Abstract
Free full text

Sweating the Small Stuff: MicroRNAs and Genetic Changes Define Pancreatic Cancer
Abstract
MicroRNAs (miRNAs) are 18- to 22-nucleotide-long, single-stranded, noncoding RNAs that regulate important biological processes including differentiation, proliferation, and response to cellular stressors such as hypoxia, nutrient depletion, and traversion of the cell cycle by controlling protein expression within the cell. Many investigators have profiled cancer tissue and serum miRNAs to identify potential therapeutic targets, understand the pathways involved in tumorigenesis, and identify diagnostic tumor signatures. In the setting of pancreatic cancer, obtaining pancreatic tissue is invasive and impractical for early diagnosis. Several groups have profiled miRNAs that are present in the blood as a means to diagnose tumor progression and predict prognosis/survival or drug resistance. Several miRNA signatures found in pancreatic tissue and the peripheral blood, as well as the pathways that are associated with pancreatic cancer, are reviewed here in detail. Three miRNA biomarkers (miR-21, miR-155, and miR-200) have been repetitively identified in both pancreatic cancer tissue and patients’ blood. Those miRNAs regulate and are regulated by the central genetic and epigenetic changes observed in pancreatic cancer including p53, transforming growth factor [beta], p16INK4A, BRCA1/2, and Kras. These miRNAs are involved in DNA repair, cell cycle, and cell invasion and also play important roles in promoting metastases.
Approximately 43,140 Americans are diagnosed with pancreatic cancer and 36,800 individuals will die of this disease this year.1 Pancreatic cancer is associated with less than a 5% 5-year survival rate. Early diagnosis is rare, and surgical treatment is most beneficial before the cancer is confirmed as being locally invasive or metastatic. There is a substantial unmet clinical need to develop diagnostic markers for early identification of pancreatic cancer. Although CA-19-9 is widely used to monitor therapy, it is most often detectable only late in disease.2 Recently, microRNAs (miRNAs), present within the tumor and in the blood, have been identified as potential quantitative measures of tumor that may be identified earlier in disease. MicroRNAs are 18- to 22-nucleotide-long, single-stranded, noncoding RNAs that regulate the expression of suites of up to 100 messenger RNA (mRNA) species at a posttranscriptional level. MicroRNA expression is very sensitive to changes within the tumor microenvironment (eg, stress, trauma, hypoxia, infection, and cancer 3–7). Pancreatic cancer tissue-derived miRNAs have been identified as signatures for diagnosis, predicting prognosis, and drug responsiveness and as potential therapeutic targets 8 (Fig. 1). Problems with using tissue miRNA as diagnostic tools are the lack of readily available samples and the invasiveness of the sample collection procedure, making it impractical for early diagnostic purposes. Some groups have now turned to assessing blood miRNAs (including whole blood, plasma, serum and peripheral blood mononuclear cells [PBMCs]) as a means to detect prostate cancer and pancreatic cancer. MicroRNA biomarkers have been identified in the blood of patients with ovarian cancer; gastric cancer; acute leukemia; lung, oral, colorectal, and pancreatic cancer; and melanoma.9–20 Based on the current literature, there is no specific miRNA that is commonly expressed in any individual cancer type. The advantages of using blood for detection include reduced invasiveness and better suitability for early detection. The assay can be included in routine blood tests for early cancer diagnoses. Here, we review multiple miRNA profiling studies from both pancreatic cancer tumor tissues and biological fluid to identify pancreatic cancer miRNA biomarkers that are commonly expressed. We also examine commonly expressed biomarkers in tissues and biofluids and the connection between the miR predicted genetic targets to glean insights into miRNA tumor biology and the reciprocal canonical genetic changes in the setting of pancreatic cancer.
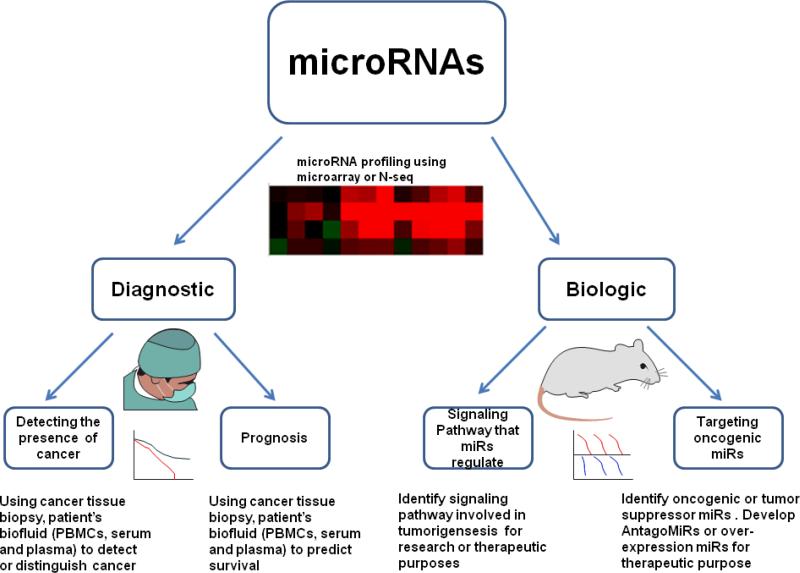
miRNA application in cancer research. miRNAs can be used for both diagnostic and biologic studies. miRNA profiling using miR microarray, qRT-PCR, or RNA-seq can identify important miRNA expression changes at various disease stages. miRNA profiling (obtained from cancer tissue biopsies or patient's blood) can be used to detect the presence of tumor and help define prognosis. miRNA profiling is widely used to identify signaling pathways involved in tumorigenesis, to develop new therapeutic strategies, or to directly target the oncogenic MicroRNAs as therapies.
BIOGENESIS OF MIRNAs AND MECHANISM OF REGULATION OF GENE EXPRESSION
MicroRNAs are transcribed by RNA polymerase II/III in the nucleus, and the primary miRNAs are then processed by Drosha into hair-loop pre-miRNAs before export to the cytoplasm by exportin 5. In the cytoplasm, pre-miRNA is cleaved by Dicer into a mature single-stranded miRNA hairpin loop, which regulates its cognate targeted gene mRNA by 2 primary mechanisms (Fig. 2). MicroRNAs utilize the RNA-induced silencing complex (RISC) to regulate target genes by binding the 3' UTR (untranslated region). When miRNA is perfectly matched with the target mRNA, it will induce cleavage, thus inhibiting gene expression. When the miRNA is imperfectly matched, it will induce translational repression. Thus, the overall mRNA remains unchanged while gene expression is inhibited. MicroRNA can induce translational repression by (i) translation initiation inhibition, (ii) postinitiation inhibition, (iii) mRNA decay in removal foci, and (iv) mRNA storage in stress granules.21 In brief, miRNA can induce translation initiation inhibition by repressing the 48S translational complex assembly,22 competing the m7G of mRNA binding site with eIF4E 23,24 (miRNA binds to the Ago2 complex to bind to m7G) or blocking poly(A)–binding protein to affect translation initiation.25 Messenger RNA can induce postinitiation inhibition by leading to higher rates of ribosome drop-off, leading to immature termination during the elongation step.26 Argonaute proteins are part of the catalytic components in RISC and are able to bind to small noncoding RNAs (including miRNAs, small interfering RNAs, and Piwi-interacting RNAs).27,28 Some of the argonaute proteins have endonuclease activity to enable degradation of perfectly complementary mRNA.29 In eukaryotes, argonaute proteins have been identified in high concentration in regional foci within the cytoplasm known as P bodies.30,31 MicroRNA induces sequestration of mRNA within P bodies.32 MicroRNAs can also induce temporary storage of mRNA in stress granules, which can either be degraded or derepressed later within the cell.33
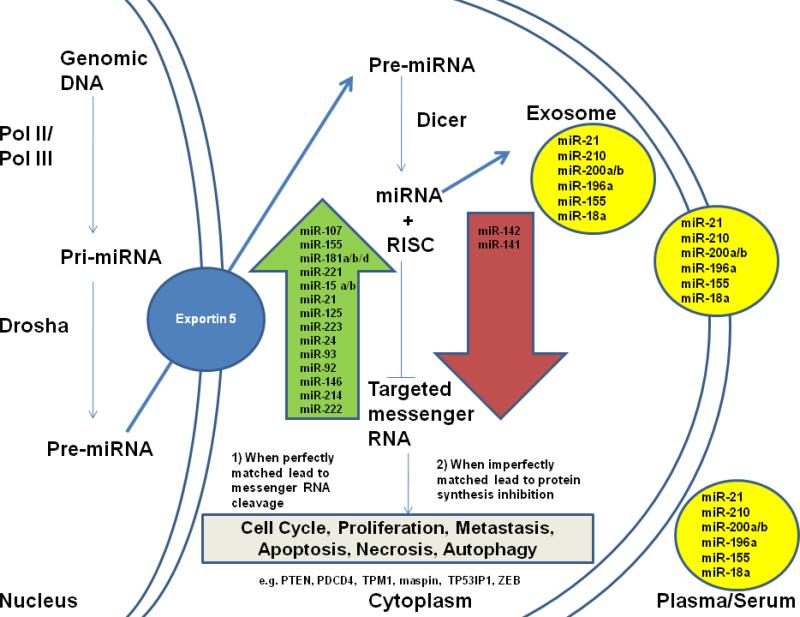
miRNA biogenesis and miRNAs that are distinctly different in pancreatic cancer tissues and blood. Primary miRNA (Pri-miRNA) is transcribed in the nucleus and then processed by Drosha into Pre-miRNA before export to the cytoplasm by Exportin 5. In the cytoplasm, the pre-miRNA, a hairpin/stem loop, is further processed by the enzyme Dicer, producing a double-stranded miRNA. One strand of the mature miRNA is then incorporated with the RISC to regulate its gene targets. There are 2 distinct regulatory mechanisms: (1) mRNA cleavage if the miRNA is perfectly matched with its targets 3’ UTR and (2) protein synthesis inhibition if the miRNA is imperfectly matched with the targets’ 3’ UTR. The targeted genes’ mRNA will be transported to P-bodies or stress granules and are thus inaccessible to the translational machinery. Some of the miRNAs found in tumor cells will, with necrosis, be released into plasma/serum or alternatively released in exosomes.
