rewiring the glucose flux from Krebs cycle towards aerobic glycolysis [28, 54]. As mentioned, recent studies indicate that MUC1 regulates the transcription of key glycolytic genes in pancreatic cell lines in a HIF-1α-dependent manner [12]. Furthermore, MUC1 stabilizes and physically interacts with HIF-1α and induces transcription of key glycolytic genes [12, 55, 56]. The crosstalk between MUC1 HIF-1α oncogenic signaling networks thus serves to facilitate aerobic glycolysis in tumors.
3.2. Modulation of flux through pentose phosphate pathway
In addition to ATP production, the generation of ribose sugars and reducing equivalents (e.g., nicotinamide adenine dinucleotide phosphate-oxidase [NADPH]) is essential for the synthesis of nucleotides in rapidly dividing tumors [57]. Moreover, NADPH not only helps in the synthesis of nucleotides, fatty acids, and sterols, but also plays a critical role in quenching ROS-associated damages by acting as a valuable component of detoxification enzyme system [58, 59]. Thus, flux in the PPP pathway, which is central to the generation of ribose sugar and NADPH, increased the amounts of glucose in transformed cells compared to non-transformed cells. PPP has two arms, oxidative and non-oxidative, which are regulated by glucose-6-phosphate dehydrogenase (G6PD) and transketolase, respectively [58]. Under basal conditions, the oxidative branch of PPP accounts for 5–30% of the total glucose flux [57]. However, in tumor microenvironments, such ratios are altered, and oxidative and non-oxidative branches equally mobilize large amounts of glucose for pentose sugar production. According to a previous study, PPP accounts for 85% of pentose sugars utilized for DNA synthesis in tumor cells [57]. Similarly, another study suggests that PPP directly controls the ribose synthesis through oxidative (<31%) and non-oxidative steps (69%) in pancreatic adenocarcinoma cells [60]. Given the crucial role of PPP in tumor growth, it is not surprising to find that key enzymes of this pathway are fine-tuned by tumor-promoting genes. For example, p53 inhibits G6PD, an important rate-limiting enzyme of the oxidative branch of PPP, through direct association [61]. This is supported by a study that shows increased glucose flux towards ribose 5-phosphate synthesis in mutant p53-associated tumors compared to their normal counterparts [61]. In the context of the non-oxidative branch, as all the steps are reversible, high levels of phosphofructokinase ensure high levels of fructose-6-phosphate and glyceraldehyde 3-phosphate that together drive the glucose flux towards PPP. Furthermore, HIF-1α is another critical regulator of the PPP that drives glucose towards the non-oxidative branch of PPP by up regulating a set of glycolytic enzymes including PGK1, aldolase A, PKM and 6-phosphofructo-2-kinase, and G6PD [22]. Given the involvement of same set of regulators (i.e., p53 and HIF-1α in PPP regulation), it is not unusual to discover that MUC1 too regulates glucose flux through these regulators. A recent study indicates that MUC1 overexpression increases the flux through pentose pathway in pancreatic adenocarcinoma cells. Specifically, by performing NMR-based metabolomics, Chaika et al. demonstrated increased synthesis of utilization of several PPP pathway metabolites [12]. Furthermore, MUC1 expression also facilitates flux through PPP by enhancing increased glucose uptake, while decreasing PKM2 activity under EGF-stimulated conditions [16].
3.3. Modulation of flux through TCA cycle
TCA cycle is a core pathway for various catabolic processes because it metabolizes pyruvate to harness reducing powers for ATP production by the respiratory chain in the cell [62]. A wealth of evidence suggests that, in accordance with glycolysis and PPP, the TCA cycle is impaired in most of tumors, where hypoxic conditions reverse the first few stages of the cycle to form citrate, which in turn produces acetyl-coA for de novo lipogenesis [63]. A regulatory interrelationship exits between TCA cycle and HIF-1α, which is directly regulated by MUC1. High levels of succinate or fumarate, caused by succinate dehydrogenase (SDH) or fumarate hydratase (FH) inhibition/mutation, respectively, stabilize HIF-1α in cancer cells [64]. Reciprocally, HIF-1α inhibits the TCA cycle by directly regulating the transactivation of PDK1, which in turn inactivates pyruvate dehydrogenase (PDH), a crucial enzyme for the conversion of pyruvate into acetyl CoA [40]. In addition to HIF-1α, p53 is another MUC1-regulated transcription factor that modulates transcription of genes involved in TCA cycle. As a result of transcriptional reprogramming by p53, flux through TCA cycle and oxidative phosphorylation is increased [42]. Thus, in line with glycolysis and PPP, MUC1 regulates the metabolic flux through the TCA cycle via HIF-1α and p53. Recent metabolomics studies confirmed such a fine-tuning mechanism in pancreatic cancer, where MUC1 expression leads to increased accumulation of TCA metabolites, indicating that MUC1 suppresses metabolite flux to and from the TCA cycle [12].
