Abstract
Free full text

Tracking stem cell differentiation in the setting of automated optogenetic stimulation
Associated Data
Abstract
Membrane depolarization has been shown to play an important role in the neural differentiation of stem cells as well as in the survival and function of mature neurons. Here we introduce a microbial opsin into embryonic stem cells and develop optogenetic technology for stem cell engineering applications, with an automated system for noninvasive modulation of embryonic stem cell differentiation employing fast optogenetic control of ion flux. Mouse embryonic stem cells (ESCs) were stably transduced with ChR2-YFP and purified by FACS. Illumination of resulting ChR2-ESCs with pulses of blue light triggered inward currents. These labeled ESCs retained the capability to differentiate into functional mature neurons, assessed by the presence of voltage-gated sodium currents, action potentials, fast excitatory synaptic transmission, and expression of mature neuronal proteins and neuronal morphology. We designed and tested an apparatus for optically stimulating ChR2-ESCs during chronic neuronal differentiation, with high-speed optical switching on a custom robotic stage with environmental chamber for automated stimulation and imaging over days, with tracking for increased expression of neural and neuronal markers. These data point to potential uses of ChR2 technology for chronic and temporally precise noninvasive optical control of embryonic stem cells both in vitro and in vivo, ranging from noninvasive control of stem cell differentiation to causal assessment of the specific contribution of transplanted cells to tissue and network function.
Introduction
Embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) potentially could be employed in the study and treatment of central nervous system (CNS) diseases such as Parkinson's disease (1-6), stroke (7-9) and spinal cord injury (10-12). Indeed, ESCs can give rise to functional neurons capable of integrating into the host after intracerebral transplantation (13-16), suggesting that stem cell therapy has the potential to become a therapeutic approach in brain disease if suitably differentiated cells can be 1) generated (17-23), 2) integrated into native circuitry (5, 24), and 3) controlled (25-28). All three of these key processes are intrinsically activity-dependent (29-32), and specific control of electrical activity in differentiating neural cells and their progeny is therefore a central goal in CNS regenerative medicine. Channelrhodopsin-2 (ChR2) is a rapidly gated blue light-sensitive cation channel suitable for noninvasive control of ion flux (33-41). Here we explore the potential of this optogenetic (42) strategy for chronic and temporally precise noninvasive optical control of electrical activity in ESCs and their progeny. For this approach it was necessary to develop a set of automated high-speed optical stimulation tools (hardware, software, and cell lines) to noninvasively and specifically modulate ESCs and their progeny with light.
Material and Methods
Mouse embryonic stem cell culturing
Mouse embryonic stem cells (CRL-1934, ATCC, Manassas, USA) were grown in DMEM medium (ATCC) containing medium conditioned by feeder cells (CRL-1503, ATCC), 15 % fetal calf serum (Gibco), 15 ng/mL leukemia inhibitory factor (LIF; Sigma-Aldrich), 0.1 mM 2-mercaptoethanol (Sigma-Aldrich), and 1 % penicillin–streptomycin (Sigma-Aldrich). The cells were cultured in 75 cm2 cell culture flasks (Falcon) with 20 mL medium at 37°C and 5 % CO2 and passaged every 3 days. Only undifferentiated cells which could be easily suspended upon brief trypsination were used for the experiments. After washing in phosphate-buffered saline (PBS; Gibco, Invitrogen), cells were counted in a Neubauer counting chamber. The viability was determined by staining with trypan blue solution (0.4 %; Sigma-Aldrich).
Transduction of ESCs with ChR2
Lentiviruses carrying the ChR2-EYFP fusion gene under the control of the EF1α promoter were generated as previously described (35). Viruses were concentrated via ultracentrifugation and redissolved in PBS at 1/1000 of the original volume. The concentrated viruses were then incubated with ESCs for 24 h and transduction efficiency evaluated using fluorescent microscopy one week after transduction. To obtain a highly and homogenously expressing ChR2-ESC colony, cells were sorted using FACS; a subpopulation consisting of the top 5 % of YFP-expressing cells was collected. A subpopulation of ChR2-ESCs was reinfected with lentiviruses upon neuronal differentiation, to test whether subsequent transduction affects viability and gene expression. Transduction of ESC-derived neurons might be of importance in future studies requiring further increase of ChR2-expression or in cell populations which cannot be sorted.
