Abstract
Free full text

Methylation Patterns in Whole Blood Correlate With Symptoms in Schizophrenia Patients
Abstract
DNA methylation, one of the main epigenetic mechanisms to regulate gene expression, appears to be involved in the development of schizophrenia (SZ). In this study, we investigated 7562 DNA methylation markers in blood from 98 SZ patients and 108 healthy controls. A linear regression model including age, gender, race, alcohol, nicotine and cannabis use status, and diagnosis was implemented to identify C-phosphate-G (CpG) sites significantly associated with diagnosis. These CpG sites were further validated using an independent data set. Sixteen CpG sites were identified with hyper- or hypomethylation in patients. A further verification of expression of the corresponding genes identified 7 genes whose expression levels were also significantly altered in patients. While such altered methylation patterns showed no correlation with disorganized symptoms and negative symptoms in patients, 11 CpG sites significantly correlated with reality distortion symptoms. The direction of the correlations indicates that methylation changes possibly play a protective mechanism to lessen delusion and hallucination symptoms in patients. Pathway analyses showed that the most significant biological function of the differentially methylated CpGs is inflammatory response with CD224, LAX1, TXK, PRF1, CD7, MPG, and MPO genes directly involved in activations of T cells, B cells, and natural killer cells or in cytotoxic reaction. Our results suggest that such methylation changes may modulate aspects of the immune response and hence protect against the neurobiological substrate of reality distortion symptoms in SZ patients.
Introduction
It has been generally accepted that both genetic and environmental factors play a significant role in schizophrenia (SZ). However, as a complex genetic disorder, the epigenetic role in the development of SZ has been recognized in recent years, after first being proposed and modeled by Petronis.1 DNA methylation as one of the main epigenetic mechanisms regulates the expression of genes across the life span. The best known mechanism involves methylation of C-phosphate-G (CpG) dinucleotides within the promoter region of a gene, which silences or downregulates the expression of the gene. It has shown functions in embryonic development,2 X-chromosome inactivation, genomic imprinting,3 and preservation of chromosome stability.4 It is not surprising that DNA methylation may mediate environmental influences on gene expression related to SZ5–8 and a epigenetic model may integrate diverse empirical data into a powerful SZ pathogenetic synthesis.9 In addition to several gene-specific methylation studies of SZ using postmortem brain tissue,10–14 a methylome-wide study of the frontal cortex strongly supported the epigenetic theory of major psychosis, in particular SZ.15
It is important to know whether, or to what extent, the easily accessible peripheral tissues such as blood can act as a surrogate to study the intersubject phenotypic variation, ie, whether methylation change in blood can indicate the status of SZ patients. A recent study by Davies et al16 analyzed DNA methylome-wide profiling across tissues and found that the interindividual differences of DNA methylation are significantly correlated across blood and brain. The correlation between blood and cerebellum was 0.76, and it was 0.66 between blood and cortex. This study together with other studies11,17–20 supports the validity for the investigation of blood DNA methylation in SZ.
Few studies have used blood DNA methylation to identify potential biomarkers for SZ disease status.21,22 Global hypomethylation in leukocytes has characterized SZ patients in 2 studies,23,24 and some genes with hyper- or hypomethylation in blood were also reported21,23 in SZ patients. Yet, the epigenetic study of SZ is still in its early phase. The mechanism by which methylation interacts with genetic and environment factors to regulate or reflect to the progress of SZ is far from clear. We believe that by incorporating methylation, gene expression, and psychopathology related to SZ we can derive informative findings to help understand the role of methylation change in SZ. In this study, we have tested SZ diagnostic effect on DNA methylation at each CpG site across the genome in 98 SZ patients and 108 healthy controls. Independent data sets were used to validate methylation changes and to investigate the corresponding gene expression changes. Furthermore, we evaluated the association of altered methylation patterns with symptoms of patients.