4. Survival under nutrient-deprived conditions
4.1. Survival under glucose deprivation
The high dependency of tumors on glucose as a major energy source, along with an acidic environment and hypoxic conditions, often limits glucose levels in the tumor microenvironment [65]. An inadequate supply of glucose, in turn, restricts tumor growth and may drive the tumors cells to apoptotic pathways. Studies indicate a direct association between glucose-related nutritional stress and ROS-associated cytotoxicity in tumors cells [1–4]. Interestingly, tumor cells escape this elimination by opting for the self-eating pathway (autophagy), through which the cell replenishes large pools of macromolecules such as nucleotides, fatty acids, and amino acids. MUC1 plays an important role in maintaining cancer cell survival under limited glucose conditions [66]. MUC1 counteracts glucose deprivation-associated ROS production and necrosis in colon cancer cells. Chemical inhibition of autophagy reverses MUC1-dependent survival under glucose starvation conditions. At a molecular level, under glucose deprivation conditions, MUC1-expressing cells demonstrate decreased activation of Akt, an inhibitor of 5′-adenosine monophosphate-activated protein kinase (AMPK), resulting in activation of AMPK [66, 67]. Thus, MUC1 promotes autophagy though stimulation of AMPK, which is also known as the energy sensing machinery of cells. Hence, MUC1 plays a key role in promoting autophagy and rescuing the tumor cells under constrained amounts of glucose.
4.2. Survival under glutamine deprivation
In addition to glucose, glutamine is another major fuel source for cancer growth and survival. Glutamine not only provides energy, but also serves as the prime nitrogen source for major biosynthetic processes in tumor cells [68]. Its significance is reflected in the fact that, along with glucose, glutamine is consumed at a higher rate by cancer cells. Under hypoxic conditions, where glucose is limited, glutamine provides carbon for lipogenesis. Moreover, glutamine relieves tumor-associated acidic stress by releasing ammonia [69]. Even though glutamine is an essential nutrient for cell survival and maintenance, cancer cells still manage to survive under glutamine depletion via autophagy and ROS-dependent pathways. In the context of cancer, a direct link exists between MUC1 and survival under glutamine deprivation [12]. MUC1 expression has been found to facilitate synthesis of glutamine, along with other amino acids, from glucose in pancreatic cancer cells [12]. Furthermore, MUC1-expressing cells demonstrate increased survival even under a limited quantity of glutamine. Such effects are even more pronounced under hypoxic conditions, which stabilize HIF-1α. MUC1-induced autophagy or p53 activation also explains this phenomenon. Future studies are expected to decipher the exact mechanism. Nevertheless, these observations suggest a crucial role of MUC1 in drafting the fate of cells under nutritional stress.
5. Regulation of cellular energy state
5.1. Modulation of reactive oxygen species
Increased production of ROS (e.g., hydrogen peroxide, superoxide radicals, and hydroxyl radicals) is a common occurrence in an array of diseases including cancer [70]. In almost all types of cancer, metabolic pathways are the prime targets of carcinogens, which drive the multiple stages of carcinogenesis by disrupting the fine balance between antioxidant enzymes and ROS [71, 72]. To maintain their growth demands and expansion, tumor cells adopt a variety of mechanisms to check ROS levels and associated necrosis pathways.
The role of MUC1 in ROS regulation is multifaceted. Not only can MUC1 redirect the metabolic flux away from the TCA cycle, which is largely responsible for ROS generation though the electron transport chain, MUC1 enforces a direct, positive feedback loop for ROS inhibition [12, 73]. ROS transiently upregulates MUC1 levels, which in turn attenuates ROS by inducing the expression of antioxidant enzymes such as catalase and glutathione peroxidase (GPx) that detoxify hydrogen peroxide by converting it into water [73]. MUC1 also inhibits ROS-induced apoptosis, which further underscores the critical role for MUC1 in stress-induced pathways [48]. MUC1 induces terminally differentiated myeloid phenotype in acute myeloid leukemia (AML) cell lines and primary AML cells in a ROS-dependent manner [48]. The importance of MUC1 in regulating ROS-induced cellular damage in tumors is reflected in part by the fact that MUC1-expressing cells have decreased metabolic flux to the TCA cycle, while simultaneously demonstrating increased flux to PPP and improved survival [12]. Increased flux through PPP in MUC1-expressing cells generates increased levels of NADPH, which can neutralize ROS (Figure 3).