ORIGIN OF MIRNAs PRESENT IN BLOOD
MicroRNAs can be isolated directly from blood (PBMCs are especially sensitive to microenvironmental changes including those arising in the setting of cancer), plasma, or serum. Studies of whole blood or PBMC miRNA expression to detect tumor (eg, ovarian cancer and melanoma) are developing rapidly 11,19 (Table 1). There are also circulating miRs normally present in the serum or plasma.35 Many scenarios have been formulated to explain how miRNA can survive endogenous ribonucleases that are present within blood. These include miRNA binding to DNA for protection from RNases and DNases,36 as well as gaining protection by envelopment within lipid or lipoprotein carriers or vesicles,37 perhaps derived from exteriorized autophagosomes (exosomes). The latter appears to be the most likely mechanism that preserves miRs in plasma and serum.35 The circulating miRs in plasma and serum might originate from tumor-derived exosomes (eg, miR-21, miR-106, miR-141, miR-14, miR-155, mir-200 family, miR-203. MicroRNA-205, miR-214, etc). Note that only miR-21 and miR-18 families are found to be up-regulated in more than 2 cancer types (Table 1). Perhaps blood miR markers may be more cancer type–specific than tissue miRNA markers.38–41 The lack of appropriate endogenous controls (a miRNA that does not change with disease stage) limits the predictive power, and further validation of the biological role of such circulating miRNAs is needed.
Table 1
miRNAs in Cancer Blood
| Type of Cancer | miRNAs Source | Down-regulated | Up-regulated | Compare with Cancer Tissue | Reference |
|---|---|---|---|---|---|
| Pancreas | Serum, Plasma | miR-18a, miR-21, miR- 210, miR-155, miR-196a, miR-200 | N/A | miR-18a, miR-21, miR-155, miR- 200, miR-196 is also over- expressed in pancreatic cancer tissue and cell line. | 11,12,52 |
| Lung | PBMCs | let-7 a,c,d,e,f,g, miR-15a, miR-20a, miR-98,miR-126, miR-195 | let-7i, miR-19a, miR-22, miR-423-5p, | miR-126, let-7 family, miR-22, miR-19 expression in PBMCs are correlated with lung cancer tissue. MiR-20a is inversely correlated in lung cancer tissue. | 8 |
| Ovary | Whole Blood | Let 7f-1, miR-28-3p, miR- 29a, mir-106b, miR-138-2, miR-146a, miR-181a, miR-181a-2, miR-192, miR-342-3p, miR-450-5p, miR-616, miR-628-5p,miR-1287 | miR-16, miR-30c-1, miR-187, miR-191, miR-191, miR-383, miR-423-3p, miR-499-3p, miR-546-5p, miR-1181, miR- 1228, miR-1253, miR-1254, miR-1289, miR-1908, miR-1915 | miR-30c-1, miR-191, miR-155, miR-16, miR-106b, miR-146a, miR-29a, and miR-383 are connected to ovarian cancer while the other miRs are not reported to be connected to the ovarian cancer. | 10 |
| Gastric | Plasma | miR-21, miR-106b | Let-7a | Inverse relationship between the plasma miRs and gastric cancer miRs expression level. | 13 |
| Acute Leukemia | Plasma | miR-92 | N/A | Acute leukemia cell might in-take miR-92 with exosome thus decreasing the miR-92 concentration in plasma. | 14 |
| Oral | Plasma | N/A | miR-31 | miR-31 is over-expressed in oral cancer tissue. miR-31 level decreased after surgical removal of cancer, very likely that the circulating miR-31 is tumor derived. | 15 |
| Colorectal | Plasma | N/A | miR-29a, miR-92a | miR-92 is also found to be up- regulated in breast cancer serum sample and in other cancer tissue. MiR-29a is up-regulated in lung and advanced colorectial neoplasia and its role in cancer biology is still unclear. | 17 |
| Melanoma | PBMC | miR-452, miR216a, miR- 17, miR-646, miR-217, miR-517, miR-593,let- 7i,miR-330-3p, miR-767-5p, miR-20b, miR-509-3-5p, miR-519b-5p, miR-518e, miR-221, miR-214,miR-106b, miR-18b, miR-108, miR-20a | Let-7d, miR-18a, miR-22, miR-30a,e, miR-99a, miR-125a-5p, miR-142-3p, miR-145, miR-146a, miR-155-3p, miR-181a-2, miR-183, miR-186, miR-199a-5p, miR-328, miR-343-5p, miR-361-3p, miR-362-5p, miR-363-3p, miR-365, miR-378, miR-422a, miR-501-5p, miR-550, miR-584, miR-625, miR-664, miR-1249, miR-1280 | miR-216a and miR-186 expression level are correlated with the melanoma tissue miRs expression level. | 18 |
For therapeutic purposes, it would be useful to identify pancreatic cancer miRNAs that are shared between clinical samples and cancer cell lines (cancer cell lines are more readily available for therapeutic target validation than clinical samples). One study compared the expression profiles between individual pancreatic cancer cell lines and clinical specimens using polymerase chain reaction (PCR) (95 miRNA primers). Eight miRNAs were found to be commonly expressed in both cell lines and clinical samples (miR-196a, mIR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b, miR-95).44 When examining the clinical specimens, 20 miRNAs were overexpressed in all 5 specimens, and 11 miRNAs were overexpressed in at least 4 specimens. The results suggest that although there are similarities between pancreatic cancer cell lines and clinical specimens, the miRNA expression patterns are not identical.
MicroRNA expression profiles in normal pancreatic tissue (referred to as pancreatic miRNome), pancreatic ductal adenocarcinoma (PDAC), pancreatitis, and pancreatic cancer cell lines have been recently examined.47 This study first created a pancreatic miRNome by clustering miRNAs that are highly expressed in pancreatic normal tissue compared with other tissues. The group used this miRNome as the parameter to measure miRNA expression changes in pancreatitis and PDAC miRNA. Twenty miRNAs were differentially expressed when comparing PDAC, chronic pancreatitis, and normal tissues. Twelve of 20 miRNAs are also differentially expressed in cancer cell lines. Furthermore, 2 potential miRNA (miR-196a and miR-217) markers are overexpressed in both primary neoplastic ductal cells and in PDAC cell lines. A similar study found that 23 (15 overexpressed and 8 underexpressed) miRNAs could be used to distinguish pancreatic cancer from pancreatitis with an extraordinary 93% accuracy.44 These similar studies identified divergent sets of miRs, possibly because of the differences in comparison strategies and the patient populations utilized by the 2 groups. One method compared expression with normal tissue, whereas the other group compared expression with a pancreatic tissue–specific gene expression file.
Pancreatic cancer–specific miRNAs are commonly expressed in both clinical specimens and pancreatic cancer cell lines, but the expression profiles are not identical to each other. Because pancreatic tumors are indeed more than just pancreatic cancer cells, examining more stage- and cell type-specific miRNA profiles should provide a more refined result.
Pancreatic cancer is a dynamic disease. Understanding the difference between stages of pancreatic cancer utilizing miRNA profiles is very important. A murine RT2 pancreatic neuroendocrine tumor model study identified pancreatic cancer miRNA markers by stage.7 The study identified primary tumor stage miRNA signatures and metastasis-specific miRNA signatures by comparing the normal islets with primary tumor, liver metastases, and tumor pools. They identified miRNA signatures for hyperproliferation and angiogenesis using flow cytometry to sort hyperproliferating islets and angiogenic islets. The result of the study provides more detail on tumor stage-specific and cell type–specific miRNA signatures in pancreatic tumors.
Two other studies compared pancreatic cancer tissue with the adjacent tissue to identify miRNA markers.43,48 One study identified 20 miRNAs that are differentially expressed in both pancreatic adenocarcinoma and cancer cell lines compared with normal pancreatic tissue miRNA.43 The in situ result showed that miR-221 and miR-376a are localized to tumor cells but not to the benign pancreatic acini or stromal cells. Deregulation of miR-15a and up-regulation of miR-214 are also potential pancreatic cancer markers.48 Microsectioning to allow in situ hybridization on epithelial cells was also compared with matched normal pancreatic tissues.45 Ten miRNAs were differentially expressed, and 2 miRNAs (miR-21, and miR-155) had the highest fold change with miR-21 and miR-155 expression correlating with precursor lesions. The results are congruent with murine RT2 studies demonstrating that miR-21 and miR-155 are overexpressed in hyperproliferating and angiogenic islets.