Methods
Participants
Participants in this study come from the Mind Clinical Imaging Consortium, a collaborative effort of 4 research sites. Site information and enrollment for SZ patients and healthy controls are explained in online supplementary text. Ninety-eight SZ patients and 108 healthy controls were analyzed here. As listed in table 1, no significant difference was observed in age and gender between patients and controls. Significantly more African Americans were enrolled as patients than controls, which we controlled for in our regression model. Alcohol and cannabis use were assessed by 2 categories: (1) abuse or dependence (not active during the month previous to the study) and (2) never abused or dependent. Nicotine use was measured by calculated Pack_Years, as done by Cullen et al.25 Summaries of symptom measures and medication status for patients are also listed in table 1. Patients symptoms were assessed by the Scale of the Assessment of Positive Symptoms and the Scale of the Assessment of Negative symptoms.26,27 Reality distortion symptom score is the sum of the global ratings of delusions and hallucinations, and disorganized symptom score is the sum of the global ratings of bizarre behavior and positive formal thought disorder. The sum of the global ratings of affective flattening, alogia, avolition-apathy, and anhedonia-asociality is the negative symptom score.
Table 1.
Demographic Information of Subjects Analyzed in This Study
| Subjects | Male/Female | Age | Race (White/Black/ Asian/Others) | Alcohol Abuse/Dependent | Cannabis Abuse/Dependent | Nicotine Pack_Year | |
|---|---|---|---|---|---|---|---|
| SZ (98) | 73/25 | 34±11 | 73/17/3/5 | 32 | 30 | 8.8±11.8 | |
| Healthy controls (108) | 70/38 | 32±11 | 97/3/4/4 | 0 | 0 | 1.0±4.8 | |
| Patient information | |||||||
 Reality distortion symptoms Reality distortion symptoms | Range: 0–10; mean: 4.84±2.80 | ||||||
 Disorganized symptoms Disorganized symptoms | Range: 0–10; mean: 1.85±1.93 | ||||||
 Negative symptoms Negative symptoms | Range: 0–20; mean: 8.11±3.97 | ||||||
 Age of onset Age of onset | Range: 13–46; mean: 22±6 | ||||||
 Antipsychotic medication Antipsychotic medication | 90 patients in use; 3 not in use; 5 with missing data | ||||||
 Use of haloperidol, risperidone, clozapine, olanzapine, quetiapine, and aripiprazole Use of haloperidol, risperidone, clozapine, olanzapine, quetiapine, and aripiprazole | 7, 32, 15, 13, 11, 18 patients for each type, respectively | ||||||
 Use of HDAC inhibitor Use of HDAC inhibitor | 6 patients on divalproex sodium; 1 on valproic acid | ||||||
Note: HDAC, histone deacetylases; SZ, schizophrenia.
DNA Methylation Assay
DNA from blood samples was assessed by the Illumina Infinium Methylation27 Assay. A methylation value, beta (β), represents the ratio of the methylated probe intensity to the total probe intensity. A series of quality controls on the beta values were applied to remove bad samples and probes (see online supplementary text). Then, 7562 “variable probes”21 in autosomes were selected for association analyses. Variable probes are defined by calculating the mean of the SD of beta values of all probes and selecting those with SD larger than the mean SD value.21
Genomic Methylation Analyses for SZ Association
Methylation has been shown to be affected by the interplay between genetic and environmental factors and has shown to differ under various conditions, including age,28,29 gender,30 race,31 diet, and lifestyle.32–34 The impact of nicotine and alcohol use on DNA methylation has been specifically studied, and both global change and changes in specific genes have been reported in various tissues.35–44 Albeit there have been no direct studies of cannabis use on methylation, it is conceivable and has been hypothesized45 that a similar effect may occur. Considering such potential confounding factors, we applied to each CpG site a linear regression model: β = α1 Age + α2 Gender + α3 Race + α4 Alcohol_Use + α5 Cannabis_Use + α6 Pack_Years + α7 Diagnosis + ε. We calculated the significance of diagnosis effect and its effect size: a delta-β (Δβ) value represents the mean difference in methylation between SZ patients and controls after correcting for the covariates. To identify the CpG sites associated with SZ diagnosis, we applied similar criteria as in the study of Dempster et al,21 requiring both significance P value <.0001 and effect size Δβ > 0.02.
Validation on Methylation Changes
A public Gene Expression Omnibus (GEO) data GSE 41037 were used for validation. These data include DNA methylation profiling of whole blood in 325 SZ patients and 394 healthy subjects, assessed by the same Illumina Infinium Methylation27 Assay. This sample comprises 446 males and 273 females with an average age of 37 ± 16 years. Without substance use information, a simple linear regression with diagnosis, age, and gender was used to validate SZ association of identified methylation sites in our data. A P value less than .05 for diagnosis effect was required for validation.
