Abstract
Free full text

A tour through the transcriptional landscape of platelets
Abstract
The RNA code found within a platelet and alterations of that code continue to shed light onto the mechanistic underpinnings of platelet function and dysfunction. It is now known that features of messenger RNA (mRNA) in platelets mirror those of nucleated cells. This review serves as a tour guide for readers interested in developing a greater understanding of platelet mRNA. The tour provides an in-depth and interactive examination of platelet mRNA, especially in the context of next-generation RNA sequencing. At the end of the expedition, the reader will have a better grasp of the topography of platelet mRNA and how it impacts platelet function in health and disease.
Introduction
Despite their small size and anucleate status, platelets have a rich repertoire of RNAs, including messenger RNAs (precursor-mRNA [pre-mRNA] and mature mRNA), structural and catalytic RNAs (ribosomal RNA [rRNA], transfer RNA, and small nucleolar RNA), and regulatory RNAs (microRNA [miRNA], long intergenic noncoding RNA, pseudogenes, and antisense RNA). Examination of RNA expression patterns in platelets has been used to identify biomarkers of disease, explain genetically or environmentally induced alterations in platelet function, and determine if genes are conserved between humans and mice.1-13 The past decade has also revealed that platelets can translate mRNA into protein or transfer RNA to recipient cells where it regulates functional processes.14-17 Because platelets are anucleate, transcriptional production of RNA (with the notable exception of mitochondria-derived transcripts) presumably occurs exclusively in the megakaryocyte. Therefore, the RNA profile in platelets also provides an accessible window into the transcriptional status of megakaryocytes, and the bone marrow signals involved, at the time of platelet production.
Several recent reviews have elegantly discussed the predictive value and functions of platelet RNAs.2,18,19 This review explores the landscape of platelet RNA as mapped by next-generation RNA sequencing (RNA-seq). Specifically, we examine what a platelet mRNA looks like at the molecular level and how specific features of the mRNA impact biological responses. Along the way, we provide a series of Web links and access instructions (see Table 1)1,20-22 for readers to follow as they explore key landmarks of platelet mRNA. Each stage of the tour (Figure 1) is discussed with reference to platelet function and in a broader biological context. Although screen shots are shown in figure format, this review is meant to be interactive and dynamic, with the goal of providing the tools necessary for researchers to explore the innumerable transcript features in platelets in the context of their own research interests.
Table 1
Links for viewing platelet RNA-seq data sets in the UCSC Genome Browser
| Site | Reference | Description: Web link > access instructions |
|---|---|---|
| 1 | 1 | Coverage track: https://hci-bio-app.hci.utah.edu/gnomex/gnomexFlex.jsp?dataTrackNumber=DT1837 > select “UCSC Browser”* |
| 2 | 1 | Bam (individual reads and sequences) track: https://hci-bio-app.hci.utah.edu/gnomex/gnomexFlex.jsp?dataTrackNumber=DT11785 > select “UCSC Browser”* |
| 3 | 20 | Coverage tracks and individual reads tracks: https://cm.jefferson.edu/data-tools-downloads/platelets-2012/ > select “A” to enter UCSC Browser* |
| 4 | 21 | Coverage tracks and individual reads tracks: https://cm.jefferson.edu/data-tools-downloads/platelets-2014/ > select “A” to enter UCSC Browser* |
UCSC, University of California, Santa Cruz.
 779
779 645-128
645-128 781
781 253”), or term of interest (“rs6065”). Scroll down to see the available tracks. The platelet tracks can be found under the header “Custom Tracks” for sites 1 and 2 or under the header “Mapping and Sequencing Tracks” for sites 3 and 4. Using the toggle boxes, set the visibility settings to “Full” for the platelet data track(s) of interest (ie, “B2_Pltlts_...”). Set visibility to “Full” for one of the Gene Predictions tracks (ie, RefSeq). “Hide” other tracks and “zoom” to view the data as presented in the figures. Select “Refresh” after making any changes to update the viewer. Other data sets, including mouse data sets, are accessible from within Gnomex by searching for “platelet” in the search box at the top. Alternatively, the .Bam and .Bai index files can be downloaded from Gnomex and opened in a genome browser such as the Integrated Genome Viewer.22 If the links don’t work initially, try a different browser. For more information on using the UCSC Genome Browser, see: https://genome.ucsc.edu/training.html.
253”), or term of interest (“rs6065”). Scroll down to see the available tracks. The platelet tracks can be found under the header “Custom Tracks” for sites 1 and 2 or under the header “Mapping and Sequencing Tracks” for sites 3 and 4. Using the toggle boxes, set the visibility settings to “Full” for the platelet data track(s) of interest (ie, “B2_Pltlts_...”). Set visibility to “Full” for one of the Gene Predictions tracks (ie, RefSeq). “Hide” other tracks and “zoom” to view the data as presented in the figures. Select “Refresh” after making any changes to update the viewer. Other data sets, including mouse data sets, are accessible from within Gnomex by searching for “platelet” in the search box at the top. Alternatively, the .Bam and .Bai index files can be downloaded from Gnomex and opened in a genome browser such as the Integrated Genome Viewer.22 If the links don’t work initially, try a different browser. For more information on using the UCSC Genome Browser, see: https://genome.ucsc.edu/training.html.How to navigate the landscape: an RNA-seq overview
To appreciate key features of platelet mRNA, one first has to understand the fundamentals of next-generation RNA-seq. RNA-seq has revolutionized medical research and, as shown in Table 2, several groups have recently used this powerful technique to profile platelet mRNA.20,21,23
Table 2
Published platelet RNA-seq (long-RNA) data sets
| Reference | 1 | 20 | 21 | 23 |
|---|---|---|---|---|
| Platform | Illumina | ABI/LT SOLiD | ABI/LT SOLiD | Illumina |
| No. samples | 4 | 4 | 10 | 4 |
| Species | 2 human/2 mouse | Human | Human | Human |
| Gender | 1 male/1 female of each species | ND | 10 males | 3 males/1 female |
| Race | ND | ND | 5 black, 5 white | ND |
| Leukocyte depletion | CD45 (Miltenyi) | CD45 (Miltenyi) | CD45 (Miltenyi) | Pan-Leuko (Dynabeads) |
| Reads | 36 bp, paired end, unstranded | 50 bp, single end, stranded (30-bp, stranded small RNA reads also available) | 50 bp, single end, stranded | 100 bp, single end, unstranded 100 bp, paired end, stranded |
| Sample prep | Poly-A+ RNA | Total RNA and rRNA-depleted RNA | Total RNA | rRNA depleted and poly-A+/5′ end–tagged RNA |
ND, no data.