Nominally specific pancreatic cancer miRNAs could be shared with other cancer types. One study compared solid tumor samples (breast, colon, lung, pancreas, prostate, stomach) miRNA expression with normal tissues (stomach, lung) from patients or individuals with no cancer (for the breast, colon, pancreas, and prostate cancer specimen).42 Twenty-one miRNAs were shared among 6 individual solid cancer types. Twenty of the pancreatic cancer miRNAs were shared with more than 1 solid tumor type. Most of the targets of these 21 shared miRs are identifiable tumor suppressors and/or oncogenes. Seventeen miRs were up-regulated, and 3 were down-regulated. A possible reason for variation between individual clinical pancreatic cancer profiling studies might be attributable to the stage of the patient sample and the type of cell that makes up the tumor. Therefore, a more refined classification of pancreatic cancer with cell type–specific isolation before miRNA profiling could be important for identifying suitable pancreatic miRNAs.
Another extensive study performed with human pancreatic cancer tissue identified miRs that are differentially expressed in individual patient groups.49 Ten miRs (miR-10a, miR-21, miR-143, miR-145, miR-155, miR-222, miR-223, miR-224, and miR-373) are up-regulated, whereas 7 miRs (miR-148, miR-216, miR-217, miR-211, miR-345, miR-596, and miR-708) are down-regulated. The study also characterized some nonoverlapping miRs: 9 miRs to distinguish tumor stage, 16 miRs to distinguish tumor grade, 4 miRs to distinguish the lymph node status, and miR-21 and miR-34a serving as survival predictive miRs.
COMPARING MIRNA EXPRESSION TO IDENTIFY PROGNOSTIC, SURVIVAL, AND CHEMORESISTANT MARKERS
Because the current 5-year survival rate for patients with pancreatic cancer is less than 5%, and surgical resection remains the most effective therapy, identifying markers to predict survival and determine chemoresistance may improve our ability to define subsets of pancreatic cancer patients most suitable for aggressive therapy. Some groups have combined clinicopathologic, follow-up data and miR expression to identify useful biomarkers to help predict survival and clinical outcome. Two independent studies found that miR-21 is a potential marker for survival.49,50 One group extracted RNA from fresh frozen samples, whereas the other group used in situ hybridization to profile the miRNA. Both groups found that pancreatic cancer patients with high miR-21 expression have a low median survival time (13.7 and 14.3 months), whereas patients with lower miR-21 expression have a longer median survival time (25.7 and 23.1 months, respectively). The first group also identified potential markers for better prognosis (high expression of miR-29c, miR-30d, and miR-34a) and determined that patients who have high miR-21 expression are more effectively treated with chemotherapy than those who have lower miR-21 expression. Pancreatic cancer patients with high miR-196a expression in their serum are correlated with poor survival with 100% sensitivity and 75% specificity (6.1 vs 12 months for the low miR-196a expression group).51 One study showed that patient tissue specimens that have high expressions of miR-142-5p and miR-204 correlate with a better patient survival rate (45 and 33 months vs 16.3 and 16.3 months for lower-expression group) when receiving gemcitabine treatment. Patients whose tumors express higher levels of miR-125a and miR-34a seemed to be more effectively treated by gemcitabine, although it did not reach statistical significance.52 The miR-200 family and miR-21 are also predictive markers for an apparent increased benefit of chemotherapy.53,54
Sadly, based on the current literature, there is thus no common pancreatic cancer signature identified among the 8 studies summarized above. Four miRNAs are commonly overexpressed; however, in 5 studies, 3 more miRNAs are commonly overexpressed in at least 4 studies, and 2 additional miRNAs are commonly overexpressed in at least 3 studies. MicroRNA-142p and miR-141 are commonly down-regulated in pancreatic cancer in at least 2 studies, whereas the expressions of 2 other miRNAs (miR-200, miR-145) are contradictory when comparing these 2 studies (Table 3). This reflects the current disarray in the field, and reproducing results is difficult based on variation in sampling of clinical specimens, platforms used to identify miRs, and bioanalytic tools.
Table 3
a. Up-regulated
| MicroRNA | No. of studies | Validated Potential Targets | Biological Significance | Reference |
|---|---|---|---|---|
| miR-107 | 5 | CDK6, DICER1, HIF-1 beta 217-219 | Proliferation, Cell Migration, Invasion, Suppressing Hypoxia Signlaing | 40,42,44,45 |
| miR-155 | 5 | TP53INP1, PU.1. SPCS1. RAST 71,72,221,222 | Suppressing Apoptosis, Inhibiting Tumor Suppressor | 40-42,44 |
| miR-181-a | 5 | TIMP3, TCL1 127,223 | Inhibit Tumor Suppressor, Suppressing Oncogene | 7, 42,44 |
| miR-181-a | 5 | TIMP3, TCL1 127, 223 | Inhibit Tumor Suppressor, Suppressing Oncogene | 7, 42, 44 |
| miR-221 | 5 | DVL2, SOCS1, p57, PTEN, p27 224-228 | Increase Cell Mobility, Inhibiting Tumor Suppressors | 40, 42, 45 |
| miR-15a,b and miR-16 | 4 | Cyclin E, BCL2 229,230 | Inhibit Tumor Suppressor, Inducing Apoptosis | 7, 42-44 |
| miR-21 | 4 | Big-h3, PTEN, PDCD4, TPM1, maspin 61, 231-236 | Inhibit Tumor Suppressor, Suppress Apoptosis, Cell invasion | 40-42,44 |
| miR-125 | 4 | Bcl1-2, p53 gene237, 238 | Suppressing Apoptosis | 40, 42, 45, 216 |
| miR-223 | 4 | C-myc, artn, LMO2-L/-S 239-241 | Repressing Estrogen receptor beta 1 expression, increase cell proliferation | 40, 41, 44, 216 |
| miR-24 | 3 | H2AX, FURIN, DND1, FAF1, DHFR, E2F2, MYC80, 242-247 | Cell Proliferation, Induce Apoptosis | 40, 42, 45 |
| miR-93 | 3 | Integrin beta 248 | Promote tumor growth and angiogenesis | 40, 41 |
| miR-181-d | 3 | TIMP3 249 | Cell Invasion | 7, 40, 41 |
| miR-92 | 2 | ERbeta1, p63 250, 251 | Repressing Estrogen receptor beta 1 expression, increase cell proliferation | 7, 42 |
| miR-146 | 2 | Up-regulated by breast cancer metastasis suppressor 1 252 | Suppresses breast cancer metastasis | 41, 216 |
| miR-214 | 2 | PTEN , ING4 43, 101 | Apoptosis, Chemotherapy resistant | 43, 45 |
| miR-222 | 2 | PUMA, AKT, p27Kip1253-258 | Cell Survival, Cell migration | 41, 216 |
MIRNA PROFILING STUDIES IN PANCREATIC CANCER PATIENTS’ BLOOD
Tissue miRNA markers could do more not only to help us understand cancer biology, but also to advance therapeutic options in treating the disease. Such markers have clear limitations as early diagnostic tools for monitoring drug response and defining disease prognosis. First, there are limited solid tumor samples available to scientists. Second, such an approach requires invasive procedures to obtain biopsies from solid tumors if they are identifiable. Thus, tissue is not an ideal approach as an early-stage diagnostic method (before symptoms develop). More importantly, it is not practical to repetitively obtain solid tumor tissue miRNA to monitor disease progression. On the other hand, patients’ blood is readily available. Blood samples can easily be obtained (pretreatment/posttreatment) and may be a more appropriate sample source to establish a miRNA based biomarker for early diagnosis of cancer, prediction of drug responsiveness, and definition of prognosis. Studies have shown promising proof of concept to utilize cancer patients’ blood miRNA profile as a diagnostic and prognostic tool in pancreatic cancer. MicroRNAs can be isolated from the PBMCs, serum, or plasma components of blood specimens. Three individual studies 12,13,34 found 6 miRNAs expressed in pancreatic cancer patients’ serum and plasma as potential biomarkers. MicroRNA-18a, miR-21, miR-210, miR-155, and miR-196a are overexpressed in a majority of the pancreatic cancer patients’ plasma examined with at least 2-fold increases.13 Sensitivity of greater than 40% and specificity of greater than 70% (Table 4) can be realized. When categorizing the patient population by age and sex, compared with healthy individuals, miR-200 a/b is overexpressed in primary pancreatic cancer and cancer cell lines, as well as pancreatic cancer patients’ serum.12 A sensitivity and specificity of 84.4% and 87.5%, respectively, for miR-200a and 71.1% and 96.9% for miR-200b were found. MicroRNA-18a (one of the miR-17-92 gene cluster families) is up-regulated in primary pancreatic cancer tissue and cancer cell lines.34 miR-18a expression in patient's serum was significantly reduced following surgical excision. Another study examined pancreatic cancer patient serum and investigated whether miR-21, miR-155, miR-196a, miR-181a, miR-181b, miR-221,and miR-222, which are differentially expressed in cancer tissues, can serve as biomarkers.51 Higher expressions of miR-21, miR-155, and miR-196a are observed in pancreatic cancer patients’ serum, but both miR-155 and miR-196a are also up-regulated in chronic pancreatitis. The group also found that patients who have higher miR-196a expression in the serum have a lower median survival (6.1 vs 12 months). Because immune cells respond to the cancer microenvironment and macroenvironment, our group hypothesizes that, in the presence of pancreatic tumor, the miRNA expression in patients PBMCs will be altered. Our laboratory has profiled pancreatic cancer patients PBMCs miRNA with Taqman Low Density Array (From Life Technologies, Grand Island, NY) in an age- and sex-matched study (unpublished data) and found that miR-125a-5p is up-regulated in patients, whereas miR-29c and miR-146b are down-regulated when compared with controls. We are currently continuing the study to determine if these individual miRs will change following receipt of chemotherapy and surgical extirpation. Most of the miRNAs circulating in the blood are thought to be present in membrane-bound vesicles or exosomes.55 Differences in plasma and serum miRNA could in part reflect liberated platelet-derived miRNAs.56
Table 4
Pancreatic Cancer Patients' Blood microRNA markers
| Type of Cancer/ Source | MicroRNA | Sensitivity | Specificity | Reference |
|---|---|---|---|---|
| Pancreatic Cancer Patients' Plasma | miR-21 | 46 | 89 | 12 |
| miR-155 | 42 | 42 | ||
| miR-196a | 53 | 78 | ||
| miR-210 | 43 | 84 | ||
| Pancreatic Cancer Patients' Serum | miR-200 a | 84.4 | 87.5 | 11 |
| miR-200 b | 71.1 | 96.9 | ||
| Pancreatic Cancer Patients' Serum | miR-18a | N/A | N/A | 52 |
| Pancreatic Cancer Patients' Serum | miR-21 | N/A | N/A | 48 |
| miR-155 | N/A | N/A | ||
| miR-196a | N/A | N/A |
Although the serum and plasma studies provide proof of concept that circulating miRNAs could be used to diagnose pancreatic cancer, we need to use caution before applying to broader application because the prevalence of pancreatic cancer adjusted to rate is 12 per 100,000 individuals, and many false positives could be expected.57 Positive and negative predictive values of 0.00016 and 0.9988, respectively, and in another study positive and negative predictive values of 0.05 and 0.9998, respectively, were found. Both studies provide a relatively good test to identify individuals who are negative for the disease, but the test is not useful to identify individuals who have pancreatic cancer.