Gene Expression Verification
A public GEO data, GSE38484, were used for verification of gene expression changes. These data include gene expression profiles in whole blood of 106 SZ patients and 96 controls. The verification was only applied to validated methylation sites, using a linear regression model with age, gender, and diagnosis as independent variables, followed by a permutation test. The details are proved in online supplementary text.
Medication and Symptoms Tests
Medication measures include the current chlorpromazine equivalent dosage,46 and 7 separate binary variables indicating the current use of haloperidol, risperidone, clozapine, olanzapine, quetiapine, aripiprazole, and histone deacetylases inhibitors (divalproex and valproic acid). In addition, duration of illness was also tested to capture the effect of chronicity. Symptoms included reality distortion symptoms, disorganized symptoms, negative symptoms, and age of onset. Correlations between methylation values of validated sites and such medication and symptom variables were calculated. A P value less than .05 was considered significant for any potential correlation. Furthermore, we conducted permutation tests, through randomly selecting a set of CpG sites equal in number to the validated sites and calculating the chance the number of correlated CpG sites was the same or higher than that found from the validated methylation sites.
Pathway and Network Analysis
Interactive pathway analyses (IPA) from Ingenuity Systems (http://www.ingenuity.com) was used to assess the validated methylation-altered genes for the enrichment of gene networks, canonical pathways, and biological processes. Enrichment for specific pathways, networks, or functions was determined relative to the Ingenuity Knowledge Base, a repository of biological interactions and functional annotations created from millions of individually modeled relationships between proteins, genes, complexes, cells, tissues, drugs, and diseases.
Results
In our data of 98 SZ patients and 108 healthy controls, we identified 20 CpG sites associated with SZ diagnosis (P < .0001, and absolute Δβ > 0.02), after controlling for age, gender, race, alcohol, cannabis, and nicotine use. During the validation test, 14 CpG sites showed patient vs control group differences with P < .05 and 2 sites showed marginal difference with P values <.06. These 16 sites presented the same altered hyper- or hypomethylation in patients in the validation data as in our discovery data, as shown by Δβ (SZ patients − controls) in table 2. Hereafter, we consider these 16 sites from 16 genes as validated methylation-altered sites, on which further gene expression verification, medication effect, symptom association, and pathway analyses were conducted. The full names of these genes are provided in online supplementary text.
Table 2.
Results From the 16 C-phosphate-G Sites Associated With SZ
| Gene | Target ID | Discovery | Validation | Gene expression | Reality Distortion Symptoms | ||||
|---|---|---|---|---|---|---|---|---|---|
| Δβa | P value | Δβa | P value | Δb | P value | r c | P value | ||
| CD244 | cg11939496 | −0.03 | 4.75E-05 | −0.04 | 6.47E-10 | −0.07 | 1.91E-02 | 0.26 | 8.50E-03 |
| LAX1 | cg12022621 | 0.02 | 4.04E-05 | 0.01 | 6.18E-02* | −0.10 | 5.67E-03 | −0.26 | 1.06E-02 |
| ESPNL | cg09039163 | −0.03 | 1.28E-05 | −0.01 | 2.52E-03 | −0.02 | 5.47E-02 | 0.11 | 2.84E-01 |
| TXK | cg20981615 | 0.03 | 3.36E-05 | 0.01 | 1.24E-04 | 0.02 | 3.34E-01 | −0.36 | 3.00E-04 |
| PRF1 | cg09914304 | 0.02 | 2.50E-05 | 0.04 | 3.69E-07 | −0.46 | 2.29E-10 | −0.20 | 4.47E-02 |
| MS4A1 | cg06806711 | 0.02 | 9.26E-06 | 0.01 | 6.10E-04 | 0.02 | 3.78E-01 | −0.15 | 1.27E-01 |
| TCN1 | cg20018806 | −0.02 | 3.65E-05 | −0.01 | 1.20E-04 | 0.23 | 2.23E-03 | 0.22 | 2.84E-02 |
| APBA2 | cg21917349 | 0.03 | 3.64E-05 | 0.01 | 1.67E-02 | −0.01 | 9.11E-01 | −0.27 | 6.50E-03 |
| FAM173A | cg09830866 | 0.03 | 9.66E-05 | 0.04 | 1.53E-07 | −0.08 | 2.72E-02 | −0.31 | 2.30E-03 |
| CBFA2T3 | cg13745346 | −0.02 | 1.11E-05 | −0.03 | 4.10E-07 | −0.09 | 1.93E-06 | 0.27 | 7.70E-03 |
| MPG | cg16003913 | −0.03 | 8.87E-05 | −0.01 | 5.89E-02* | 0.00 | 7.32E-01 | 0.31 | 1.60E-03 |
| CD7 | cg02473123 | 0.02 | 6.05E-05 | 0.02 | 1.11E-03 | −0.22 | 2.41E-05 | −0.22 | 2.76E-02 |
| MPO | cg04988978 | −0.03 | 9.15E-05 | −0.01 | 6.93E-05 | 0.08 | 2.54E-01 | 0.18 | 7.30E-02 |
| SLC25A10 | cg07845392 | −0.02 | 2.59E-05 | −0.04 | 9.50E-10 | 0.01 | 5.48E-01 | −0.03 | 7.51E-01 |
| CKM | cg19154438 | 0.02 | 3.33E-05 | 0.03 | 6.63E-09 | 0.00 | 4.75E-01 | −0.21 | 4.18E-02 |
| H1F0 | cg07141002 | −0.03 | 8.38E-06 | −0.04 | 2.92E-06 | −0.08 | 1.22E-01 | 0.14 | 1.58E-01 |
aΔβ of SZ patients—controls.