RNA-seq libraries are prepared from either total RNA or a subfraction of RNA (ie, polyadenylated, rRNA-depleted, 5′ cap–containing small RNA, etc.) by random shearing of RNA into 100- to 400-bp fragments. Barcodes and sequencing primer binding sites are then added, and the RNA is briefly amplified, followed by size selection of 100- to 400-bp fragments (also called inserts). This size selection captures most protein-coding RNAs and long noncoding RNAs. For small RNA-seq, which captures mature mRNAs, piwi-interacting RNAs, and other small RNAs, fragmentation is skipped, and inserts <40 bp are selected. Following insert selection, ~30 bp up to >150 bp of 1 end (single-end read) or both ends (paired-end reads) of each fragment are sequenced. Sequences (reads) are then computationally mapped by finding unique (usually) matches between the annotated genome/transcriptome and the sequenced end of the RNA fragment. If paired ends are sequenced, then each end is matched with the expected distance constrained to the approximate expected fragment size. If libraries are stranded, then the read can be mapped directionally to either the positive or negative strand of DNA. RNA-seq methodology is reviewed in more detail elsewhere.24-27
Four long RNA-seq data sets (ie, protein coding and long noncoding RNA) from human platelets and 1 from mouse platelets have been published.20,21,23 Other small RNA-seq data sets (ie, mature miRNAs) have also been published but are not discussed here.20,28 As shown in Table 2, each RNA-seq data set offers some complementary features not provided by the other. For example, in our experience, shorter (36 bp) reads map better to poorly annotated transcripts or novel small exons, whereas longer reads (101 bp) aid in isoform discrimination and map better to lower-complexity regions. Paired reads are useful for determining splicing and exon exclusion patterns. Stranded reads discriminate between genes overlapping in the genome, but transcribed from opposite strands. Oligo-dT enriches for protein-coding transcripts, whereas ribosomal depletion of total RNA will capture more noncoding transcripts. In 1 of their samples, Kissoupoulou et al23 use a 5′ end–tagging strategy that marks the extreme 5′ terminus of transcripts. The aligned reads from 3 of these data sets are available for rapid navigation via Web links. Instructions to access and visualize the aligned data in the UCSC Genome Browser29 are given in Table 1.
A panoramic view of a platelet transcript as depicted by RNA-seq
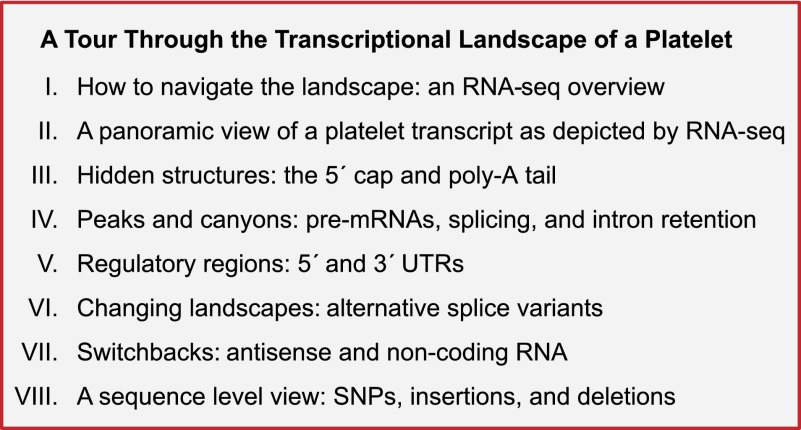
RNA-seq creates snapshots of mRNAs that provide a wealth of information, from gene expression to regional features and detailed sequence.24 Before exploring subdomains of platelet mRNA, it is helpful to understand the topography of snapshots that are used to represent RNA-seq results. Figure 2A represents a visualization of RNA-seq alignments to glycoprotein IX (GP9). In this example, GP9 is annotated as it appears in platelets, a mature message that does not contain intronic sequences. Note that individual paired reads, which in this case are ~36 bp in length, map across the entire GP9 annotated transcript. The reads that cover each nucleotide along the transcript are summed, and the count (y-axis) is displayed in the histogram-like graph (coverage or read depth graph) above the reads. Various methods, ranging in robustness, for calculating expression exist and are reviewed by Dillies et al and Garber et al.30,31 One simple and commonly used index of abundance is the read (or fragment) per kilobase normalized to a million.27 This is calculated from the sum of all the reads across a transcript, normalized to the transcript length and to the total number of reads mapping to the entire transcriptome. Variability in fragmentation, sequencing, and mapping efficiency for subsequences within the transcript affects the read depth at each loci, hence the “hills” and “valleys” of expression across the transcript.31 Thus, RNA-seq can quantify per-base, per-exon, per–annotated region, or per-transcript expression on a genome-wide scale without prior knowledge of the transcript.24
General features of an mRNA transcript as visualized by RNA-seq. This figure shows RNA-seq read coverage plots and individual reads for platelet GP9 mRNA as it appears (ie, with introns removed) in the platelet (A) or as viewed along genomic coordinates in a genome browser (B). The blue annotations along the bottom are the consensus RefSeq annotations of the gene structure. Individual exons are represented by the blue bars, and introns (only in panel B) by the connecting blue lines. The thicker regions of the bars distinguish the coding region from the 5′ and 3′ untranslated regions (UTRs). Arrows indicate the strand orientation (forward [+] or reverse [−]) of the annotation. For GP9, there are 3 exons and 2 introns. In GP9, the 5′ UTR ends just beyond the start of the third exon (at the ATG start codon of the coding region), and the 3′ UTR starts near the end of the third exon (after the TAA stop codon). Above the annotation are the individual reads that align to the transcript. For simplicity, not all reads are shown here. For visualization purposes, the individual reads are colored according to the exon to which they align. Above the reads is the read coverage map. This is generated by plotting the sums of mapped reads that overlap (cover) each nucleotide position along the genomic coordinates (x-axis). The y-axis represents the number of reads. The examples are as illustrated in Integrated Genome Viewer.22 See it yourself: To see GP9 in platelets via the UCSC Genome Browser, follow the Web link > access instructions for site 1 (see Table 1). For individual reads, follow the Web link > access instructions for site 2 (see Table 1). Search for GP9 in the browser.