Interestingly, most of those pancreatic cancer serum/plasma miRNAs have also been found in pancreatic cancer tissues in more than 1 study. MicroRNA-21, miR-155, and the miR-200 family are differentially expressed in both pancreatic cancer tissue and blood in more than 1 study. These particular miRs are involved in cell cycle, cell migration, and cell survival (Fig. 3) and are also clinically significant in cancer biology. The following section discusses the significance of these 3 best validated miRs.
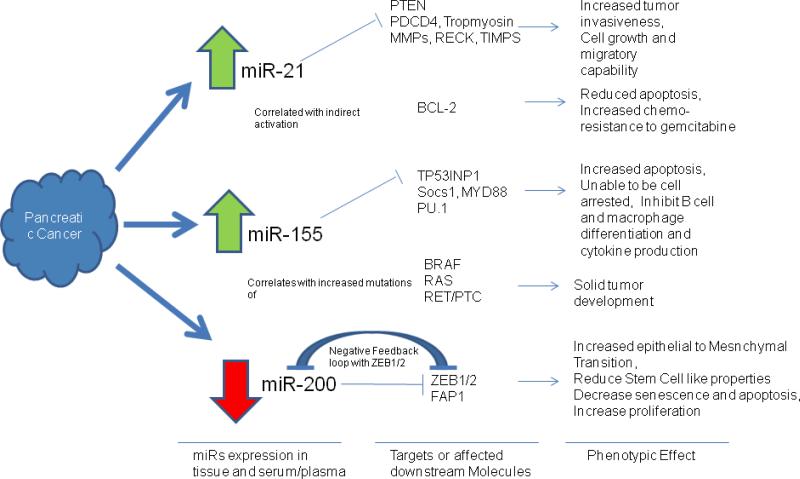
Three commonly differentially expression MicroRNAs in pancreatic cancer tissue and patient blood: MicroRNAs 21, 155, and 200. miRNA 21, 155, and 200 are commonly differential expressions in pancreatic cancer tissue and patient blood. They play important roles in tumorgenesis and drug resistance by repressing their targeted genes. miR-21–regulated genes are involved in apoptosis, tumor invasion, metastasis, and cell cycle progression. Up-regulation of miR-21 is commonly found in cancers and the blood. Here, a schematic diagram of predicted miR-21 target genes important in cancer biology is presented. 218 miR-21 targets PTEN (regulating expression of PI3K, FAK, Akt, matrix metalloproteinases, p53, and caspase 8), BCL2 (via PI3K targeting), PDCD4, tropomyosin, matrix metalloproteinases, RECK, and TIMPs, which promotes tumors resistant to apoptosis, metastatic growth, and increase their migratory capability. miR-21 also correlated with activation of BCL2, which reduced pancreatic cancer cell apoptosis and chemosensitivity to gemcitabine.miR-155 is found often up-regulated in cancer tissues and/or patient's serum. Potential targets of miR-155 include TP53IP1 (via caspase-3), PU.1, SOCS1, MYD88 (via JAK/STAT), which are involved in apoptosis, cell cycle progression, immune cell differentiation, and proinflammatory cytokine production all of which are important for tumor development. miR-155 overexpression is also associated with increased mutation in BRAF, RAS, and RET/PTC, genes that are commonly dysregulated in solid tumors. miR-200 family members include miR200 b/c, miR-429 (subgroup2), and miR-141. miR-200a (subgroup 1) forms a negative feedback loop with ZEB1/2, involved in the TGF[beta] signaling pathway. ZEB1 is a critical EMT activator in human cancers . ZEB2 interact with p300, pCAF to induce EMT. miR-200 family regulate EMT by regulating ZEB1/2. ZEB 1/2 also plays an important role in control stem cell–like properties, senescence, apoptosis, and proliferation (by repressing cyclin D1). miR-200 family members also target FAP1, one of the apoptosis inhibitors.219
PATHWAYS OF COMMONLY EXPRESSED PANCREATIC CANCER MIRNA IN BOTH TISSUE AND BLOOD
Currently there is no unique set of miRNA biomarkers contrasting pancreatic cancer tissue and blood miRNA profiling studies from other cancer profiles. However, there are potential miRNA biomarkers (miR-21, miR-155, and miR-200) that are identified in both pancreatic cancer tissue and patients’ blood. Are there any unique characteristics shared between those miRNAs that make them potential markers for both tissue and blood? Following the pathways that those miRNAs are involved in may provide clues to explain why these individual miRNAs can serve as suitable biomarkers.
MicroRNA-21
MicroRNA-21 is located on chromosome 17. The mature sequence is 21 base pairs long. MicroRNA-21 regulates genes involved in apoptosis, proliferation, migration, and metastasis (Fig. 3). Several groups have shown up-regulation of miR-21 in pancreatic cancer cells. Higher miR-21 expression in pancreatic cancer tissues is correlated with higher invasiveness and lower survival rates.58 One validated target of miR-21 is the PTEN (phosphatase and tensin homolog) tumor suppressor gene that is commonly mutated or lost in many human cancers. PTEN regulates cell death by inhibiting the AKT signaling pathway through dephosphorylation of phosphatidylinositol (3,4,5)-triphosphate.59 This promotes apoptosis and tumor suppression. Inhibition of PTEN by miR-21 inhibits apoptosis and therefore promotes tumorigenesis. Another validated target of miR-21 is the tumor suppressor gene PDCD4 (programmed cell death 4). Decreased PDCD4 expression correlates with increased miR-21 expression in pancreatic cancer cells.60 The PDCD4 gene plays a role in apoptosis, and inhibition of PDCD4 can promote tumorigenesis. Interleukin 10 production in macrophages is mediated by miR-21 and PDCD4, playing a role in inflammation and cancer formation.61 Yet another validated target of miR-21 is the tumor suppressor gene TIMP3 (tissue inhibitor of metalloproteinase). Decreased expression of TIMP3 correlates with elevated expression of miR-21 in PDAC.60 Other potential targets of miR-21 that are also involved in cell death and apoptosis are TPM1 (tropomyosin 1) and maspin.62,63 Two proteins that show increased activity, correlating with higher expression of miR-21, are MMP2 (matrix metalloproteinase 2) and VEGF (vascular endothelial growth factor), which are important for invasion and angiogenesis.64 Interestingly, increased expression of miR-21 is noted in gemcitabine-resistant cells.65 Exposure to gemcitabine increases miR-21 expression in pancreatic cancer cell lines.64 These findings suggest a link between the targets of miR-21 and acquired drug resistance in pancreatic cancer. In addition to pancreatic cancer tissue and blood (serum and plasma), miR-21 is overexpressed in other cancer types including hepatic, renal, colorectal, breast, and small cell lung, as well as in metastatic cancer.7,66 Higher expression of miR-21 is associated with increased invasiveness and lower survival rates in these cancer types. Increasing evidence is thus emerging that miR-21 is a key biomarker and therapeutic target for invasive tumors. MicroRNA-21 is highly expressed in more invasive tumors and blood compared with less invasive tumors and is associated with poor survival. Because miR-21 is commonly deregulated in various cancers, it may be useful as a prognostic marker for more invasive versus less invasive cancers, but it does not provide specific cancer type detection.