bΔ, a gene expression difference of SZ patients—controls.
c r, correlation between reality distortion symptom scores and methylation values.
*Marginal significant P values of ~.06.
From the gene expression verification test, 7 genes showed marked expression changes with P < .05 (shown in bold in the gene expression column in table 2) with CD244, LAX1, PRF1, FAM173A, CBFA2T3, and CD7 upregulated and TCN1 downregulated in patients. According to the 100 000 permutation test on randomly selected 16 genes, the chance of having 7 or more genes with significant SZ-related up- or downregulation is less than 0.05. Five of these changes followed the direction of hypomethylation to upregulation and hypermethylation to downregulation, eg, SZ patients showed hypomethylation at the CpG site in TCN1 and corresponding upregulation of TCN1 gene expression. Two genes, CD244 and CBFA2T3, presented downregulation with hypomethylation.
When testing medication effects, the methylation of gene MS4A1 presented a significant association with current chlorpromazine equivalent dosage (P = .01). Higher dos age correlated to higher methylation values, leading to the same direction as SZ-related hypermethylation. Thus, MS4A1 hypermethylation observed in patients may be due to medication. Therefore, we removed MS4A1 from the genetic pathway analyses of SZ methylation-altered genes. Out of 7 types of psychiatric medications, only quetiapine showed an effect on methylation patterns of CD244, PRF1, APBA2, FAM173A, CBFA2T3, and CD7, where use of quetiapine was linked to methylation change counteracting the SZ related hyper- or hypomethylation. A post hoc test showed that the use of quetiapine was not correlated to any symptom scores (P > .38). Regarding illness duration, genes MPG and SLC25A10 showed significant correlations with P = .015 and P = .003, respe ctively. The longer the duration of illness, the higher the methylation value. This hypermethylation effect is opposite to the hypomethylation observed in patients, suggesting that illness duration may reflect a mixture of prolonged medication exposure and/or a saturation effect.
Out of the 16 sites, none were correlated with disorganized symptoms or negative symptoms. In contrast, 11 CpG sites were significantly correlated with reality distortion symptoms, which are shown as P values in bold in the last column in table 2. The 100 000 permutation test showed that the chance of having 11 or more sites associated with reality distortion symptoms is less than 1.0 × 10−6. We plotted the slopes between reality distortion symptom scores and methylation beta values in figure 1, as well as the mean values for both patients and controls. Seven sites from genes LAX1, TXK, PRF1, APBA2, FAM173A, CD7, and CKM, showed that SZ patients were on average hypermethylated with higher mean beta values than controls; the higher the beta values patients had, the lower the reality distortion symptom scores were. In the other 4 CpG sites from genes CD244, TCN1, CBFA2T3, and MPG, SZ patients were on average hypomethylated with lower mean beta values than controls, and the lower the beta values, the lower the reality distortion symptom scores. Therefore, from the relative position of mean differences and the slope direction, it can be seen that the altered hyper- or hypomethylation observed in SZ patients relates to lower reality distortion symptom scores. In addition, methylation of CBFA2T3 also correlated to age of onset (P = .028); late onset was linked to low methylation values, suggesting that hypomethylation in patients delays symptom onset.