Neuronal differentiation of embryonic stem cells
Neuronal differentiation was performed as previously described (17, 22). For optical stimulation, ESCs were plated on matrigel-coated dishes in embryoid body stage in complete ESC medium (see above). After 24 hours, medium was changed to ESC medium lacking LIF and including 0 - 5 μM retinoic acid, and changed every second day for 5 days.
Immunohistochemical staining of cultured cells
Cells were fixed with 4% paraformaldehyde in PBS for 30min at room temperature. Fixation was stopped by washing cells three times with 0.1 M glycine/PBS. Cells were permeabilized and blocked (4 % BSA/0.4 % saponin/PBS) for 30min and incubated in primary antibody solution at 4°C overnight. Cells were washed 4 times and incubated with secondary antibody at room temperature for 2 h. Cells were washed 3x with PBS, and at the final washing step DAPI was added (1:50,000). Coverslips were mounted using anti-quenching Fluoromount. Primary antibodies were mouse anti-SSEA1 (Chemicon 1:300), mouse anti-nestin (Chemicon 1:200), chicken anti-βIII tubulin (Chemicon 1:200), mouse anti MAP2ab (Sigma 1:500), rabbit anti vGlut 2 (Chemicon 1:200), and rabbit anti-α 1C, -α 1D, -α 1G, and –α 1H (all Alomone labs; 1:200). Cy3 or Cy5 conjugated donkey anti mouse, chicken and rabbit secondary antibodies (Jackson) were all used at 1:200.
RT-PCR
Cells were homogenized by Homogenizer (Invitrogen). RNA isolation was performed using Micro-to-Midi Total RNA Purification System (Invitrogen). Prior to RT-PCR, RNA samples were pretreated with DNaseI (Invitrogen) and reverse transcription conducted per manufacturer's protocol. Negative controls without reverse transcriptase did not result in amplified sequences. Mouse hippocampal total RNA was purchased from Clontech and the resulting cDNA served as a positive control. For PCR analysis, primers targeted to coding regions of two subunits each from both the L- and T-type VGCC families were used, as follows (see Table 1 for primer sequences): L-type α1C; L-type α1D; T-type α1G; T-type α1H; Housekeeping gene (Actin). PCR products of actin and L-type and T-type subunits were cloned and sequenced to confirm identity. For analysis of stemness and neural markers, total RNA was extracted from cells using Mini RNeasy kit (Qiagen). Primers targeted to the coding regions of following markers were used (see Table 1 for primer sequences): Pluripotency marker Oct4; Sox2; Nestin; NCAM1; MAP; housekeeping gene GAPDH.