MicroRNA-155
MicroRNA-155, located on chromosome 21, has a mature sequence that is 24 base pairs long. In pancreatic cancer, miR-155 is up-regulated in both tissue and the patient's blood, making it a potential pancreatic cancer marker.13,34,67 MicroRNA-155 is overexpressed in pancreatic intraepithelial neoplasia 45 and is associated with increased invasiveness in colorectal cancer as well.68 MicroRNA-155 represses suppressor of cytokine signaling 1,69 a tumor suppressor that functions as a negative feedback regulator of JAK/signal transducer and activator of STAT signaling 70; inhibits MYD88 71 a key proinflammatory cytokine signaling pathway; and targets TP53INP1 (tumor suppressor gene),a proapoptotic stress-induced p53 target gene 72 (Fig. 3). MicroRNA-155 is overexpressed in various cancers (eg, leukemia,73–75 breast, colon, cervical, and pancreatic cancers 42,43,47,76–83). MicroRNA-155 also plays important roles in hematopoiesis,84,85 inflammation,86–88 T- and B-cell activation,89 cardiovascular diseases,90,91 and viral infection.92,93TP53INP1 is down-regulated during pancreatic cancer development, and miR-155 represses expression of TP53INP1.72 Inhibiting miR-155 expression in pancreatic cancer cell lines enhances TP53INP1 and increases apoptosis. High miR-155 expression in pancreatic cancer and colorectal cancer patients’ tissue is associated with lower survival (23.86% vs 76.14%),58 but not in those patients with small lung cancer.68,94 MicroRNA-155 expression is higher in later stages of pancreatic cancer,58 and this is also true for breast cancer tissue and sera.95 MicroRNA-155 is a potential miRNA biomarker within tumor tissue as well as blood. Similar to miR-21, miR-155 dysregulation is apparent in individual cancer types but is thus not specific to pancreatic cancer. Because miR-155 plays an essential role in inflammatory regulation 71 and tumor suppression, miR-155 could be a potential tissue/blood biomarker for patients with pancreatic and other epithelial neoplasms.
MicroRNA-200a/b
The miR-200 family includes miR-200a/b/c, miR-400, and miR-141, which are located on chromosomes 1 and 12. MicroRNA-200c is also overexpressed in pancreatic cancer cell lines (CAPAN-1, SW1990, CFPAC-1, and H48N). Furthermore, this overexpression inhibits invasion of pancreatic cancer cells, but promotes their proliferation.96 MicroRNA-200a, miR-200b, and miR-200c are down-regulated in gemcitabine-resistant pancreatic cancer cells. MicroRNA-200 down-regulation is implicated in the epithelial-to-mesenchymal transition (EMT) phenotype of gemcitabine-resistant cells.97 The miR-200 family targets ZEB 98–100 (a key transcriptional factor that represses E-cadherin). MicroRNA-200 down-regulation is associated with early metastasis (Fig. 3). The overall expression levels of the miR-200 family in pancreatic cancer as well as other cancer types vary greatly depending on the stage of the tumor.101–106 MicroRNA-200 expression is down-regulated in early metastatic tumors. In late-stage metastasis, however, miR-200 expression sometimes is unchanged or even up-regulated when compared with normal tissues. Low miR-200 expression level in ovarian cancer is correlated with poor complete response rate to paclitaxel-based treatment.107 MicroRNA-200 is also found to be overexpressed in pancreatic cancer patients’ sera by 2- to 3-fold.12 The miR-200 family is a potential dynamic biomarker for tumor progression because its expression in pancreatic cancer patients’ tissue and blood depends on the progression of the tumor. MicroRNA-200 is down-regulated in early metastasis but is unchanged or even up-regulated in late metastasis.
MicroRNA-21, miR-155, and miR-200a/b are deregulated in both tumor tissue and pancreatic cancer patients’ blood. Although particular miRNA biomarkers do not regulate the same pathway in cancer biology, they are all correlated with more invasive/metastatic tumors in clinical studies. These 3 miRNAs markers are often found to be overexpressed in more invasive tumor tissue and in some cancer patients’ blood. Functional validation of those miRs in knockout (or overexpression) systems in mice confirms their role in cancer development.108 MicroRNA-155 is important to maintain immune system function and plays a crucial role in B-cell malignancy in murine models.89,109–111 Overexpression of miR-21 in the mouse induces pre–B-cell lymphoma.35,112,113 Overexpression of miR-21 is found in constitutively activated Kras involved in late stage of tumorigenesis, whereas it has no effect in the absence of Kras.112 MicroRNA-21 expression is associated with apoptosis and cell proliferation.114 MicroRNA-200 deregulation is required to induce metastatic tumor in KrasLA1;Trp53R72/H[DELTA]G mice.115 Taken together, overexpression of miR-21/miR-155 and down-regulation of miR-200a/b in patients’ tissue and blood might serve as a biomarker panel for invasive pancreatic cancer.
Caution is warranted before using miR-21, miR-155, and miR-200a/b as type-specific cancer biomarkers. There are still no unique cancer type–specific miRNA biomarkers that are commonly differentially expressed among individual clinical studies. In pancreatic cancer, only 11 miRNAs (miR-107, miR-125, miR-15b, miR-21, miR-24, miR-155, miR-181a, miR-221, miR-92, miR-181-d, and miR-223) are commonly deregulated in various studies. Furthermore, the commonly deregulated miRNAs are not just found in pancreatic cancer, but also in other tumor types.42
CONNECTIONS BETWEEN MIR-21, MIR-200a/b, MIR-155, AND DEFINED GENETIC LESIONS IN PANCREATIC CANCER
Pancreatic cancer progression is associated with several defined genetic mutations or loss, and because miRNAs can regulate oncogene and tumor suppressor genes, these can in turn be also regulated by other genes. It is of interest to examine if there is any connection between commonly altered pathways, such as transforming growth factor [beta] (TGF-[beta])/SMAD4, Kras, BCRA, p53, and p16,116 and miRNAs. In our estimation, molecules released from necrotic tumor cells, especially damage-associated molecular pattern (DAMP) molecules may also alter the miRNA expression in pancreatic cancer tissue/blood. We discuss the linkage between known alterations in pancreatic cancer genetic pathways and these differentially expressed miRNAs in the following sections.
Transforming Growth Factor [beta]
Transforming growth factor [beta] (TGF-[beta]) has a dual role in cancer biology: an antitumor role and tumor promoter role.117 Transforming growth factor [beta] is a potent tumor suppressor that signals via the SMAD pathway and intersects with the Wnt-[beta] catenin signaling pathway in normal cells. It regulates the cell cycle (both SMAD dependent and SMAD independent) by inhibiting cyclin-dependent kinases and E2F and histone deacetylases during the G1 phase of the cell cycle. In pancreatic cancer cells, SMAD4 (the co-SMAD that cooperates with SMAD3 and SMAD2 promoting TGF-[beta]'s inhibitory function) is often mutated or lost, especially in cells with a propensity for distant metastases.118–121 Pancreatic cancer cells do not respond to TGF-[beta] signaling even in the presence of high-level expression of TGF-[beta] receptors, which limits its ability to inhibit cell growth and metastasis.122 The loss/mutation of SMAD4 in the TGF-[beta] pathway in pancreatic cancer cells attenuated the inhibitory function of TGF-[beta]. Furthermore, TGF-[beta] is also associated with cancer invasiveness (and metastasis), regulating extracellular matrix expression, angiogenesis, and immunosuppression.117 Transforming growth factor [beta] is regulated by several miRNAs including miR-15/16, miR-224, miR-106b, the miR-200 family, miR-155, miR-181b/d, miR-21, miR-17-92, and miR-24.123 MicroRNA-15/16 negatively controls TGF-[beta]'s downstream responsive element, Acvr2a with resultant induction, and patterning of mesoderm germ layer during embryo development.124 MicroRNA-224 enhances TGF-[beta]–induced Germinal Center proliferation by inhibiting SMAD4.125 MicroRNA-106b overexpression impairs the TGF-[beta] tumor suppressor pathway.126 Transforming growth factor [beta] increases miR-181b/d, thereby decreasing TIMP3-associated hepatocarcinogenesis.127 MicroRNA-17-92 impairs gene activation by TGF-[beta].128,129 MicroRNA-24 indirectly reduces SMAD protein expression attenuating TGF-[beta] signaling by targeting Trb3.130 Compared with tissue and biofluid miRNA markers in pancreatic cancer patients, miR-21, miR-200 family, and miR-155 are commonly deregulated. MicroRNA-21 up-regulation is mediated by TGF-[beta] via a SMAD4-independent pathway (but SMAD3 is required), which leads to down-regulation of PDCD4, resulting in turn in a decrease in apoptosis and less tumor-suppressive activity. Increases in SMAD3 activity is found in cancer.131 MicroRNA-200 is regulated by TGF-[beta] via ZEB, and prolonged autocrine TGF-[beta] suppresses miR-200, which in turn promotes the EMT.132 Transforming growth factor [beta] can up-regulate miR-155 via SMAD4; knocking down miR-155 suppresses TGF-[beta]'s ability to induce EMT, cell migration, and invasion.133 Both miR-155 and miR-21 are linked, via a SMAD3-dependent pathway. MicroRNA-155 inhibits SMAD2, which leads to a more potent SMAD3-dependent TGBF [beta] signal that in turn up-regulates miR-21 expression and drives EMT. As cancer cells become more mesenchymal, ZEB1/2 is up-regulated and represses expression of the miR-200 family. Therefore, miR-21, miR-155, and the miR-200 family may be biomarkers for metastatic cancer that have the TGF-[beta] signaling pathway disrupted.