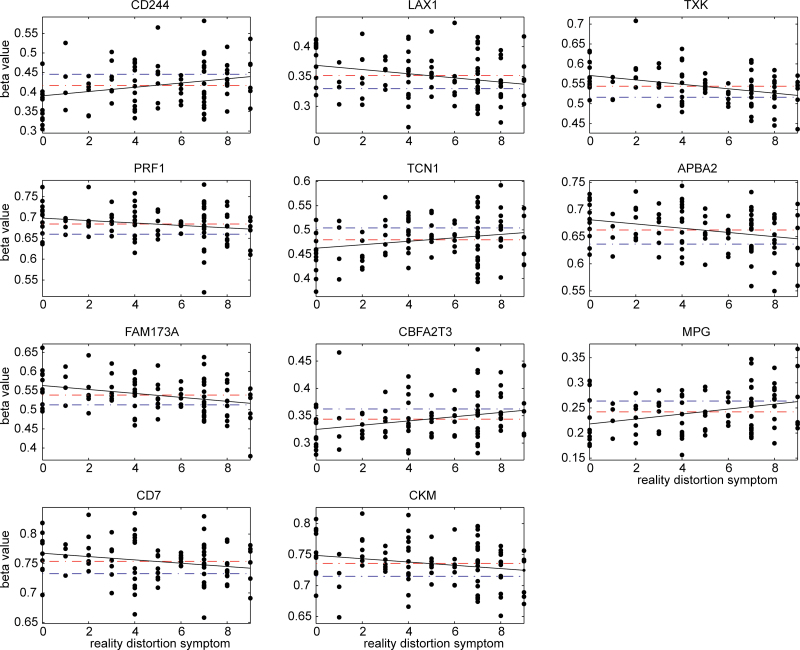
Methylation beta values of 11 C-phosphate-G (CpG) sites in schizophrenia (SZ) patients sorted by their reality distortion symptom scores; x axis plots reality distortion symptoms and y axis plots the measured beta values. Red dashed lines are the mean beta values of patients, while blue dashed lines are the mean beta values of healthy controls. The black line presents the slope between reality distortion symptom score and methylation beta value in patients.
Removing 1 gene, MS4A1, IPA revealed the most significant network of hematological development and function from methylation-altered genes, involving activation of lymphocytes (CD244, CD7, LAX1, and PRF1), activation of T cells (CD7, LAX1, PRF1, and TXK), activation of natural killer cells (CD224 and PRF1), activation of B cells (LAX1), and differentiation of granulocytes (CBF2T3). These genes together with genes MPO and MPG, involved in cytotoxic reaction, formed the most significant biological function, the inflammatory response (P = 7.0 × 10−6).
Discussion
Given the known effect on methylation from age, gender, ethnicity, substance use, and comorbidity between SZ and substance abuse, we implemented a full regression model to account for possible confounding factors. The 16 CpG sites not only presented significant association with SZ diagnosis after controlling for confounding factors in our data but also presented the same hyper- or hypomethylation in patients in an independent data set with a significance P value close to or less than .05. The significance criteria used in our discovery data are not strictly Bonferroni corrected. Instead, it is a combination of P < .0001 and effect size Δβ > 0.02. This practice21 provides a balanced control for false-positive and false-negative results. Using our data as an example, 2 CpG sites associated with diagnosis in the discovery group (P = 4.6 × 10−6 and 5.5 × 10−6, passing Bonferroni correction, and Δβ < 0.02) could not be replicated with the validation group with P = .28 and .39. These 2 sites are likely false positives. In contrast, 1 CpG site (cg03636183) residing in F2RL3 showed an association with Pack_Years of P = 2.1 × 10− 5, failing Bonferroni correction, but its connection with smoking has been reported and replicated in different studies.38,40 Thus, strict Bonferroni correction will likely cause a false-negative finding in this case. We believe that by using combined P value and effect size as criteria, plus validation tests, we are able to identify with confidence the altered methylation associated with SZ diagnosis.