Signaling downstream of MUC1 regulates the expression levels of ROS-attenuating enzymes (SOD1/2, CAT, and GPx) and increases the flux through the pentose phosphate pathway (PPP) in order to fine tune ROS levels in cancer cells. The PPP pathway produces high levels of NADPH that can neutralize intracellular ROS. MUC1 also enhances expression of PDK1 and LDHA that cause decreased carbon flux to the TCA cycle and hence, decreased ROS production. Enzymes upregulated by MUC1 are indicated by an upward red arrow. All the enzymes are written in blue color. Metabolic pathways upregulated by MUC1 are indicated by an upward purple arrow, whereas a downward purple arrow indicates metabolic pathways downregulated by MUC1. Abbreviations include the following: glucose transporter1 (GLUT1), pentose phosphate pathway (PPP), glutathione reductase (GR), glutathione peroxidase (GPx), catalase (CAT), superoxide dismutase (SOD), acetyl coenzyme A (AcCoA), MCT (Monocarboxylate transporter), TCA cycle (Tricarboxylic Acid cycle), pyruvate dehydrogenase (PDH), pyruvate Dehydrogenase Kinase 1 (PDK1), lactate dehydrogenase A (LDHA), α-Ketoglutarate (α-KG), and solute carrier family 1, member 5 (SLC1A5).
5.2. Regulation of ATP production by MUC1
Maintaining cellular energy is of prime importance for the rapidly dividing tumor cells, as low levels of ATP can slow cell growth and division [74]. This is supported by the fact that ATP levels govern folding and expression of growth factor receptors, thus ensuring limited growth under energy crunch [75]. More recently, a role of ATP in driving glucose uptake and PFKFB activity has been highlighted [76]. Furthermore, the ATP/AMP ratio determines cell survival, and if this ratio is significantly low, cells undergo apoptotic and necrotic pathways [77]. Under normal conditions, glycolysis followed by oxidative phosphorylation manages the cellular energy currency. However in cancer cells, where the demand-to-supply ratio is low, glycolysis takes a center stage for ATP production [77]. Though inefficient in energy production, a very high glycolytic rate overpowers oxidative phosphorylation by producing ATP over 100 times faster in cancer cells [65].
In addition to glucose, glutamine is another major fuel source for cancer growth and survival [68]. HIF-1α, c-myc, PI3K/Akt and mTOR are some of the critical metabolic regulators regulating glucose and/or glutamine flux in order to modulate ATP levels [62]. MUC1 exerts its effect on ATP production both indirectly via HIF-1α, PI3K/Akt and directly via its association with PKM2, a key enzyme that catalyzes ATP production using phosphoenolpyruvate and ADP [12, 16, 51]. Depending on its binding, MUC1 can positively or negatively regulate PKM2 activity [16]. For example, MUC1 binding at the Cys-474 of PKM2 induces PKM2 activity, which in turn promotes aerobic glycolysis, as reported in breast cancer cells [16]. Under EGF-stimulated conditions, MUC1 inhibits PKM2 activity. This inhibition results in increased flux via PPP and increased biosynthesis of nucleotides/amino acids. Another crucial association of MUC1 with ATP production is the involvement of MUC1 in maintaining electron transport chain and mitochondrial membrane integrity [48]. Markedly, MUC1 inhibition by GO-203, a cell permeable peptide that mimics MUC1 juxtamembrane region and blocks MUC1 function, is associated with the loss of mitochondrial transmembrane potential in multiple myeloma cells [48]. Furthermore, MUC1 inhibition leads to increased expression of F1-FO-ATPase, a proton-pumping ATPase, which effluxes more protons instead of producing ATP under conditions of reduced membrane potential and thus leads to ATP depletion in myeloma cells [49]. In addition to enhancing ATP production, MUC1 promotes autophagy in response to glucose deprivation in order to maintain ATP levels in cancer cells [66]. Inhibition of autophagy abrogates MUC1-mediated sustenance of ATP levels under a limited supply of glucose. Thus, MUC1 exerts its prosurvival role by relieving cells from bioenergetic stress.