Kras
Kras is the most frequently mutated gene (>95%) in PDAC.134 Mutation in Kras disables GTPase to hydrolyze GTP, resulting in a constitutively activated protein. As PDACs progress, Kras mutated tumor cells may accumulate mutations in other genes such as p53 and SMAD4. The Kras mutation occurs in the early stage of pancreatic cancer development and is associated with the loss of tumor suppressor genes in late stages.135–141 Ras regulates cellular proliferation, differentiation, migration, and apoptosis via activation of the MAP kinases cascade (AKT and the P13K pathway). Ras is deregulated in many cancer types,142 leading to decreased apoptosis, increased cell invasion, and metastasis. Activating mutations of Ras are found in 90% to 95% of all pancreatic tumors (and a quarter of all other tumors). Thus, Kras is one of the most frequent mutations in pancreatic cancer. Alteration in codons 12 or 13 causes Ras to be constitutively active.143 Several miRNAs are involved in the Kras pathway including miR-143/145, miR-217, miR-155, let-7a, and miR-200a. Kras signaling represses the expression of miR-143/145. In addition, Kras and RREB1 (Ras responsive element protein binding 1) are targets of the miR-143/145 cluster.144 This results in a feed forward mechanism that potentiates Kras signaling. MicroRNA-21 and miR-155 45 also play a role in the Kras signaling pathway by repressing their targets PTEN (phosphatase and tensin homolog) and activating the AKT pathway. MicroRNA-21 and miR-155 also repress PCDC4 playing a role in the Kras signaling cascade. MicroRNA-217 145 and miR-96 146 both target Kras and function as tumor suppressors to down-regulate Kras signaling. These miRs are down-regulated in pancreatic ductal carcinoma tissue samples. Let-7a and miR-200a play an important role in Kras signaling in conjunction with DCAMKL-1 (double-cortin–like and CAM kinase–like 1). DCAMKL-1 is a pancreatic stem cell marker, and inhibits expression of miR-200a and Let-7a.147 These miRNAs target Kras and c-Myc, and ZEB1 and ZEB2 inhibit tumorigenesis and EMT. When DCAMKL-1 is overexpressed in pancreatic stem cells, these miRNAs are repressed and lead to increased Kras signaling. Overexpression or underexpression of these specific miRNAs can play a role in constitutive Kras signaling leading to increased cellular proliferation, decreased apoptosis, and promotion of EMT.
Breast Cancer Susceptibility Protein
Breast cancer 2 susceptibility protein (BRCA2) is essential for cell proliferation, differentiation, and DNA repair.148–150 BRCA2 mutation is commonly associated with breast and ovarian cancer but also increases the risk of pancreatic cancer.151 In murine models, BRCA2 mutation in concert with other mutations (eg, Kras, p53) defines a role for BRCA in PDACs.152 When p53 is intact, BRCA2 mutation alone is not sufficient to drive PDAC, whereas double mutations can enhance PDAC development. Double mutation of BRCA and Kras in p53 intact cells cannot fully drive PDAC, but when p53 is also mutated, mice rapidly develop PDAC. Pancreatic cancer patients with BRCA2 mutations are found to be sensitive to DNA cross-linking agent therapy, and some conversion from sensitive to resistance is occasionally due to the secondary mutation that restores expression of wild-type BRCA2.153,154 Although there are no direct studies on how miRNA might play a role in BRCA mutated pancreatic cancer, some miRs are differentially expressed in BRCA mutated tumor cells. For example, a polymorphism in miR-146a increases the risk of breast cancer, and the variant C allele in miR-146a has a stronger binding capacity in the 3' UTR of BRCA1/2 mRNA.155 In ovarian cancer, miR-29a/b is up-regulated in BRCA1/2 loss tumors when compared with those without loss.156 MicroRNA-200a and miR-21 are up-regulated in high-grade/low-grade ovarian cancer when compared with normal tissues. BRCA1 epigenetically represses miR-155. Tumor growth is attenuated by knocking down miR-155.157 Perhaps in the 3 common pancreatic cancer miRs (miR-21, miR-200a, miR-155) that we have focused on, loss or mutation of p53 and Kras mutation is also required for BRCA mutated cells to develop PDAC, and further investigation is required to explore this in this subset of patients.
p53
p53 Is one of the most frequently mutated tumor suppressor genes in human tumors 158–160 that plays an important role in activating DNA repair, inhibiting autophagy, and promoting cell cycle arrest as well as apoptosis to limit transformation.161 It is also frequently mutated in pancreatic adenocarcinomas; p53 162 and its gene product TP53INP1 regulate the cycle though pretranscriptional, transcriptional, and posttranscriptional actions.163 We have shown that p53 directly interacts with high-mobility group box 1 (HMGB1),164 and together these molecules may regulate some aspects of miRNA expression. p53 Regulates or is regulated by miRNAs to form a regulatory network as a tumor suppressor.165 MicroRNA-29, miR-122, and miR-125 together regulate the p53 inhibitor p85a/Cdc42 and cyclin G1 or directly inhibits p53.166–168 p53 Up-regulates miRs such as miR-34, miR-215, and miR-16-1, which in turn target downstream messages encoding BCL2, p21, CDK4/6, and cyclin D1 by controlling their maturation.169–172 MicroRNA-155 can repress expression of TP53INP1 in pancreatic tumors. Restoring TP53INP1 expression helps inhibit pancreatic tumor growth 71. p53 Mutation also leads to higher miR-21 expression via p68/p72 miRNAs processing, which results, in turn, in more EMT and chemoresistance.67,173 Interestingly, the potential miR markers miR-21, miR-155, and miR-200 interact with each other via the p53 pathway. Up-regulation of miR-155 can repress TP53INP1, which also leads to higher expression of miR-21. p53 Mutant cells also have higher miR-21 expression levels. MicroRNA-21 is associated with higher EMT, leading to down-regulation of miR-200 (a key repressor for ZEB1 in EMT pathway). Therefore, up-regulation of miR-21 and miR-155 and down-regulation of miR-200 family may serve as a potential marker for metastatic tumors with p53 mutation.
p16
p16 Is a tumor suppressor protein also known as cyclin-dependent kinase inhibitor 2A (CKDN2A) p16Ink4A and multiple tumor suppressor 1 (MTS1). p16 Proteins regulate cell cycle progression, apoptosis, and DNA repair, and the genes that encode p16 are lost in 80% to 95% of cases of pancreatic cancer 174 being observed in even early stage of pancreatic intraepithelial neoplasia lesions.175 p16 Mutations in combination with Kras, p53, and SMAD4 mutations have also been observed in advanced pancreatic cancer.176–178 p16 Inhibits cyclin-dependent kinases 1, 4, and 6 (CDK1/4/6) and also helps to stabilize p53.179 These functions in addition to repression of transcription factors such as c-Myc and nuclear factor [kappa]B all contribute to p16's ability to control the G1 stage of the cell cycle. Recent studies have also indicated a novel role for p16 in regulating oxidative stress through the MAPK pathway.180 p16 Induces overexpression of miRNAs 410 and 650 by changing the equilibrium of specific transcription factors. These miRs interact with the CDK1–3' UTR and cause posttranslation inhibition of CDK1. CDKN2A (p16) is a target of miR-10b. Inhibition of miR-10b induces cell cycle arrest and apoptosis reducing tumor size.181 In addition, miR-20a increases p16 protein levels and plays a role in senescence.182 Therefore, a mutation in p16 causing decreased levels of miRs 410 and 650, up-regulation of miR-10b, or inhibition of miR-20a can lead to increased cellular proliferation and a greater likelihood of tumorigenesis. Although p16 plays a role in p53 signaling pathway, the known miRNAs involved in p16 regulation do not link to miR-155, miR-21, and miR-200 family.