To further probe the biological effect of altered methylation patterns, we tested their corresponding gene expression changes. Seven genes were significantly up- or downregulated in SZ patients with P < .05. Five CpG sites were located near to transcription start sites (TSS), ranging from 144 to 705 base pairs away, and their corresponding gene expression changes followed the direction of hypermethylation to downregulation and hypomethylation to upregulation, which reflects a common methylation regulation effect. The other 2 CpG sites in genes CD244 and CBFA2T3, located 915 and 1121 base pairs away from their TSS, respectively, were hypomethylated in patients, and their gene expression were downregulated. Given only 1 CpG site for each gene was measured in our data, we were unable to explore the exact mechanisms of gene expression change. However, the changes that we observed provides further evidence that altered methylation patterns in patients may affect downstream cellular change and further influence behavioral or clinical symptoms.
Our correlation tests with symptoms showed pronounced associations of methylation with reality distortion symptoms, while no connection with disorganized and negative symptoms. Eleven CpG sites were correlated with reality distortion symptoms, which occurred well below chance (P < 1.0 × 10−6). As illustrated in figure 1, for the 7 sites, which were hypermethylated in patients, increased methylation was associated with lower reality distortion symptom scores. For the 4 sites, which were hypomethylated in patients, decreased methylation was associated with lower reality distortion symptom scores. The 11 sites present that the altered methylation is related to a lower reality distortion symptom score. Though we cannot prove causality, the results are consistent with methylation changes being a responsive or protective biological reaction to counteract delusion and hallucination symptoms. The same suggestion can be drawn from the correlation of CBFA2T3 hypomethylation with age of onset, whereby the SZ-related hypomethylation delays illness onset.
This suggestion is further strengthened by the genetic function analyses. In particular, genes involved directly in the inflammatory response provide a possible biological pathway to react to or participate in the pathological changes of SZ. Our findings that the methylation of these genes, related to inflammatory processes and reality distortion symptoms in the SZ group, converges with several lines of evidence regarding the importance of inflammatory processes in SZ. Doorduin et al,47 using positron emission tomography, documented increased microglial activation in the hippocampus of SZ subjects with significant positive symptoms. Moreover, antipsychotic drugs have been reported to modulate microglial activation and inhibit the production of various cytokines in the brain.48,49 In addition, a variety of anti-inflammatory drugs, like celocoxib, minocycline, and aspirin, appear to have antipsychotic properties.50–53 It is possible that the immune-modulating effects on psychotic symptoms are in part through gene expression change (specifically downregulation confirmed by our study) regulated by hypermethylation of LAX1, TXK, PRF1, CD7, and CKM and hypomethylation of CD244, CBFA2T3, and MPG. It is also noteworthy that gene TCN1 encodes a vitamin B12-binding protein that facilitates the transport of B12 into cells. Our data suggest that upregulation of TCN1 expression by hypomethylation in SZ patients reduces reality distortion symptoms, consistent with the observation of B12 deficiency-induced SZ-like symptoms.54,55
Regarding medication influence, our test showed that hypermethylation of MS4A1 in patients was significantly influenced by medication dosage. Therefore, it was removed for genetic analyses. The use of quetiapine in patients decreased the SZ-associated hyper- or hypomethylation and was not related to symptoms. Therefore, we believe that the hyper- or hypomethylation acting against reality distortion symptoms in patients is not an effect of the medication.
The cross-sectional nature of this study limits causal inferences. Recently, Davies et al16 reported a set of genes in neurodevelopment and neuronal differentiation functional pathways differentially methylated across blood, brain, and regions of brain in a healthy population.16 We tested whether this set of genes better relates to SZ or SZ symptoms. We found no significant results (for details see online supplementary text), suggesting that these genes are more sensitive to healthy brain development than to SZ. We speculate that the methylation patterns we identified in blood related to inflammation may correspond to similar methylation patterns in brain tissue, as demonstrated by high correlation of interindividual variation across brain and blood.16 Future postmortem methylation studies and longitudinal studies assessing the relationship between symptoms and methylation would provide important mechanistic information.
