Abstract
Free full text

OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences.
Abstract
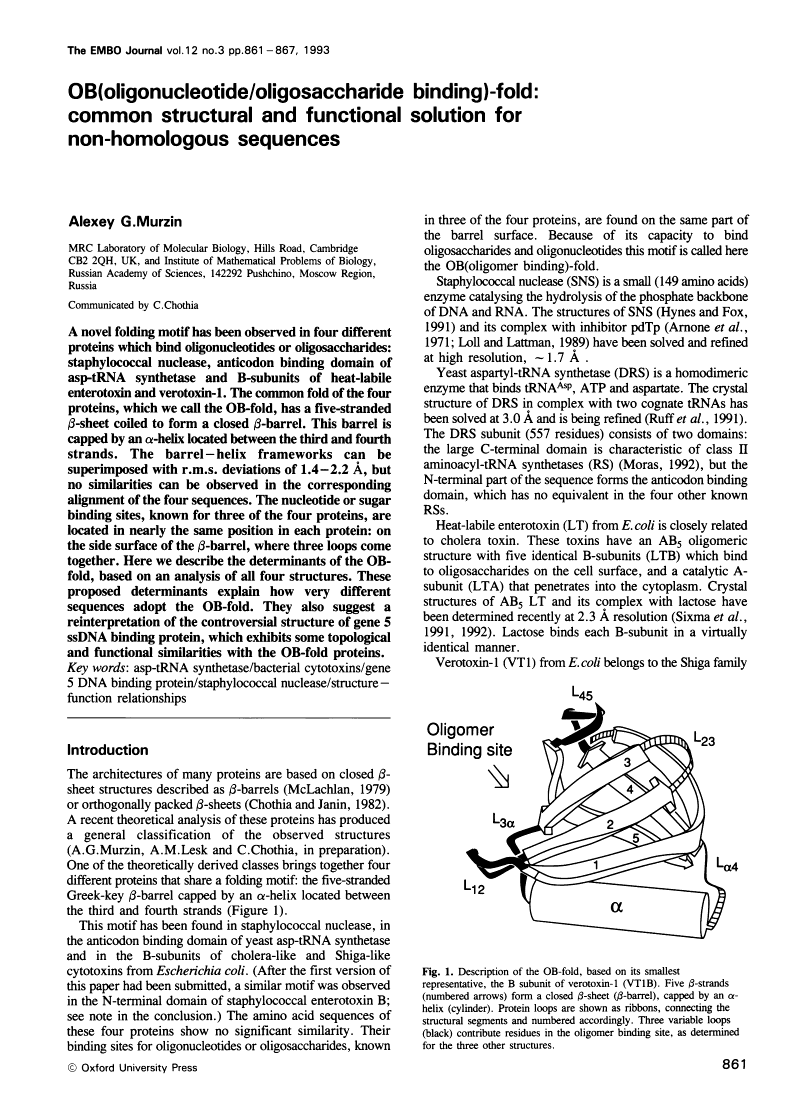
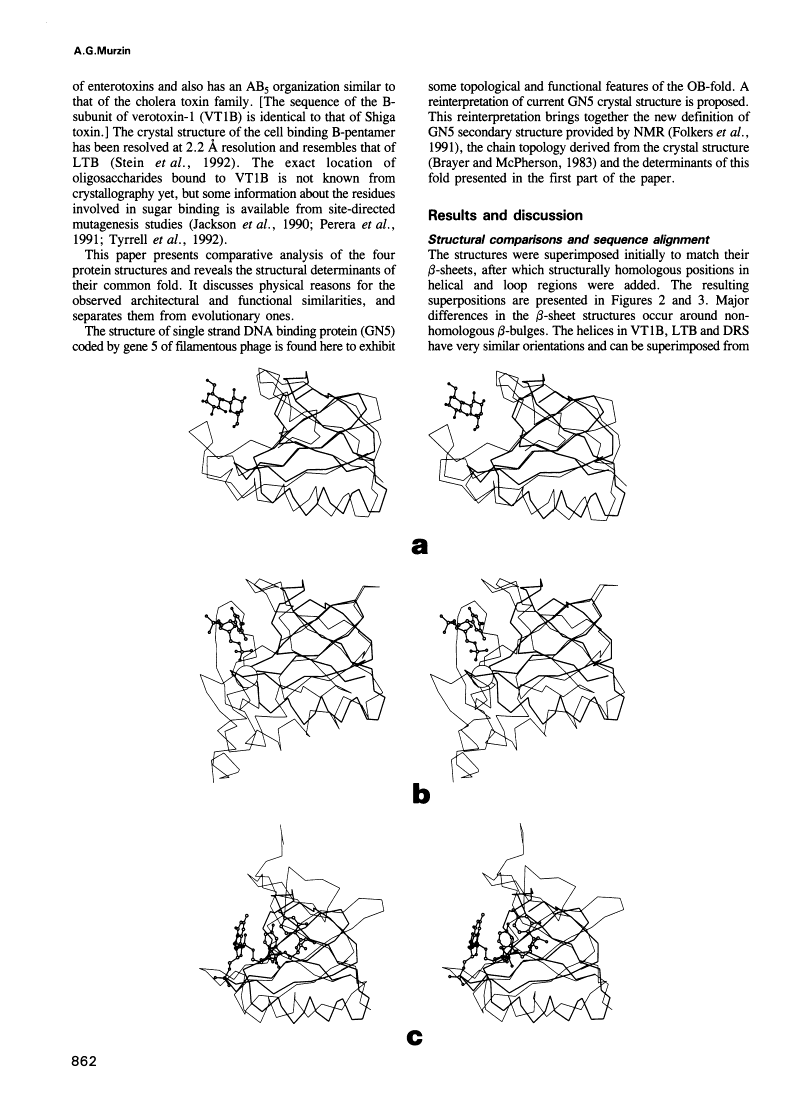
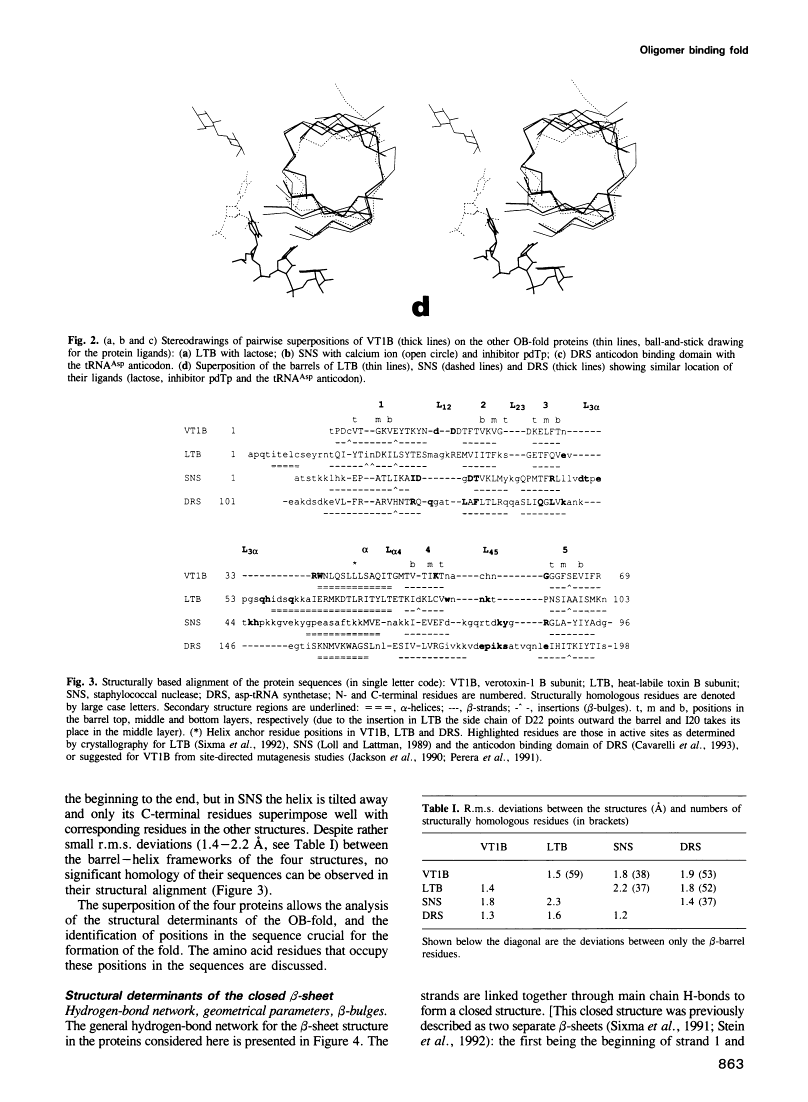
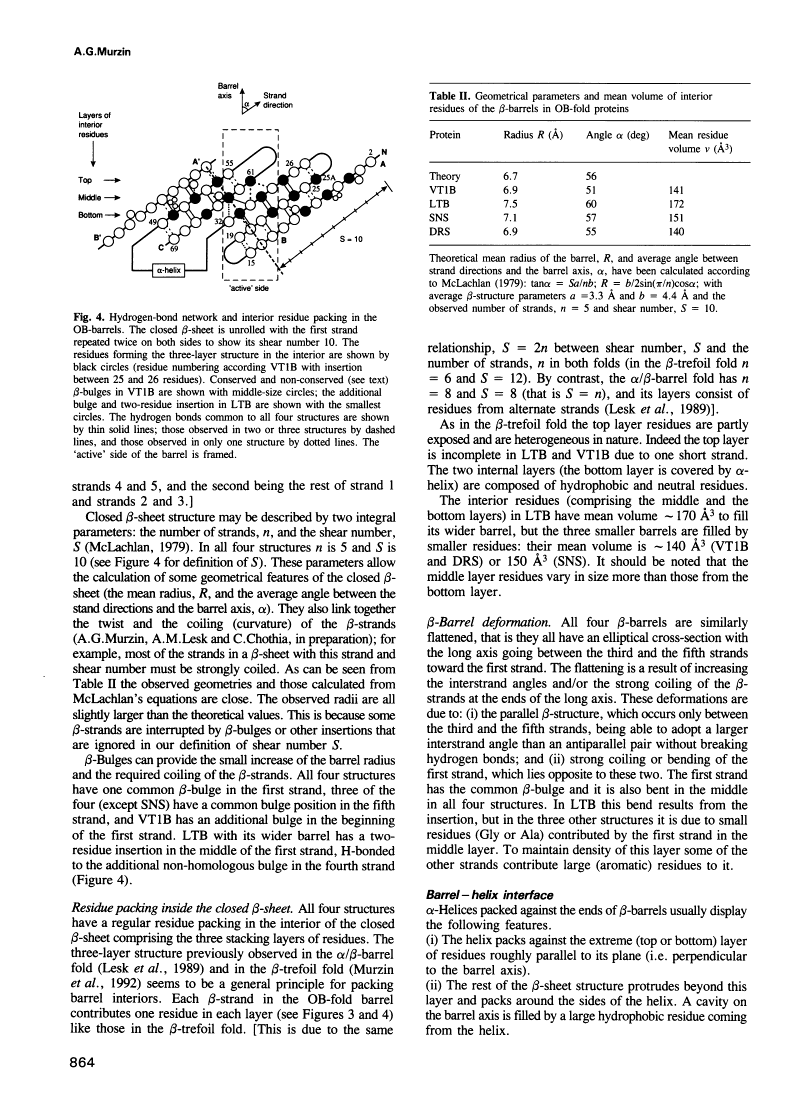
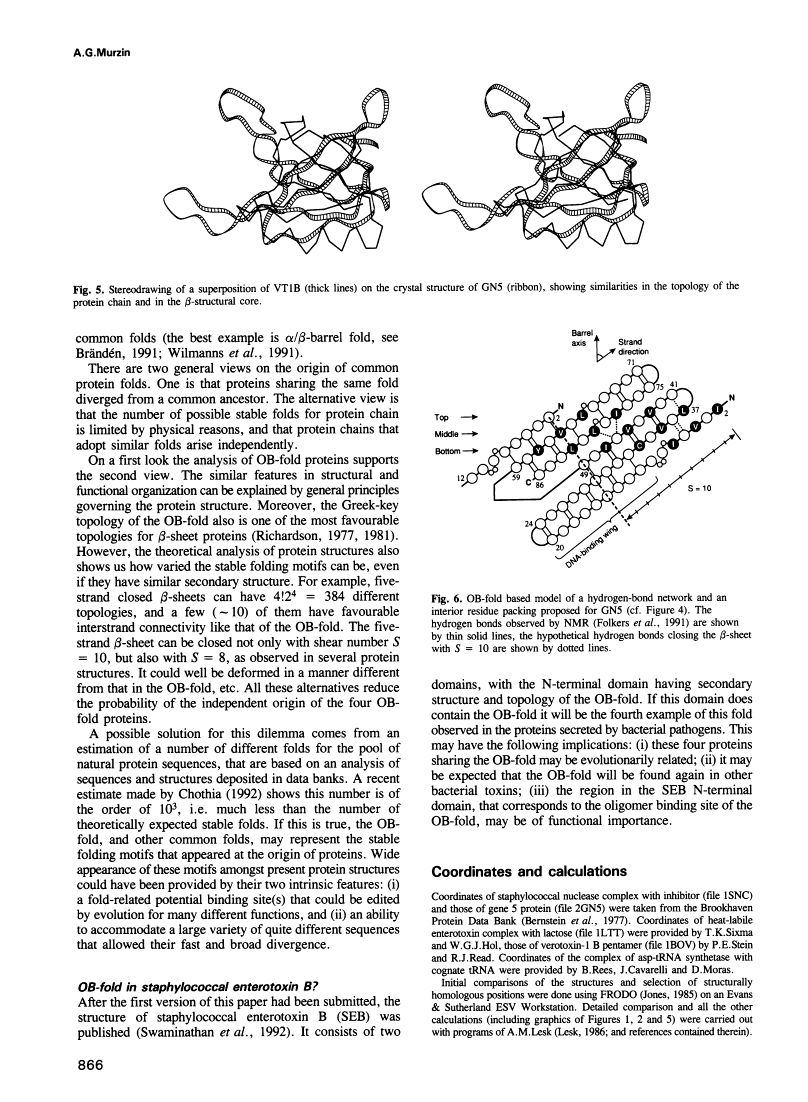
A novel folding motif has been observed in four different proteins which bind oligonucleotides or oligosaccharides: staphylococcal nuclease, anticodon binding domain of asp-tRNA synthetase and B-subunits of heat-labile enterotoxin and verotoxin-1. The common fold of the four proteins, which we call the OB-fold, has a five-stranded beta-sheet coiled to form a closed beta-barrel. This barrel is capped by an alpha-helix located between the third and fourth strands. The barrel-helix frameworks can be superimposed with r.m.s. deviations of 1.4-2.2 A, but no similarities can be observed in the corresponding alignment of the four sequences. The nucleotide or sugar binding sites, known for three of the four proteins, are located in nearly the same position in each protein: on the side surface of the beta-barrel, where three loops come together. Here we describe the determinants of the OB-fold, based on an analysis of all four structures. These proposed determinants explain how very different sequences adopt the OB-fold. They also suggest a reinterpretation of the controversial structure of gene 5 ssDNA binding protein, which exhibits some topological and functional similarities with the OB-fold proteins.