Table 1
| gene | primer forward | primer reverse |
|---|---|---|
| L-type α1C | GTGGTTAGCGTGTCCCTCAT | GTGGAGACGGTGAAGAGAGC |
| L-type α1D | AATGGCACGGAATGTAGGAG | GACGAAAAATGAGCCAAGGA |
| T-type α1G | CTGAGCGGATCTTCCTAACG | TGAAAAAGGCACAGCAGATG |
| T-type α1H | TGGGAACGCTTCTTCTCT | TGGGCATCCATGACGTAGTA |
| Oct4 | CTCCTGAAGCAGAAGAGGATCAC | CTTCTGGCGCCGGTTACAGAACCA |
| Sox2 | TGCAGTACAACTCCATGACCA | GTGCTGGGACATGTGAAGTCG |
| Nestin | CAGCGTTGGAACAGAGGTTG | GCTGGCACAGGTGTCTCAAG |
| NCAM1 | TATCCCAGTGCCACGATCTC | TGGCTTCCTTGGCATCATAC |
| MAP | GGTGGCAAGGTGCAGATAAT | CTTTGGCATTCTCCCTGAAG |
| Actin | GGCATTGTGATGGACTCCGG | TGCCACAGGATTCCATACCC |
| GAPDH | CCATCACCATCTTCCAGGAG | GTGGTTCACACCCATCACAA |
Long-term optical stimulation of ESCs
Key components of the hardware interface include (a) Oasis4i Controller (Objective Imaging) (hardware for x-y-z 3-axis and focus control) (http://www.objectiveimaging.com/Download/OI_Download.htm - software development kit (SDK) for the Oasis4i Controller). (b) DG4 Ultra High Speed wavelength switcher (Sutter), (c) Retiga SRV Camera (Qimaging), and (d) Leica DM6000 Microscope controlled by AHM (Abstract Hardware Model) controller. The parallel port is controlled using DLPORTIO library file (www.driverlinx.com/DownLoad/DlPortIO.htm - Dlls to for parallel port control) and camera parameters (gain, exposure) set using QCam SDK (Ver. 5.1.1.14) (http://www.qimaging.com/support/downloads/ - SDK to control the Retiga SRV/ Exi Cameras). The custom software user interface to the optogenetic stimulation setup was developed using the Microsoft Foundation Library (MFC; Ver. 8.0) and is available on request. Briefly, regions of interest (for example, an embryoid body or a small well in a multiwell plate) to be stimulated and/or imaged are selected using the Oasis4i Controller, and their locations saved using the MFC interface. Stimulation parameters (excitation filter wavelength, the duration of the excitatory pulse, and the frequency and duty cycle of excitation) are then set in the custom GUI. To allow stimulation space to be mapped, each region of interest can be readily programmed to receive a different stimulus pattern to operate over the many days of stimulation and imaging. Similarly, imaging parameters can also be varied for selected regions, including number of images per region and exposure, gain, excitation and emission filters.
Undifferentiated cells were seeded on matrigel (BD) coated coverslips in 24-well plates in complete ESC medium at a density of 100,000 cells/well. Both native ESCs and ChR2-expressing ESCs were used in different wells on the same plate. 24 hours after seeding, medium was changed to the various experimental conditions including complete ESC medium, ESC medium lacking both LIF and conditioned media from feeder cells (differentiation medium), differentiation medium with 1μM retinoic acid (RA) (Sigma), and differentiation medium with 2.5 μM RA. Optical stimulation was conducted using the described custom hardware and software (Fig. 4). Up to 30 regions of interest (ROIs) were defined per well, ensuring that all cell-containing regions on the coverslip were stimulated. ROIs were illuminated every hour around the clock over 5 days with blue light (470 nm) pulsing at 15 Hz for 10 s, using a 10x objective (NA 0.3). Every 8 hours, a photomicrograph was programmed to be taken of the selected ROIs. At the end of the experiment, coverslips were removed from the plates and immediately fixed with paraformaldehyde in PBS and stained as described above. Mounted slides were labeled with coded numbers by a colleague so that the investigators conducting confocal analysis were blind to treatment condition.