Our study comes with a number of limitations. One main limitation is that methylation value derived from whole blood presents a mixture of various leukocyte subtypes. As reported by Reinius et al,56 methylation differs between leukocyte subtypes, and methylation patterns in whole blood can be different from that in specific cell types. Our findings regarding methylation in whole blood can be partially confounded by various proportions of leukocyte subtypes. Further study is necessary to confirm the relation between methylation of inflammation genes with SZ. Second, measures from Illumina Methylation27 Assay were not confirmed by a different platform due to inadequate DNA quantity. However, Breitling and colleagues38 utilized Sequenom MALDI-TOF mass spectrometry to verify the pattern detected by Illumina Methylation27 Assay and arrived at similar results. Alternatively, we used an independent sample to validate the methylation pattern. Third, gene expression data to verify the methylation functional effect were drawn from different samples. It may reduce the direct connection between methylation and gene expression change, but the expression change we have observed in a different sample still provides, to an extent, evidence of methylation regulation effect. Fourth, we investigated DNA methylation of peripheral blood sample, where methylation of brain tissue may be more directly related to schizophrenic symptoms. The last limitation is that methylation can be potentially affected by many environmental factors, such as diet, medication, and lifestyle. We do not have accurate chronic medication dosage measures to precisely assess chronic medication effect though we have used current medication dosage and illness duration to provide some indications. We have tested the effect of body mass index (BMI) by adding BMI into the regression model, which did not change our results. We also tested the interaction terms between alcohol, cannabis, and nicotine use and diagnosis, but neither interaction term changed results.
In summary, DNA methylation of some genes, mainly related to inflammation, in blood is significantly altered in SZ patients, and this hyper- or hypomethylation pattern also protects against reality distortion symptoms. Our data provide evidence for the functional impact of methylation in SZ, and a potential pathway through epigenetic regulation, which has been hypothesized earlier.
Supplementary Material
Supplementary material is available at http://schizophrenia bulletin.oxfordjournals.org.
Acknowledgment
The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
Articles from Schizophrenia Bulletin are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/schbul/sbt080
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/schizophreniabulletin/article-pdf/40/4/769/16705871/sbt080.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/schbul/sbt080
Article citations
Epigenetic profile of the immune system associated with symptom severity and treatment response in schizophrenia.
J Psychiatry Neurosci, 49(1):E45-E58, 01 Jan 2024
Cited by: 2 articles | PMID: 38359932 | PMCID: PMC10890792
DNMT1 downregulation as well as its overexpression distinctly affect mostly overlapping genes implicated in schizophrenia, autism spectrum, epilepsy, and bipolar disorders.
Front Mol Neurosci, 16:1275697, 06 Dec 2023
Cited by: 0 articles | PMID: 38125006 | PMCID: PMC10731955
Analysis of DNA methylation at birth and in childhood reveals changes associated with season of birth and latitude.
Clin Epigenetics, 15(1):148, 11 Sep 2023
Cited by: 1 article | PMID: 37697338 | PMCID: PMC10496224
Endocannabinoid signaling and epigenetics modifications in the neurobiology of stress-related disorders.
Neuronal Signal, 7(2):NS20220034, 25 Jul 2023
Cited by: 2 articles | PMID: 37520658 | PMCID: PMC10372471
Review Free full text in Europe PMC
YBX1-Mediated DNA Methylation-Dependent SHANK3 Expression in PBMCs and Developing Cortical Interneurons in Schizophrenia.
Adv Sci (Weinh), 10(20):e2300455, 21 May 2023
Cited by: 3 articles | PMID: 37211699 | PMCID: PMC10369273
Go to all (69) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE38484
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Association of DNA Methylation Differences With Schizophrenia in an Epigenome-Wide Association Study.
JAMA Psychiatry, 73(5):506-514, 01 May 2016
Cited by: 99 articles | PMID: 27074206 | PMCID: PMC6353566
Identification of schizophrenia-associated loci by combining DNA methylation and gene expression data from whole blood.
Eur J Hum Genet, 23(8):1106-1110, 26 Nov 2014
Cited by: 26 articles | PMID: 25424713 | PMCID: PMC4795100
Methylome-wide association study of first-episode schizophrenia reveals a hypermethylated CpG site in the promoter region of the TNIK susceptibility gene.
Prog Neuropsychopharmacol Biol Psychiatry, 106:110081, 24 Aug 2020
Cited by: 4 articles | PMID: 32853717
Epigenome-wide association study of suicide attempt in schizophrenia.
J Psychiatr Res, 104:192-197, 20 Jul 2018
Cited by: 12 articles | PMID: 30103066
Review
Funding
Funders who supported this work.
NIBIB NIH HHS (4)
Grant ID: R01 EB005846
Grant ID: R01 EB006841
Grant ID: R01 EB020407
Grant ID: R01EB005846
NIDA NIH HHS (1)
Grant ID: R33DA027626
NIMH NIH HHS (2)
Grant ID: R01 MH104680
Grant ID: R01 MH094524