Unlike the depiction in Figure 2A, RNA-seq data are typically visualized in genomic space using a genome browser such as the UCSC Genome Browser.29 Figure 2B depicts the same transcript, GP9, in a genome browser. Note now that the exons are separated by the introns encoded in the genome, and like canyons between peaks of expression, the introns are devoid of aligned reads. This is because GP9 is completely spliced. Reads connected by thin lines represent either connected paired or split reads. Although the intron spanned by paired reads is much longer than the expected 200-bp fragment size, when the transcript is spliced the paired reads map at the expected distance.
Hidden structures: the 5′ cap and poly-A tail
Although anucleate, platelet mRNAs mirror mRNAs found in nucleated cells. Eukaryotic RNAs are prepared for nuclear export and translation by the addition of a 5′ 7-methylguanosine (7mG) cap, the addition of a 3′ poly-A tail of variable length, and the splicing out of introns.32 The 5′ cap facilitates ribosome loading for protein translation via recognition and recruitment by eukaryotic initiation factor 4E (eIF4E).33,34 The poly-A tail helps circularize mRNAs through interaction with the scaffold protein eIF4G, which initiates cap recognition by eIF4E.33 Both ends protect mRNA from degradation.33 The poly-A tail can be extended or shortened over the life of the RNA, but gradual poly-A shortening followed by decapping is the eventual fate of most RNAs.35,36 In other mammalian cells, typically only 5′-capped, 3′-polyadenylated, spliced mRNA will be exported from the nucleus and used for translation.32 The majority of the unspliced cytoplasmic message is rapidly degraded by nonsense mediated decay (NMD).37
The customary use of oligo-dT to prime reverse transcription leaves little doubt that the majority of platelet transcripts are polyadenylated, consistent with descriptions of poly-A+ RNA in platelets reported by Roth and colleagues in 1989.38 Platelets contain poly-A binding proteins such as activated-platelet protein-139 that may support poly-A–stimulated protein translation. Evidence that a subset of transcripts is 5′ capped comes from reports that protein translation can be initiated in platelets.40 The poly-A tail is not genomically encoded, and stretches of As cannot be effectively aligned from RNA-seq reads. Therefore, poly-A reads are hidden in typical RNA-seq work flows. The 5′ cap is also not detected by normal RNA-seq reactions.
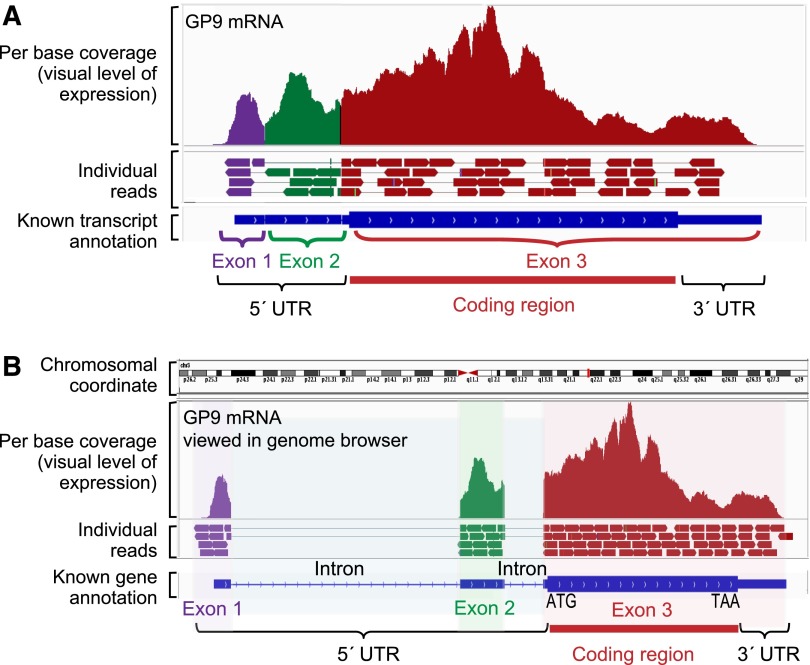
To characterize capping and polyadenylation in platelets, we have performed RNA-seq on transcripts captured with oligo-dT–conjugated beads or by a hyperaffinity mutant eIF4E protein that enriches for transcripts with a 7mG cap.41 Figure 3A offers a pictorial of this strategy. When comparing RNA-seq of poly-A–isolated mRNA with cap-captured RNA, poly-A isolation captures the majority of annotated protein-coding transcripts, whereas some transcripts with a very short or no poly-A tail are missed. Such is the case for many histone-coding transcripts with characteristically short or no poly-A tails,42 which are missed by poly-A capture, yet readily observed when isolated by the 5′ cap–based strategy (Figure 3B). Conversely, a handful of mRNAs detected by poly-A–based methods are not captured by 5′-based methods (not shown). Aside from these exceptions, the bulk of annotated platelet transcripts that are captured by oligo-dT are also captured by cap-binding strategies (J.W. Rowley and A.S. Weyrich, unpublished data). A prime example is PAR1, whose mRNA was captured by both 3′- and 5′-based methods (Figure 3B). Other transcripts, in particular noncoding RNA, are undoubtedly present in platelets that contain neither a 5′ cap nor a 3′ poly-A tail.20
Platelet transcripts are capped, tailed, and spliced. (A) The techniques we have used to capture and concentrate poly-A mRNA or 5′-capped mRNA from platelets. The captured RNAs are concentrated by immunoprecipitation techniques. For poly-A pull-down, the bead-conjugated oligo-dT sequence binds to the poly-A tail. For the 5′-cap pull-down, a bead-conjugated hyperaffinity mutant enzyme binds to the 7mG cap at the 5′ end of the mRNA. (B) Bar graph of the RNA-seq expression estimates (RPKM) of a histone transcript HIST1H4B and the transcript coding for the platelet thrombin receptor protease-activated receptor 1 (F2R) following mRNA isolation by a 5′-cap or poly-A tail pull-down. (C) RNA-seq coverage graph of FBJ murine osteosarcoma viral oncogene homolog B (FOSB), which appears mostly unspliced in platelets. Note that reads align to both exons and introns. Compare this with GP9 in Figure 2, which has very few reads mapping to introns. RPKM, read (or fragment) per kilobase normalized to a million.