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (1.1M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnone A, Bier CJ, Cotton FA, Day VW, Hazen EE, Jr, Richardson DC, Yonath A, Richardson JS. A high resolution structure of an inhibitor complex of the extracellular nuclease of Staphylococcus aureus. I. Experimental procedures and chain tracing. J Biol Chem. 1971 Apr 10;246(7):2302–2316. [Abstract] [Google Scholar]
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. [Abstract] [Google Scholar]
- Brayer GD, McPherson A. Refined structure of the gene 5 DNA binding protein from bacteriophage fd. J Mol Biol. 1983 Sep 15;169(2):565–596. [Abstract] [Google Scholar]
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992 Jun 18;357(6379):543–544. [Abstract] [Google Scholar]
- Chothia C, Janin J. Orthogonal packing of beta-pleated sheets in proteins. Biochemistry. 1982 Aug 17;21(17):3955–3965. [Abstract] [Google Scholar]
- Folkers PJ, van Duynhoven JP, Jonker AJ, Harmsen BJ, Konings RN, Hilbers CW. Sequence-specific 1H-NMR assignment and secondary structure of the Tyr41----His mutant of the single-stranded DNA binding protein, gene V protein, encoded by the filamentous bacteriophage M13. Eur J Biochem. 1991 Dec 5;202(2):349–360. [Abstract] [Google Scholar]
- Hynes TR, Fox RO. The crystal structure of staphylococcal nuclease refined at 1.7 A resolution. Proteins. 1991;10(2):92–105. [Abstract] [Google Scholar]