INTERPLAY OF DAMP MOLECULES AND MIRNA IN PANCREATIC CANCER
Many studies have focused on investigating the mutations that are directly responsible for cancer development. However, recent evidence demonstrates that changes in the microenvironment such as inflammation also play an important role in tumorigenesis.183 Tumor cells have limited apoptosis and emergent pronounced increases in autophagy that results in necrosis under conditions of heightened stress. In this instance, they release so-called DAMP molecules that trigger inflammation and immunity. Damage-associated molecular pattern molecules such as HMGB1 and expression of one of its cognate receptors, RAGE (receptor for advanced glycation end products), play important roles in cancer biology.184,185 In brief, HMGB1 is a highly conserved nuclear protein that bends DNA and promotes access to transcriptional protein assemblies on specific DNA targets.186,187 High-mobility group box 1 can also serve as an extracellular signaling molecule during inflammation, cell differentiation, cell migration, and wound repair driving acute inflammatory response and tumor metastasis.186–189 High-mobility group box 1 is released from necrotic and stressed autophagic cells and is actively secreted by inflammatory cells binding with receptors including RAGE, Toll-like receptor family (TLR2, TLR4, TLR9), and CD24 mediating response to infection or injury to regulate inflammation.186–190 High-mobility group box 1 is also induced by chemotherapy and radiotherapy, and the release of HMGB1 contributes to the disordered tumor microenvironment. RAGE is a member of the immunoglobulin superfamily encoded within the class III major histocompatibility complex that can be activated by advanced glycation end products and several DAMP molecules. RAGE ligands including DNA, HMGB1, S100B, Mac1, and S100A6 activate intracellular signaling molecules (eg, nuclear factor [kappa]B, MAP kinases) to induce proinflammatory response.191,192 Overexpression of RAGE lowers cell proliferation, and down-regulation promotes development of advanced-stage lung tumors.193 However, in non–lung cell tumors, RAGE ligands are overexpressed.194 Currently, there are few miRNA studies on HMBG1 and RAGE, but those miRNAs being investigated share some common pathways with the 3 potential pancreatic cancer miRNA markers (miR-200, miR-155, and miR-21). MicroRNA-16 is down-regulated by S100b (one of the RAGE ligands) in THP-1 monocyte cell lines.195 MicroRNA-16 targets BCL2,196 an antiapoptotic gene, which is differentially expressed in many tumors.197 Wild-type p53 in diffuse large-B-cell lymphoma can target overexpressed BCL2 and induce cell arrest and apoptosis, but cell death is reduced when p53 is inhibited.198 Our laboratory is currently investigating “DAMP-miRs” with freeze-thaw cell lysates from HMBG1 wild-type cells and HMGB1 knockout cells. MicroRNA-34c has been identified as up-regulated in human PBMCs following stimulation.199 MicroRNA-34 family members are transactivation targets of p53,200 and miR-34 targets various cell cycle and apoptosis proteins including BCL2 and c-Myc.201 Ectopic miR-34 expression induces apoptosis and, in the absence of miR-34c, promotes apoptosis induced by p53 activating agents.202 Kras and the DAMP/RAGE pathway are connected by the p53 signaling pathway, which forms a signaling network with these 3 potential pancreatic cancer miRNA markers (Fig. 4).
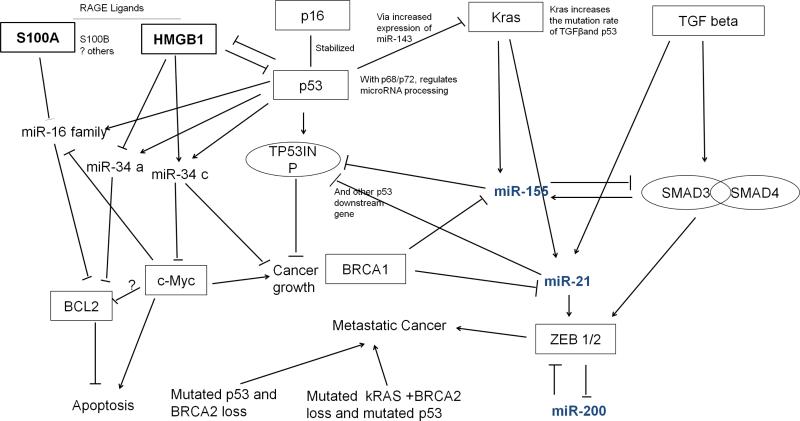
Predicted network linking pancreatic cancer miRNA markers, known pancreatic cancer genetic lesions, and DAMPs. miR-21, miR-155, and miR-200 family members are connected to known genetic lesions found in pancreatic cancers including mutant K-ras, BRCA1, TGF[beta], and DAMPs (HMGB1) and DAMP-receptors (RAGE) as well as the p53 pathway. Note that the role played by c-myc in apoptosis regulation is paradoxical; under some conditions, c-myc promotes proliferation, and under other condition c-myc promotes apoptosis.221 c-myc is crucial for apoptosis and requires p53 to activate apoptosis.
UTILITY OF HYPOMETHYLATED OR HYPERMETHYLATED MIRNA GENES AS SPECIFIC EARLY DIAGNOSTIC MARKERS FOR PANCREATIC CANCER
The identification of specific miRNA markers is important for the early diagnosis of pancreatic cancer. DNA methylation is a process that involves the addition of a methyl group to the 5 position of the cytosine pyrimidine ring or the number 6 nitrogen of the adenine purine ring. Although methylation is essential for normal cell development and gene transcription, aberrant methylation is associated with carcinogenesis. Unmethylated CpGs are often grouped in clusters called CpG islands, which are present in the 5' regulatory regions of many genes. In many disease processes, such as cancer, gene promoter CpG islands acquire abnormal hypermethylation, resulting in transcriptional silencing that can be inherited by daughter cells following cell division.
Hypermethylation of miRNA genes leads to decreased expression of the associated mature miRNAs, whereas hypomethylation leads to enhanced expression. Hypermethylation is one of the major epigenetic modifications that repress transcription via the promoter region of tumor suppressor genes.204 The majority of miRNAs in tumors are repressed, indicating that they play essential tumor suppressor functions.205 Such aberrantly methylated miRNAs could serve as early diagnostic markers in multiple cancer types, especially in pancreatic cancer.
Multiple miRNAs have been reported to be hypermethylated and hence repressed in pancreatic cancer. One of these is miR-148a, which is down-regulated in early PDAC 206 and can potentially be used as an early diagnostic marker. Another epigenetically silenced miRNA in pancreatic cancer is miR-107.207 In contrast to repressed miRs in pancreatic cancer patients, both miR-200a and miR-200b are hypomethylated and hence are significantly elevated in patient sera with respect to healthy controls.12 The methylation status of all the previously mentioned miRs could be utilized as early diagnostic markers for pancreatic cancer.
CURRENT CHALLENGES IN MIRNA EXPRESSION STUDIES
Many challenges remain in the field for establishing pancreatic cancer biomarkers. First, it is quite a challenge to compare the miRNA expression between individual studies. Microarray and quantitative reverse transcriptase (qRT)–PCR techniques were used in the pancreatic tissue and biofluid miRNA profiling studies, but the techniques have limitations. Prior knowledge about individual miRNAs is often required, suggesting that novel miRNAs are often omitted. Background levels may be high owing to cross-hybridization, a low dynamic expression range, and complicated normalization methods to compare individual studies.208 The next-generation sequencing (NGS) technology can overcome some of these limitations. Next-generation sequencing has greater expression range compared with microarray 209,210 and has higher reproducibility between experiments.211 Furthermore, NGS provides digital readouts that make direct comparisons between studies from different laboratories possible. Some groups have used the NGS technology to discover novel miRNAs species that are not in current microarray platforms.212,213 Therefore, applying the NGS technology to pancreatic cancer tissue and patients’ biofluid may result in a more quantifiable and comparable miRNA biomarker signature when compared with microarrays. Specific pancreatic cancer miRs could be the ones that have low copy numbers and are not expressed in any other cancer types. The NGS technology may be used as a sensitive tool to reveal these low-copy-number, cancer-specific miRs.
Second, some miRNAs are released from tumors into the serum and plasma by tumor-derived exosomes,38 which may serve as another diagnostic tool. However, the mechanism by which tumor miRNAs are released by exosomes and identifying which miRNAs are released remain unclear. Studies have shown that 46% of the miRNAs in the tumor-derived exosomes are correlated with the tumor tissue miRNAs. Why the rest of 54% tumor tissue miRNAs are not released into the serum/plasma remains unclear.38–41 It is possible that all miRNAs in the tumor are released into the serum, but the abundance is too low for qRT-PCR and microarray detection. Perhaps using the NGS technique (such as RNA-seq) can overcome some of the limitations to identify suitable miRNA markers. Currently, it is still unclear what mechanism controls the release of miRNAs from the tumor into the patients’ serum. It is possible that only metastatic tumors release the miRNAs via exosomes. Based on the current serologic miRNA studies, 2 of 3 miRNAs markers are found to be associated with metastasis. If we take a closer look at the commonly overexpressed miRNA markers in pancreatic tissues, miR-107, miR-221, and miR-16 also play a role in metastasis.214–216 It is possible that they are indeed present in the patients’ blood, below the detection range of microarrays.
Optimizing the comparison strategy may improve the development of pancreatic cancer miRNA biomarker. Currently there are 3 widely used comparison strategies: (1) comparing to other healthy individuals, (2) comparing to adjacent normal tissues, and (3) comparing the gene expression to a pancreatic tissue–specific profile. One might argue that comparing with other healthy individuals to find differentially expressed pancreatic cancer miRNAs will identify genetic variations due to differences between individuals. On the other hand, comparisons with normal adjacent tissue removes shared common genetic variations; it also introduces problems associated with miRNA expression patterns associated with disease, as stressors derived from the tumor can promote genetic and phenotypic alterations within the surrounding tissues.184,217 Comparing gene expression with a pancreatic tissue– specific profile can provide a more tissue-specific miRNA biomarker but could fail to identify the biomarkers that are commonly expressed in multiple cancer types.42 Therefore, optimizing comparison strategies is needed to improve pancreatic cancer miRNA biomarker development.