5.3. Regulation of Autophagy
Autophagy, also known as self-eating, maintains cellular integrity by degrading long-lived proteins and cellular organelles as a part of the normal developmental process or under nutritional and metabolic stress [78]. This process is marked by the formation of a double-membrane vesicle, which incorporates cytoplasm along with organelles and empties the contents into lysosomes [79]. In the context of cancer, many studies have supported the role of autophagy in cell maintenance and survival of tumor cells as autophagy allows the dividing cells to respond to the changing milieu [80]. In cases of nutrient and metabolic stress, autophagy furnishes the proliferating cells with macromolecule precursors, such as amino acids, nucleotides, and carbon, thus preventing cell death to occur through apoptotic and necrotic pathways [81]. Moreover, autophagy programs chemoresistance in tumor cells [82]. Being a critical pathway for tumor growth and survival, autophagy is regulated by multiple signaling molecules, including mTOR, PDK1, PTEN, Akt, and TSC1/2 [74]. As expected, MUC1 also regulates autophagy. In particular, MUC1 promotes autophagy in response to glucose deprivation, which is reversed upon silencing ATG7 (Autophagy-related 7), an E1-like activating enzyme involved in ubiquitin-like systems and required for the formation of autophagic vacuoles [66]. Furthermore, MUC1 suppresses PI3K/Akt pathways, while facilitating AMPK activation, which in turn promotes autophagy under conditions of glucose starvation [51, 83]. HIF-1α, an interaction partner of MUC1 and a known regulator of autophagy, may further facilitate MUC1-mediated autophagy induction in the hypoxic environment of tumors.
6. Regulation of lipid metabolism
Carbohydrate, energy, and lipid metabolism are closely related. Thus, any change in one of these metabolic processes will alter the course of other pathways as well. This holds true for lipid metabolism where a plethora of evidence indicates the involvement of altered lipid metabolism in the pathogenesis of cancer [84, 85]. Studies demonstrate that most tumors augment de novo fatty acid synthesis, irrespective of available lipids resources [86]. This, in turn, helps tumors rapidly expand their cells by providing them with more building blocks, crucial signaling molecules, and valuable energy sources in response to limited glucose availability. Thus, along with synthesis, the beta-oxidation pathway is also altered to cope with the changes in cellular energetics of cancer cells [87]. Most tumors highly express enzymes and factors involved in lipid biosynthesis, such as acetyl-CoA carboxylase (ACC), ATP citrate lyase (ACL), fatty acid synthase (FAS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and sterol regulatory element-binding protein (SREBP1) [88–90]. Not surprisingly, these enzymes are downstream to many tumor suppresser genes and oncogenes, including PI3K, MAPK, p53, c-myc, and EGFR [91]. Regulation of lipid metabolism by MUC1 came into the limelight following a study by Pitroda et al. that investigated genes associated with MUC1-induced tumorigenesis [15]. This study found that in breast cancer cells, MUC1 induces a set of lipid metabolic genes, called MUC1-induced lipid metabolism signature (MLMS) [15]. MLMS constitute a set of 38 genes, which fall under lipid-metabolizing enzymes and transporters. Importantly, the list includes commonly upregulated genes in various malignancies and transformed phenotypes, such as ACL, FASN, and SERBF1 (Sterol Regulatory Element-Binding transcription Factor 1); ACL helps synthesize acetyl-CoA, a precursor for fatty acid and cholesterol synthesis. SREBP1 (SREBF1 gene product) controls the genes involved in fatty acid, cholesterol, and triglyceride synthesis. This process also involves FAS, which catalyzes the synthesis of palmitate from acetyl-CoA and malonyl-CoA in the presence of NADPH. MLMS predicts the recurrence and metastasis in tamoxifen-treated breast cancer patients, thus indicating the role of alterations in MUC1-induced lipid metabolism in the process [15]. Furthermore, MUC1-mediates HIF-1α stabilization/activation, which in turn can promote transcription of FASN and SREBP, whereas excessive lactate secretion-induced acidosis can promote FAS induction through epigenetic mechanisms, as reported in breast cancer cells [92, 93].
7. Therapeutic implications of the metabolic functions of MUC1
MUC1 signaling impinges upon multiple aspects of tumor cell biology. Such signaling cross-talk makes tumor cells more aggressive and resistant to current chemotherapeutic regimens. MUC1-mediated metabolic alterations are, at least in part, responsible for MUC1-induced phenotypes in tumor cells. Hence, targeting metabolic adaptations induced by MUC1 can potentially impact multiple facets of tumor biology and, hence, the patient outcome. Here, we describe some aspects of tumor biology that are regulated by MUC1 and can be exploited to devise therapies against tumor burden.