- Jackson MP, Wadolkowski EA, Weinstein DL, Holmes RK, O'Brien AD. Functional analysis of the Shiga toxin and Shiga-like toxin type II variant binding subunits by using site-directed mutagenesis. J Bacteriol. 1990 Feb;172(2):653–658. [Europe PMC free article] [Abstract] [Google Scholar]
- Hendrickson WA. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. [Abstract] [Google Scholar]
- Lesk AM, Brändén CI, Chothia C. Structural principles of alpha/beta barrel proteins: the packing of the interior of the sheet. Proteins. 1989;5(2):139–148. [Abstract] [Google Scholar]
- Loll PJ, Lattman EE. The crystal structure of the ternary complex of staphylococcal nuclease, Ca2+, and the inhibitor pdTp, refined at 1.65 A. Proteins. 1989;5(3):183–201. [Abstract] [Google Scholar]
- Lüthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992 Mar 5;356(6364):83–85. [Abstract] [Google Scholar]
- McLachlan AD. Gene duplications in the structural evolution of chymotrypsin. J Mol Biol. 1979 Feb 15;128(1):49–79. [Abstract] [Google Scholar]
- Moras D. Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem Sci. 1992 Apr;17(4):159–164. [Abstract] [Google Scholar]
- Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992 Apr;12(4):345–364. [Abstract] [Google Scholar]
- Murzin AG, Lesk AM, Chothia C. beta-Trefoil fold. Patterns of structure and sequence in the Kunitz inhibitors interleukins-1 beta and 1 alpha and fibroblast growth factors. J Mol Biol. 1992 Jan 20;223(2):531–543. [Abstract] [Google Scholar]