The beginning of the poly-A tail can be inferred from the reads by a sharp decline in read depth at the end of the 3′ UTR accompanied by 3′-end mapped reads containing strings of As.23,43 This type of analysis was used by Osman and coworkers to identify 2 polyadenylation start sites for transcript pro-platelet basic protein.23 On the other hand, the length of poly-A tails in platelets and whether tail shortening occurs over time are not known. This is in part because new and old populations of platelets are difficult to separate in vivo.
In vitro studies suggest that some platelet mRNAs are not stable over time. Reticulated platelet counts potentially decrease after platelet storage.44,45 mRNA for P-selectin and glyceraldehyde-3-phosphate dehydrogenase disappear in stored platelets at 22°C but decline only 40% over 5 days when stored at 4°C.46 Sulfotransferase family cytosolic 2B member 1b (SULT2B1B) mRNA, which affects platelet functional responses via catalysis of the sulfonation of cholesterol,47 also diminishes rapidly at 37°C.48 SULT2B1B mRNA levels decrease by 40% within just 30 minutes at 37°C but are stable at 4°C. Interestingly, addition of high-density lipoprotein to the platelets slowed the decay. Of note, SULT2B1B protein levels and its product level of cholesterol sulfate tracked with mRNA levels suggest continued protein translation and functional implications of mRNA stability in platelets. Whether the poly-A tail and cap are involved in the stability and translation of these and other transcripts in platelets merits further investigation.
Peaks and canyons: pre-mRNAs, splicing, and intron retention
Surprisingly, platelets contain a subset of pre-mRNAs that have escaped the nucleus and NMD without splicing.49 Platelet activation induces splicing and enables protein production from the newly formed mRNA. Unspliced pre-mRNA can be observed in RNA-seq data visually or bioinformatically by examining the ratio of intronic to exonic reads. Interleukin-1β, a prototypical unspliced mRNA in platelets,49 contains intronic reads in relatively high proportion to exonic reads.2 According to our own analysis of platelet RNA-seq data sets, transcripts like interleukin-1β, with a large fraction of completely unspliced pre-mRNA, are infrequent. Despite this, additional examples of pre-mRNA splicing followed by protein production in platelets have surfaced in the literature.50,51 In addition to these, using RNA-seq we have bioinformatically identified a handful of putatively unspliced transcripts in platelets that retain 1 or more introns. One example shown in Figure 3C is FOSB. FOSB is a transcription factor that regulates a wide array of cellular functions.52 Whether FOSB can be spliced within platelets remains to be determined.
Regulatory regions: 5′ and 3′ UTRs
Serial analysis of gene expression in platelets previously demonstrated that the set of transcripts in platelets have longer UTRs than the set of transcripts found in other cells.53 UTRs can also be assessed at the individual level. Figure 4A-B shows examples of transcripts in platelets where the actual UTR does not match the predicted UTR. The 5′ and 3′ UTRs regulate RNA stability and protein translation.54-56 Within the 5′ UTR, secondary structure and primary sequence motifs direct ribosome scanning and initiation of translation.57 A kozak sequence within the 5′ UTR that contains the AUG start codon flags the ribosome to start translation of the open reading frame (ORF). Premature upstream ORFs regulate the efficiency of initiation at the coding ORF, and secondary structures in 5′ UTRs further affect ribosome loading and scanning efficiency.58 Proteins and small RNAs (ie, miRNAs) bind to sequence motifs within the 5′ UTR and 3′ UTR to regulate translation or degradation.54,55,57,59 In platelets, for example, the 5′ UTR of B-cell lymphoma 3 controls translation of the mRNA in an mammalian target of rapamycin–dependent manner.60 Given the many sequence-dependent effects of the UTR on RNA stability and translation, the predicted biological relevance of an annotated UTR sequence vs the actual UTR expressed in platelets could be profoundly different.