7.1. Desmoplasia and chemoresistance
Desmoplasia refers to the growth of fibrous and connective tissues [94]. Irrespective of the type of cancer, desmoplasia results from the proliferation of alpha-smooth muscle actin (α-SMA)-positive fibroblasts, which cause excessive deposition of extracellular matrix proteins [95]. Desmoplastic growth provides tumors a unique environment that favors growth and expansion along with resistance against chemotherapy [95]. Multiple studies suggest a direct role of MUC1 in facilitating desmoplasia and chemoresistance [96–98]. At a metabolic level, MUC1 expression leads to metabolite secretions that are known to facilitate stromal cell metabolism, growth, and/or recruitment. High amounts of lactate secreted into the extracellular milieu by MUC1-expressing cells may serve as a nutrient for the stromal cells, which possibly in turn convert lactate into pyruvate and utilize it for energy generation through the TCA cycle [12, 16, 99]. Thus, the lactate secreted potentially facilitates strong stromal cell recruitment and desmoplasia, which may diminish access of tumor cells to chemical agents and result in chemoresistance [100]. This evidence presents MUC1 as a valuable therapeutic target for effective drug treatment in tumors.
7.2. Anti-apoptotic/prosurvival functions
Defective apoptosis is one of the hallmarks of cancer, and a number of in vitro and in vitro studies have provided ample evidence to support this claim [17]. Under homeostasis, a sudden insult/change in the genetic material or metabolic machinery signals for apoptosis and favors elimination of the damaged cells [101]. However, these “death signals” are skewed in the tumor cells and enable them to escape elimination and sustain unchecked growth [102]. Multiple studies suggest anti-apoptotic functions of MUC1 [13, 51, 103]. In addition to the direct regulation of apoptotic pathways, MUC1 also facilitates survival under conditions of glucose and glutamine deprivation [12, 66]. Furthermore, in the absence of growth stimuli, MUC1-PKM2 interaction enhances activity of the PKM2 to switch the carbon flux from biomass production to ATP production and thus prevent apoptosis [16]. Hence, these data suggest a prominent role of MUC1 in preventing apoptosis and promoting survival response in tumor cells.
7.3. Recurrence and metastasis
Even after treatment, cancer often reappears either at the same/adjacent site or at a distant location [104]. This ability of cancer to spread to distant places via lymphatic or blood circulation is referred as metastasis [105]. The metastatic properties of tumors is a major hurdle in the effective treatment of cancer [106]. While MUC1 is expressed in both primary and metastatic tumor tissues, increased expression of MUC1 in primary tumors correlates with metastasis in patients [8, 9, 107–109]. From a metabolic standpoint, MUC1-expressing tumor cells secrete significant amounts of lactate into the extracellular milieu, thereby decreasing the extracellular pH and making the tumor cells motile and invasive [12, 16, 110]. Furthermore, MUC1 expression in the tumor cells allows them to survive harsh/nutrient-deprived conditions during their transit though different tissues [12, 66]. Also, signaling interactions of MUC1 with growth factor pathways, such as interactions of the EGFR-phosphorylated MUC1 cytoplasmic tail with PKM2, facilitate carbon flux to biomass production pathways under growth stimuli [16]. These metabolic adaptations due to MUC1 enable the tumor cells to successfully establish lesions at distant metastatic sites, and may facilitate tumor recurrence from any dispersed cells even after the primary tumor has been resected.
8. Conclusions and Future Perspectives
Together these studies highlight the role of MUC1 in tumorigenesis and offer therapeutic implications for targeting this transmembrane protein. For example, MUC1 signaling facilitates a variety of cellular changes in tumor cells. Due to loss of apical-basal polarity, the aberrant signaling interactions of MUC1 with RTKs and other signaling molecules induce oncogenic signaling by MUC1. Transcriptional reprogramming of metabolic genes by MUC1 along with alterations in metabolic enzymatic activities lead to alterations in the carbon flux. Such metabolic orchestration by MUC1 allows cells to fully harness the nutritional resources of tumor microenvironments and support growth and invasiveness while facilitating survival even under conditions of nutritional stress. How such metabolic adaptations reorient the stroma to further support tumor cell survival, growth, and resistance to chemotherapy requires further investigated. Future studies are also needed to fully understand the metabolic consequences MUC1-mediated signaling in response to various extracellular cues. It also remains to be determined how phosphorylation and other post-translational modifications modulate metabolic functions of MUC1. Nonetheless, MUC1 presents a unique target to overcome the metabolic adaptions of tumor cells that facilitate tumor growth, metastasis, and chemotherapy resistance in cancer patients.