- Perera LP, Samuel JE, Holmes RK, O'Brien AD. Identification of three amino acid residues in the B subunit of Shiga toxin and Shiga-like toxin type II that are essential for holotoxin activity. J Bacteriol. 1991 Feb;173(3):1151–1160. [Europe PMC free article] [Abstract] [Google Scholar]
- Richardson JS. beta-Sheet topology and the relatedness of proteins. Nature. 1977 Aug 11;268(5620):495–500. [Abstract] [Google Scholar]
- Richardson JS. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. [Abstract] [Google Scholar]
- Ruff M, Krishnaswamy S, Boeglin M, Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry JC, Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. [Abstract] [Google Scholar]
- Sixma TK, Pronk SE, Kalk KH, Wartna ES, van Zanten BA, Witholt B, Hol WG. Crystal structure of a cholera toxin-related heat-labile enterotoxin from E. coli. Nature. 1991 May 30;351(6325):371–377. [Abstract] [Google Scholar]
- Sixma TK, Pronk SE, Kalk KH, van Zanten BA, Berghuis AM, Hol WG. Lactose binding to heat-labile enterotoxin revealed by X-ray crystallography. Nature. 1992 Feb 6;355(6360):561–564. [Abstract] [Google Scholar]
- Stein PE, Boodhoo A, Tyrrell GJ, Brunton JL, Read RJ. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature. 1992 Feb 20;355(6362):748–750. [Abstract] [Google Scholar]