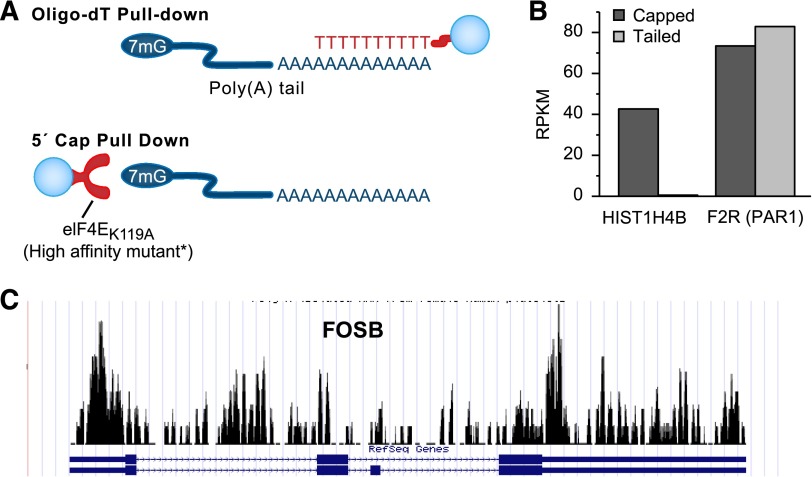
Platelet transcripts have variable 5′ or 3′ UTRs. (A) Shown are coverage graphs of the 5′ end of GFI1B in platelets. The top panel represents coverage of all reads mapping to the 5′ end of GFI1B. The red box is drawn around the region that maps beyond the known transcript annotation (blue). The bottom panels show only those reads that contain the sequence tag ligated onto the 5′ end at time of sample preparation (for details, see Osman et al23). The 5′-tagged reads are only the extreme 5′-terminal reads and therefore mark the transcription start site. (B) Coverage graph of the 3′ end of purinergic receptor P2Y, G-protein coupled 1 (P2RY1) in platelets. The red box is drawn around the region that maps beyond the known 3′ UTR annotation. miRNA target analysis of this additional sequence identified several miRNA binding sites. A representative subset of the target sites is marked underneath the graph. See it yourself: To see GFI1B and P2RY1 in platelets via the UCSC Genome Browser, follow the Web link > access instructions for site 1 (see Table 1). For individual reads, follow the Web link > access instructions for site 2 (see Table 1). Search for GFI1B or P2RY1 in the browser. Note that the 5′-terminal reads in the bottom panels of A were processed from the data set published by Osman et al23 and are therefore not yet available in a browser-friendly format.
Growth factor independent 1B transcription repressor (GFI1B) controls megakaryocyte development,61 and GFI1B mutants can cause diseases of abnormal platelet formation and function like gray platelet syndrome (GPS).62 In Figure 4A, the annotated 5′ UTR of GFI1B covers ~350 bp of exons 1 and 2. RNA-seq reads in platelets suggest that transcription begins an additional 300 bp upstream from the annotated transcripts (Figure 4A, upper panel). The preponderance of 5′-terminally tagged reads (Figure 4A, lower panel) that map nearly exclusively to the start of the extended region suggests a dedicated transcription start site rather than aberrant run-on transcription. The additional 5′ sequence contains 3 short upstream ORFs.
miRNAs regulate RNA stability and protein translation by binding nearly complementary target sequences in the 3′ UTR of transcripts.63 In platelets, miRNAs are abundant and stable.64,65 They have been reported as biomarkers and as functional modifiers of platelets.18,19,66 Interestingly, platelets can transfer miRNAs to regulate targets in other cells.15-17 miRNA studies in platelets are increasingly popular and are the subject of several reviews.18,19,67,68
miRNA studies incorporate target prediction algorithms based on 3′-UTR annotations to identify potential miRNA-mRNA pairs.69 Differences in 3′-UTR annotations may affect the outcome of such studies. P2RY1 accounts for ~1/3 of adenosine diphosphate receptors on platelets.70 Stimulation of P2RY1 by adenosine diphosphate induces intracellular calcium fluxes and platelet aggregation.71,72 P2RY1 is a potential target of antithrombotics,70,72 and differences in P2RY1 expression alter bleeding times and thrombosis in mice.73 In Figure 4B, the annotated 3′ UTR of P2RY1 is nearly 1 kb, whereas the actual 3′ UTR in platelets is considerably longer. According to miRNA target prediction algorithms (miRanda74), this additional 2.5 kb harbors additional miRNA binding sites (Figure 4B). This indicates that actual transcript data, not predicted annotation data, should be used for miRNA target site predictions in platelets. Sequences in view can be extracted directly in the UCSC Genome Browser by selecting “DNA” in the “view” menu.
Changing landscapes: alternative splice variants
Single nucleotide polymorphism (SNP) association studies75 and mechanistic studies of platelet endothelial aggregation receptor 1 (PEAR1) link it strongly to an aggregatory role in platelets.76 As illustrated in Figure 5A, RNA-seq distinguishes between multiple annotations for PEAR1. Alternative splicing affects nearly every multiexon eukaryotic transcript.77 Exon skipping, intron retention, alternate exon usage, and addition or truncation of exons are examples of alternative splicing.77,78
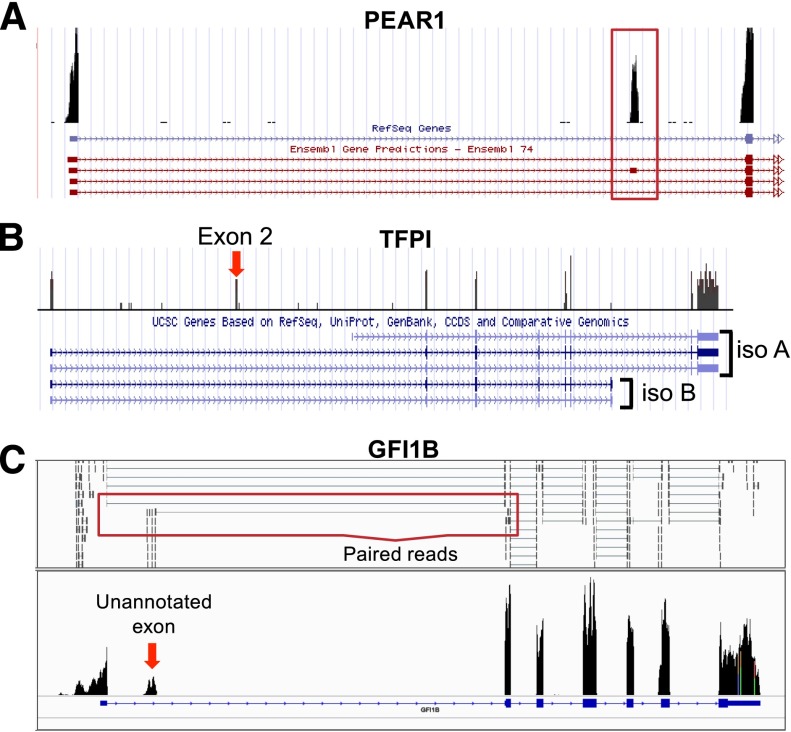
Platelets express mRNA splice variants. Shown are coverage graphs of PEAR1 (A), tissue factor pathway inhibitor (TFPI) (B), and GFI1B (C) in platelets. (A) Note that there are 4 different Ensembl annotations listed underneath PEAR1. The red box is drawn around the second exon, which distinguishes the second annotation from the other 3 annotations. (B) The annotations for 2 major isoforms of TFPI, α (iso A) and β (iso B), are distinguished by their 3′ ends. Note that platelets are rich in isoform α but not isoform β. Exon 2 (exon 2 is very small, so it is difficult to see the annotation here), which can also be removed by alternative splicing, is also present in the platelet. (C) The arrow points to a putative novel exon of GFI1b. Annotations for an exon at this region are not present in any of the Ensembl, UCSC, or RefSeq databases. The red box outlines examples of paired reads that map from the unannotated novel exon to the known second exon suggesting that it is part of the GFI1B transcript. See it yourself: To see PEAR1, TFPI, and GFI1B in platelets via the UCSC Genome Browser, follow the Web link > access instructions for site 1 (see Table 1). For individual reads, follow the Web link > access instructions for site 2 (see Table 1). Search for PEAR1, TFPI, or GFI1B in the browser.