Lastly, developing a noninvasive early diagnostic strategy is crucial for patients with pancreatic cancer. Early diagnosis is rare, and surgical extirpation is thought to be most beneficial before the cancer becomes locally invasive or metastatic. Although currently there are many potential biomarkers distinguishing normal pancreatic tissues and cancer, it is not useful as an early diagnostic tool. A few circulating miRNA biomarkers are being validated and developed to distinguish healthy individuals from pancreatic cancer patients. Because the prevalence of pancreatic cancer is 12 of 10,000 in the United States, it is very difficult to develop a high positive predictive value test to screen for pancreatic cancer patients. It is necessary for any test to have at least 0.99995 specificity and a 95% positive predictive value. If individuals are screened and tests show that they are negative for pancreatic cancer with the current circulating miRNA assays available, there is only 0.2% likelihood that they have pancreatic cancer. Therefore, if individuals are screened and results show that they are positive for pancreatic cancer, although the positive predictive value is only between 0.016% and 5% that they do have pancreatic cancer, it can allow them to undergo further examination to confirm if they have the disease as an early diagnostic test. MicroRNA-18a and miR-200a/b might serve as biomarkers to monitor the disease following treatment as they show promising sensitivity and specificity when the individual is confirmed to have pancreatic cancer.
SUMMARY
Pancreatic cancer miRNA biomarker signatures appear to be a protean area of investigation for future diagnostic or therapeutic purposes. Although there are potential pancreatic miRNA biomarkers in pancreatic tissue and patients’ blood, those biomarkers are not pancreatic cancer–specific, but could be quite useful in studying recurrence or progression. It is possible to establish a miRNA cancer biomarker signature, but distinguishing the site of origin of the cancer also remains challenging. Furthermore, because cancer is a dynamic disease, presorting the patients’ sample based on disease stages, ethnicity, and age before miRNA profiling might facilitate the identification of unique pancreatic cancer signatures for individual stages of cancer. It will also be interesting to apply the NGS technology to profile the cancer tissue and biofluid miRNAs to develop a more quantifiable and comparable, cancer type–specific miRNA signature for pancreatic cancer diagnosis and therapeutic target development. What is quite clear is that as our deeper understanding of the tumor microenvironment and macroenvironment reveals complexities of genetic and epigenetic control mechanisms, the frequent occurrence of aberrant forms of cell death in response to chronic stress requires that more holistic approaches integrating the known genetic changes and miR expression patterns now be considered.218–224 Integrating the host response with the panoply of genetic changes the occurrence within the tumor are now necessary for a full explication of cancer biology and the development of effective diagnostic tests and therapies.
Table 2
Pancreatic Cancer Tissue miRNAs
| Sample | Up-regulated | Down-regulated | Biological Significance | Ref. |
|---|---|---|---|---|
| Mouse | miR-15b, miR-92, miR-124a, miR-129, miR-132, miR-137, miR-181a/d, miR-184, miR-344, miR-376b, miR409-3p, miR410, miR-434-3p, miR-449 | miR-126, miR-141, miR-142-3-p, miR-142-5p, miR-143, miR-150, miR-182, miR-184, miR-200a/b/c, miR-335, miR-429 | The miRs signatures are different depending on the stage of the cancer. | 7 |
| Human Pancreatic Cancer Tissue | miR-16-2, miR-23a, miR-24-1,2, miR-26a, miR-29b-1,2, miR-99b, miR-100, miR-103-1,2, miR-107, miR-125a, miR-125b-1,2, miR- 126, miR-130a, miR-145, miR- 146, miR-191, miR-214, miR-221 | miR-155 | 20 of the pancreatic cancer miRs (e.g. miR-21, miR-17-5p, miR-24-2, miR-181b-1, miR- 107,miR-221 etc) are also commonly over-expressed in breast, colon, lung, prostate and stomach cancer. | 45 |
| Human Pancreatic Cancer Tissue | Let-7d,f, miR-15b, mR-16,miR- 21,miR-24-1,2, miR-92-1, miR- 107, miR-125b-1, mIR-155, miR- 181a,c, miR-212, miR-221, miR301, miR-376a, miR-424 | miR-139, miR-142, miR-345 | MiR-376a and miR-301 is deregulated in pancreatic cancer while not reported in other cancer tissue. | 42 |
| Human Pancreatic Cancer and Pancreatic Cancer Cell Line | miR-10b, *miR-15a,b, *miR-17-3p, miR-17-5p, miR-20a, miR-21, miR-24, miR-27a, miR-93, miR103, miR-106a, miR-107, miR-*125a,b, *miR-135, *miR-142-3p, miR-143, miR146b, miR-155, miR-181d, *miR-185, miR-191, miR-196, *miR-200b, miR-221, *miR-222, miR-223, miR-224 | miR-141, miR-219, miR-373 | 8 miRNAs (miR-196a, miR-190, miR-221, miR-22, miR-200b, miR-15b,and miR-95) are commonly expressed in both clinical sample and cell lines. Those miRs might be attractive miRNA-based therapeutics target. | 40 |
| Human Pancreatic Neoplasia | miR-16, miR-21, miR-100, miR- 107,miR-155,miR-181 a,miR-181c, miR-223, miR-210 | N/A | miR-155 and miR-21 are shown to be the potential markers for intraductal papillary mucinous neoplasms | 44 |
| Human Pancreatic Adenocarcinoma | miR-10-a/b, **miR-21, miR-23-a/b. miR-99. miR-100-1/2, miR-103-2, miR-107, miR-125a, miR-125-b-1, miR-143, miR-146, **miR-155, **miR-181a/b/d, miR-199a-1, miR-205, **miR-210,miR-213, **miR-221, miR-222, miR-223 | **miR-148a/b, **miR-375 | 12 microRNAs that are differentially expressed between pancreatic cancer to normal tissue is distinguished from pancreatitis** | 216 |
| Human Pancreatic Ductal Adenocarcinoma | miR-18c, miR-29c, miR-31, miR-93, miR-96, miR-130b, miR-141, miR-143, miR-145, miR-146a, miR-148-a/b, miR-150, miR-155, miR-196a, miR-196b, miR-203, miR-205, miR-210, miR-216, miR-217, miR-221, miR-222, miR-223, miR-224, miR-375 | N/A | Direct measure of miR-196 and miR-217 can provide a simple index to identify pancreatic ductal adenocarcinoma from normal tissue | 41 |
| Human | miR-15a, miR-27a, miR-100, | N/A | miR-21 might be related to poor | 43 |
| Pancreatic Cancer Tissue | miR-125b, miR-181a, miR-200a, miR-214 | response of pancreatic cancer cells to chemotherapy | ||
| Human Pancreatic Ductal Adenocarcinoma | miR-10a, miR-21, miR-143, miR-145, miR-155, miR-222, miR-223, miR-224, miR-373 | miR-148, miR-216, miR-217, miR-211, miR-345, miR-596, and miR-708 | miR-21 and miR-34a as survival predictive miRs | 46 |
Acknowledgments
This work was supported by the National Institutes of Health (R01CA160417 to D.T.).
Footnotes
Disclosure: The authors have no conflicts of interest or funding to disclose.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1097/mpa.0b013e3182854ab0
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4086428?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1097/mpa.0b013e3182854ab0
Article citations
Impact of exosome therapy on pancreatic cancer and its progression.
Med Oncol, 40(8):225, 05 Jul 2023
Cited by: 4 articles | PMID: 37405480 | PMCID: PMC10322774
Review Free full text in Europe PMC
Programming inactive RNA-binding small molecules into bioactive degraders.
Nature, 618(7963):169-179, 24 May 2023
Cited by: 22 articles | PMID: 37225982 | PMCID: PMC10232370
Research progress on microRNAs as molecular markers in pancreatic cancer: a narrative review.
J Gastrointest Oncol, 13(4):2048-2056, 01 Aug 2022
Cited by: 1 article | PMID: 36092335 | PMCID: PMC9459207
Review Free full text in Europe PMC
miRSCAPE - inferring miRNA expression from scRNA-seq data.
iScience, 25(9):104962, 17 Aug 2022
Cited by: 3 articles | PMID: 36060076 | PMCID: PMC9437856
Use of Biomarkers and Imaging for Early Detection of Pancreatic Cancer.
Cancers (Basel), 12(7):E1965, 19 Jul 2020
Cited by: 21 articles | PMID: 32707720 | PMCID: PMC7409286
Review Free full text in Europe PMC
Go to all (22) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Insights into the Role of microRNAs in Pancreatic Cancer Pathogenesis: Potential for Diagnosis, Prognosis, and Therapy.
Adv Exp Med Biol, 889:71-87, 01 Jan 2015
Cited by: 34 articles | PMID: 26658997 | PMCID: PMC5706654
Emerging roles of miRNAs in the development of pancreatic cancer.
Biomed Pharmacother, 141:111914, 10 Jul 2021
Cited by: 32 articles | PMID: 34328099
Review
Plasma miRNAs in diagnosis and prognosis of pancreatic cancer: A miRNA expression analysis.
Gene, 673:181-193, 18 Jun 2018
Cited by: 75 articles | PMID: 29913239
Circulating microRNAs as Potential Biomarkers in Pancreatic Cancer-Advances and Challenges.
Int J Mol Sci, 24(17):13340, 28 Aug 2023
Cited by: 13 articles | PMID: 37686149 | PMCID: PMC10488102
Review Free full text in Europe PMC