Acknowledgments
The authors thank Melody Montgomery and Dr. Laura Simone at UNMC for help in the professional editing of this manuscript. This work was supported in part by R01 (R01 CA163649, NCI) to P.K.S, American Association for Cancer Research (AACR)-Pancreatic Cancer Action Network (PanCAN) Career Development Award (30-20-25-SING) to P.K.S., the Specialized Programs for Research Excellence (SPORE, P50 CA127297, NCI) Career Development Award to P.K.S., SPORE (P50 CA127297, NCI) Developmental Research Project Award to P.K.S., Pancreatic Tumor Microenvironment Research network (U54, CA163120, NCI) to P.K.S., LB506 (2014-37, DHHS-NE) to P.K.S., DOD BCRP Idea Award (BC122040, U.S. Army/USAMRAA/CDMRP) to P.K.S. and Cancer Prevention and Control Nutrition seed grant (15618, GSCN) to P.K.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.bbcan.2014.01.001
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4045475?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.bbcan.2014.01.001
Article citations
Salivary Transmembrane Mucins of the MUC1 Family (CA 15-3, CA 27.29, MCA) in Breast Cancer: The Effect of Human Epidermal Growth Factor Receptor 2 (HER2).
Cancers (Basel), 16(20):3461, 12 Oct 2024
Cited by: 0 articles | PMID: 39456554 | PMCID: PMC11506585
Hypoxia induces ROS-resistant memory upon reoxygenation in vivo promoting metastasis in part via MUC1-C.
Nat Commun, 15(1):8416, 28 Sep 2024
Cited by: 0 articles | PMID: 39341835 | PMCID: PMC11438863
Up-Regulation of Non-Homologous End-Joining by MUC1.
Genes (Basel), 15(6):808, 19 Jun 2024
Cited by: 0 articles | PMID: 38927743
MUC1 promotes cervical squamous cell carcinoma through ERK phosphorylation-mediated regulation of ITGA2/ITGA3.
BMC Cancer, 24(1):559, 03 May 2024
Cited by: 0 articles | PMID: 38702644
Evaluation of mucin-1, nuclear factor κB, and hemoglobin A1c levels in obese and non-obese individuals.
Rev Assoc Med Bras (1992), 70(4):e20231214, 03 May 2024
Cited by: 0 articles | PMID: 38716942 | PMCID: PMC11068388
Go to all (62) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer.
Proc Natl Acad Sci U S A, 109(34):13787-13792, 06 Aug 2012
Cited by: 139 articles | PMID: 22869720 | PMCID: PMC3427054
Targeting MUC1-C inhibits the AKT-S6K1-elF4A pathway regulating TIGAR translation in colorectal cancer.
Mol Cancer, 16(1):33, 02 Feb 2017
Cited by: 29 articles | PMID: 28153010 | PMCID: PMC5290603
MUC1: a multifaceted oncoprotein with a key role in cancer progression.
Trends Mol Med, 20(6):332-342, 22 Mar 2014
Cited by: 406 articles | PMID: 24667139 | PMCID: PMC5500204
Review Free full text in Europe PMC
MUC1 protein induces urokinase-type plasminogen activator (uPA) by forming a complex with NF-κB p65 transcription factor and binding to the uPA promoter, leading to enhanced invasiveness of cancer cells.
J Biol Chem, 289(51):35193-35204, 04 Nov 2014
Cited by: 21 articles | PMID: 25371209 | PMCID: PMC4271208
Funding
Funders who supported this work.
American Association for Cancer Research (AACR)—Pancreatic Cancer Action Network (PanCAN) Career Development Award (1)
Grant ID: 30-20-25-SING
Cancer Prevention and Control Nutrition (1)
Grant ID: 15618
DOD BCRP Idea Award (1)
Grant ID: BC122040
LB506 (1)
Grant ID: 2014-37
NCI NIH HHS (4)
Grant ID: P50 CA127297
Grant ID: U54 CA163120
Grant ID: P30 CA036727
Grant ID: R01 CA163649
Pancreatic Tumor Microenvironment Research network (1)
Grant ID: U54, CA163120
R01 (1)
Grant ID: R01 CA163649
SPORE Developmental Research Project Award (1)
Grant ID: P50 CA127297
Specialized Programs for Research Excellence (SPORE) Career Development Award (1)
Grant ID: P50 CA127297