Many alternative splice variants lead to NMD.37 Other splice variants regulate protein synthesis or code for alternate protein products. Splice variants of the same gene can effect opposing functions, and some variants may alter physiological responses to drugs.79 Proteomics detects many novel protein isoforms in platelets. In one proteomics study,80 a number of proteins with novel exon–skipping events were identified. Three of these, integrin (ITG) A2, fumarate hydratase, and aminopeptidase puromycin sensitive, were further validated by polymerase chain reaction at the RNA level.80 Interestingly, activation altered the number of exon-skipping events.80
We have seen how alternate initiation and termination can affect regulatory elements within UTRs. Alternative splicing is another way to alter the 5′ and 3′ UTR. As shown in Figure 5B, RNA-seq can distinguish TFPI splice variants in platelets. TFPI inhibits tissue factor activity and thereby limits excessive coagulation.81 Multiple splice variants have been characterized for TFPI. Megakaryocytes and platelets express TFPIα, 1 of 2 major well-characterized isoforms81 produced by alternative splicing of the 5′ and 3′ end of the mRNA. Part of the 5′ UTR in exon 2, which can be removed by alternative splicing,81,82 represses TFPI translation. On the other hand, the 3′ UTR of the TFPIα isoform in platelets relieves exon 2–mediated repression.82 The proteins produced by TFPI splice variants are also different. The C termini of the TFPI isoforms differ in tissue factor inhibitory activity.83 Furthermore, TPFIα is soluble and not linked to the cell surface like TFPIβ.83
The production of a soluble vs membrane-bound form of proteins is a recurring theme of alternative splicing in platelets. Soluble P-selectin is shed from platelets after activation, mediated in part by extracellular cleavage.84 Alternative production of soluble P-selectin may also be involved. Complementary DNA sequencing predicted the presence of a soluble splice variant of P-selectin.85-87 Correspondingly, soluble P-selectin is found in the nonmembrane fraction of platelets, totaling up to 10% of total P-selectin levels.88 Splice variants of ITGB389 code for a truncated protein, with the transmembrane and cytoplasmic domains missing. Protein assays have confirmed the expression of truncated ITGB3 protein in platelets.89 Platelets contain multiple forms of the low-affinity immunoglobulin Fc region receptor IIa mRNA. Variants with and without the transmembrane exon are detected at approximately equal proportions in platelets.90
Several other notable splice variants with relevance to platelet function have been reported. A variant of cyclooxygenase 2a is increased 200-fold after coronary artery bypass grafting.91 Variants of phospholipase C, β2 code for 2 different proteins in platelets.92 An angiopoietin-1 splice variant that opposes its parent gene is found in platelet α-granules.93 These examples only scratch the surface of the extensive collection of splice variants found in platelets. Some platelet-specific splice variants, including novel exons, have yet to be annotated within major databases. Such is the case with GFI1B (Figure 5C).
Switchbacks: antisense and noncoding RNA
Noncoding RNAs regulate the production of target mRNAs or the translation of target mRNAs into protein, or they directly modulate the target proteins themselves. Platelets contain all major classes of noncoding RNAs including miRNAs (see “Regulatory regions: 5′ and 3′ UTRs”), long intergenic noncoding RNAs, pseudogenes, and antisense RNA. Of these, platelets are particularly enriched in miRNAs, pseudogenes, and antisense RNAs.20
In eukaryotic cells, a major fraction of expressed genes are accompanied by an antisense.94 Antisense transcripts regulate their complementary counterpart in numerous ways. Antisense RNAs mask miRNA binding sites, destabilize transcripts, repress translation, and promote or repress transcription.94,95 They mediate both cytoplasmic and nuclear RNA-RNA and RNA-DNA interactions.94 Figure 6 depicts an example antisense RNA found at the 5′ end of CD109. CD109 is a highly expressed human platelet alloantigen.96 In this example, the antisense to CD109 begins at the 5′ end of CD109.