- Swaminathan S, Furey W, Pletcher J, Sax M. Crystal structure of staphylococcal enterotoxin B, a superantigen. Nature. 1992 Oct 29;359(6398):801–806. [Abstract] [Google Scholar]
- Tyrrell GJ, Ramotar K, Toye B, Boyd B, Lingwood CA, Brunton JL. Alteration of the carbohydrate binding specificity of verotoxins from Gal alpha 1-4Gal to GalNAc beta 1-3Gal alpha 1-4Gal and vice versa by site-directed mutagenesis of the binding subunit. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):524–528. [Europe PMC free article] [Abstract] [Google Scholar]
- Weber DJ, Gittis AG, Mullen GP, Abeygunawardana C, Lattman EE, Mildvan AS. NMR docking of a substrate into the X-ray structure of staphylococcal nuclease. Proteins. 1992 Aug;13(4):275–287. [Abstract] [Google Scholar]
- Wilmanns M, Hyde CC, Davies DR, Kirschner K, Jansonius JN. Structural conservation in parallel beta/alpha-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry. 1991 Sep 24;30(38):9161–9169. [Abstract] [Google Scholar]
Associated Data
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1002/j.1460-2075.1993.tb05726.x
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/j.1460-2075.1993.tb05726.x
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Protein Fold Usages in Ribosomes: Another Glance to the Past.
Int J Mol Sci, 25(16):8806, 13 Aug 2024
Cited by: 0 articles | PMID: 39201491 | PMCID: PMC11354259
Structural basis of tRNA recognition by the widespread OB fold.
Nat Commun, 15(1):6385, 29 Jul 2024
Cited by: 0 articles | PMID: 39075051 | PMCID: PMC11286949
Challenges and Opportunities Arising from Host-Botrytis cinerea Interactions to Outline Novel and Sustainable Control Strategies: The Key Role of RNA Interference.
Int J Mol Sci, 25(12):6798, 20 Jun 2024
Cited by: 1 article | PMID: 38928507
Review
Inhibition of SARS-CoV-2 Nsp9 ssDNA-Binding Activity and Cytotoxic Effects on H838, H1975, and A549 Human Non-Small Cell Lung Cancer Cells: Exploring the Potential of Nepenthes miranda Leaf Extract for Pulmonary Disease Treatment.
Int J Mol Sci, 25(11):6120, 01 Jun 2024
Cited by: 0 articles | PMID: 38892307 | PMCID: PMC11173125
Mono-ADP-ribosylation, a MARylationmultifaced modification of protein, DNA and RNA: characterizations, functions and mechanisms.
Cell Death Discov, 10(1):226, 11 May 2024
Cited by: 0 articles | PMID: 38734665 | PMCID: PMC11088682
Review Free full text in Europe PMC
Go to all (587) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
OB-fold: growing bigger with functional consistency.
Curr Protein Pept Sci, 4(3):195-206, 01 Jun 2003
Cited by: 33 articles | PMID: 12769718
Review
NMR structure of a stable "OB-fold" sub-domain isolated from staphylococcal nuclease.
J Mol Biol, 250(2):134-143, 01 Jul 1995
Cited by: 34 articles | PMID: 7608966
Crystal structure of a new heat-labile enterotoxin, LT-IIb.
Structure, 4(6):665-678, 01 Jun 1996
Cited by: 50 articles | PMID: 8805549