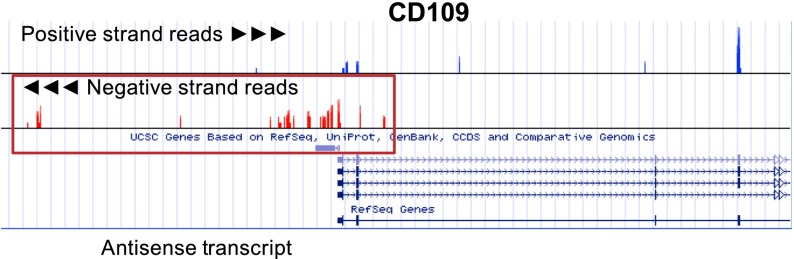
Antisense transcripts in platelets. Shown are positive and negative strand coverage graphs of the 5′ end of CD109 and of AK124950, an antisense transcript in platelets. Positive strand reads are on the top panel in blue, and negative strand reads are in the bottom panel in red. Both the expression of CD109 and the expression of an antisense transcript (AK124950, surrounded by the red box) transcribed from the opposite strand of DNA at the 5′ end of CD109 can be inferred when the read orientation is taken into account. See it yourself: To visualize CD109 and its antisense transcript in platelets via the UCSC Genome Browser, follow the Web link > access instructions for site 3 or 4 (see Table 1). Search for CD109 or AK124950 in the browser. Note that only stranded data (only site 3 or 4 of Table 1) can distinguish antisense from sense transcripts.
A sequence level view: SNPs, insertions, and deletions
By zooming to the sequence level, individual SNPs, insertions, and deletions that are within the expressed transcript can be identified by individual RNA-seq reads. Searching for “platelet” within the “clinical synopsis” field within Online Mendelian Inheritance in Man (http://omim.org/, April 4, 2014 update) retrieves 79 different inherited platelet disorders, corresponding to 59 different defective genes. The inherited platelet disorders include well-characterized platelet gene/phenotypes such us ITGA2B/Glanzmann thrombasthenia97 or neurobeachin-like 2/GPS.98-100 Clicking on the genome coordinates (GRCh37) listed within any of these disorders while the RNA-seq platelet data UCSC Genome Browser window is open will bring you directly to the location listed overlaid with read sequence information. Of note, visualization of mutations in RNA-seq read sequences from a patient with GPS helped characterize a splicing defect in their neurobeachin-like 2 transcripts.98
Genome-wide association studies (GWASs) have identified SNPs near >40 different genes associated with platelet size, count, or function.101-105 Meta-analyses have extended this number to ~80 (for a convenient list, see Bunimov et al102). In GWASs, a trait-associated SNP serves as a regional marker of allelic association without necessarily being the cause of the trait.106,107 Figure 7 depicts a reported GWAS common variant associated with platelet counts, rs6065,101,108 which is found within the RNA-seq read sequences of GP1BA compared with another individual without the variant. As we understand more about the meaning of GWAS SNPs, the ability to link RNA transcripts with marker SNPs and the causative mutations of trait variation may become important.
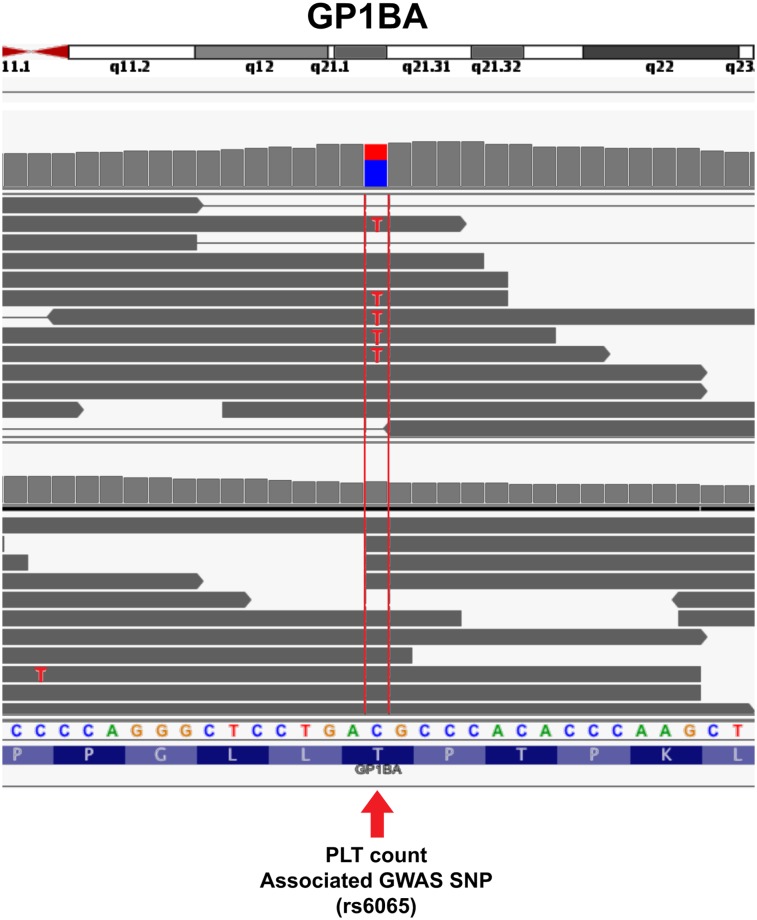
Single nucleotide resolution of glycoprotein Ib alpha (GP1BA) in platelets. Individual reads from platelets from 2 different individuals are shown. The arrow points to the location of the SNP rs6065, which associates with platelet counts in GWASs. Looking at the individual nucleotide sequences compared with the reference sequence indicates that the reads from the bottom individual match the reference cyotosine (reference matches are grayed out), whereas approximately half of the reads from the top individual contain the C to T polymorphism. See it yourself: To visualize rs6065 in GP1BA expressed in platelets via the UCSC Genome Browser, follow the Web link > access instructions for site 2 (see Table 1). Search for rs6065 and zoom in to the center of the region, or alternatively, type chr17:4 836
836 371-4
371-4 836
836 392 in the search box.
392 in the search box.
Perspective
After completing this tour, it should be obvious that platelets have a complex mRNA signature that reflects and affects their function. Indeed, mRNA expression profiling was recently used to identify the molecular basis for differences in platelet reactivity between blacks and whites.7 The study found by microarray that phosphatidylcholine transfer protein (PCTP) mRNA is fourfold higher in blacks than in whites. This correlated with PCTP protein levels and platelet reactivity to PAR4. Another RNA, miR-376c, inversely correlated with platelet reactivity and regulated PCTP expression. This study is mentioned to underscore how platelet mRNA can be leveraged to identify and understand the molecular mechanisms that control human disease. Of note, the RNA-seq reads from 5 black and 5 white individuals’ platelets were subsequently published.21 In this data set (which is site 4 of Table 1 in this review), consistent with their microarray findings, PCTP mRNA is sevenfold higher in blacks than in whites.21
The intrinsic functional capacity and variability of platelets are determined by environmental signals and genetic factors seen by the megakaryocyte. As has been done for other cells,109 integrating platelet and megakarycoyte RNA-seq with whole-genome sequencing or genome-wide microarray genotyping will provide valuable information regarding the relationship between the environment and genetics of altered gene expression within platelets. In addition to expression, RNA-seq analysis provides a tool for examining transcripts, and their plausible roles in platelet function and disease, at several other layers within the platelet terrain including the 5′ cap and poly-A tail, the 5′ and 3′ UTRs, intron retention, alternative splicing, strand, and single nucleotide sequences (summarized in Figure 8). In conjunction with published reports, we predict that forthcoming RNA-seq data sets will provide unprecedented opportunities to assess both “what’s in a platelet” and “what’s different about platelets in disease.”
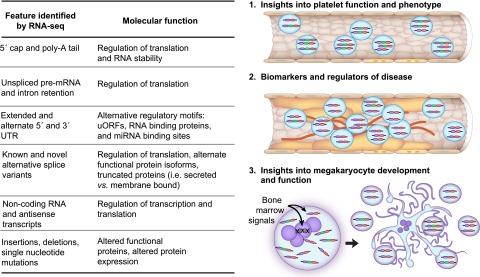
RNA-seq facilitates discovery in platelets. The table summarizes the various transcript features, and their respective functions, that are identifiable through RNA-seq in platelets. Each feature in platelets can be analyzed to provide mechanistic insights into platelet function (1), as biomarkers or modulators of disease (2), or for their role in megakaryocyte development and function (3). As an example, the vessel in (1) depicts platelets that express 2 different splice variants: a predominant 3-exon variant (purple-green-red) and a minor 2-exon variant (purple-red). In vessel (2), which is atherosclerotic, the predominant isoform in platelets is shifted to the 2-exon isoform that may serve as a biomarker for atherosclerosis and may additionally contribute to disease pathogenesis. As depicted in (3), a combination of genetics and environmental signals reaching the bone marrow megakaryocyte dictate the expression of the RNAs, and the features of RNA, that are captured as the platelets are formed.
Acknowledgments
We are grateful for the contributions of Diana Lim in preparation of the figures.
This work was supported by National Institutes of Health, National Institute of General Medical Sciences awards 1U54 HL112311-01, 1K01 GM103806-01, and 2R01 HL066277-11, as well as by the German Research Foundation (SCHU 2561/1-1).
Authorship
Contribution: S.S. and J.W.R. drafted and prepared the manuscript; and A.S.W. reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jesse W. Rowley, Department of Internal Medicine, University of Utah School of Medicine, Eccles Institute of Human Genetics, Building 533, Room 4260, 15 North 2030 East, Salt Lake City, UT 84112; e-mail: [email protected].
References
Articles from Blood are provided here courtesy of The American Society of Hematology
Full text links
Read article at publisher's site: https://doi.org/10.1182/blood-2014-04-512756
Read article for free, from open access legal sources, via Unpaywall:
http://www.bloodjournal.org/content/bloodjournal/124/4/493.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1182/blood-2014-04-512756
Article citations
Liquid biopsy: paving a new avenue for cancer research.
Cell Adh Migr, 18(1):1-26, 01 Sep 2024
Cited by: 0 articles | PMID: 39219215 | PMCID: PMC11370957
Review Free full text in Europe PMC
A Platelet Reactivity ExpreSsion Score derived from patients with peripheral artery disease predicts cardiovascular risk.
Nat Commun, 15(1):6902, 20 Aug 2024
Cited by: 1 article | PMID: 39164233 | PMCID: PMC11336089
Plasma growth factors maintain constitutive translation in platelets to regulate reactivity and thrombotic potential.
Blood Adv, 8(6):1550-1566, 01 Mar 2024
Cited by: 3 articles | PMID: 38163324 | PMCID: PMC10982986
Megakaryocyte- and erythroblast-specific cell-free DNA patterns in plasma and platelets reflect thrombopoiesis and erythropoiesis levels.
Nat Commun, 14(1):7542, 20 Nov 2023
Cited by: 3 articles | PMID: 37985773 | PMCID: PMC10662131
The Novel Role of Noncoding RNAs in Modulating Platelet Function: Implications in Activation and Aggregation.
Int J Mol Sci, 24(8):7650, 21 Apr 2023
Cited by: 0 articles | PMID: 37108819 | PMCID: PMC10144470
Review Free full text in Europe PMC
Go to all (65) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs
- (2 citations) dbSNP - rs6065
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Platelets deliver small packages of genetic function.
Blood, 122(2):155-156, 01 Jul 2013
Cited by: 13 articles | PMID: 23847185
Platelet mRNA: the meaning behind the message.
Curr Opin Hematol, 19(5):385-391, 01 Sep 2012
Cited by: 85 articles | PMID: 22814651 | PMCID: PMC3670814
Review Free full text in Europe PMC
Analysing the platelet transcriptome.
Vox Sang, 87 Suppl 2:42-46, 01 Jul 2004
Cited by: 10 articles | PMID: 15209877
Review
Activated platelets can deliver mRNA regulatory Ago2•microRNA complexes to endothelial cells via microparticles.
Blood, 122(2):253-261, 07 May 2013
Cited by: 256 articles | PMID: 23652806
Funding
Funders who supported this work.
NHLBI NIH HHS (4)
Grant ID: 2R01 HL066277-11
Grant ID: 1U54 HL112311-01
Grant ID: U54 HL112311
Grant ID: R01 HL066277
NIGMS NIH HHS (2)
Grant ID: 1K01 GM103806-01
Grant ID: K01 GM103806
 1,2
1,2