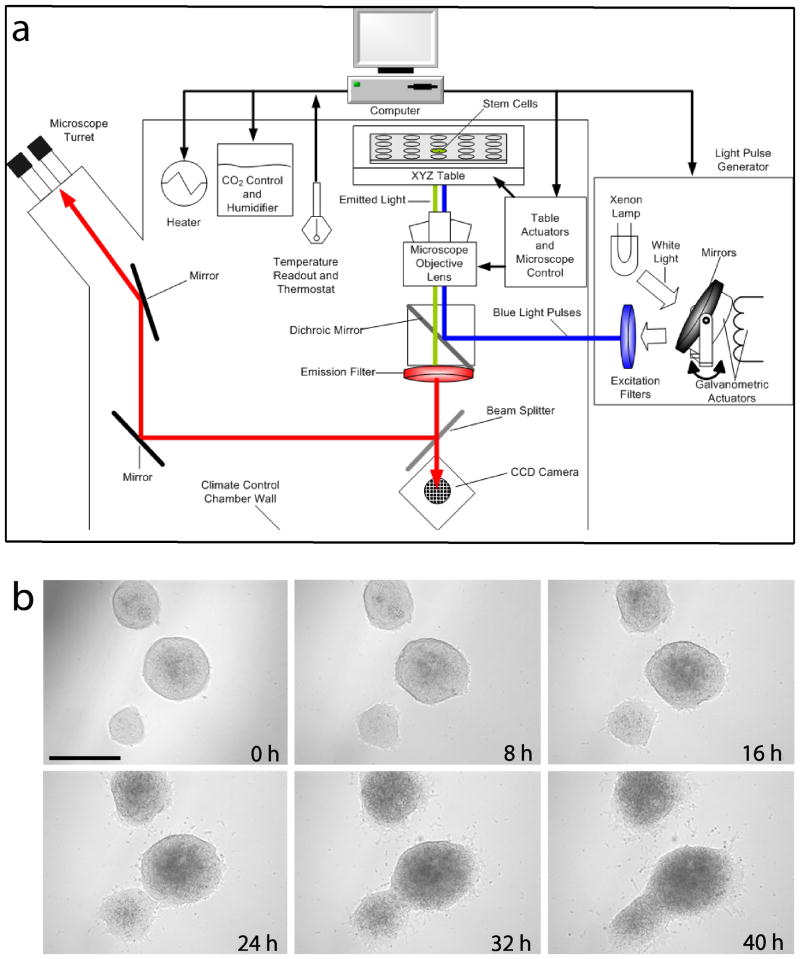
(a) Schematic of automated optical stimulation setup. The multiwell plate containing cells is placed on the stage of a fluorescence microscope rig with robotic stage, millisecond-scale optical switching, autofocus, and environmental control of temperature and CO2. Custom software controls all functions of the stage and microscope, as well as the CCD camera and the light source (methods). (b) Photomicrographs obtained by automated setup of identical ROI every 8 hours. Time course of differentiation and survival of mESCs upon incubation with 2.5 μM RA can be monitored. Scale bar: 300 μm.
Confocal microscopy and image analysis
Confocal imaging was conducted using the Leica SP2 confocal microscope with a 40x oil objective (NA 0.75) and an Olympus FV1000 for analysis of neuronal differentiation, with a 60x oil objective (NA 1.42). For DAPI excitation, a 402 nm diode laser was used; Cy5-nestin was excited using a 633 nm HeNe laser. 6 ROIs were randomly and blindly selected for analysis per coverslip, and 1024×1024 8-bit confocal images were obtained. For each ROI, a z-stack with 8–12 x-y-sections and a z step size of 0.98 μm were collected, thereby including all cells present in the ROI. Data analysis was conducted using ImageJ (NIH, USA) software, and after unblinding, confocal images of all ROIs of all coverslips of each condition (e.g. ChR2-ESCs, optically stimulated, 2.5 μM RA) were converted into a single z-stack. Fluorescence intensity histograms were calculated (43) for DAPI and nestin channels. DAPI histograms reflecting the cell numbers allowed for a normalization of nestin histograms. All nestin voxel numbers have been divided by this DAPI factor. For analysis of neuronal differentiation, Cy3-β-3-tubulin was excited using a 559 nm laser. Again, 6 ROIs were randomly and blindly selected for analysis per coverslip, and 1024×1024 8-bit confocal images were obtained. β-3-tubulin expressing cells constituted a monolayer, individual cells could be easily discriminated. Therefore, a z-stack with 3 x-y-sections and a z-step size of 1.4 μm was collected in each ROI, covering all β-3-tubulin expressing cells. The Cy3-channel was merged with the DAPI channel using ImageJ software. β-3-positive cells were counted manually for all ROIs. After unblinding, cell numbers of all ROIs of one condition were summed up and normalized to the condition native ESC, 2.5 μM RA. Statistical analysis was conducted using SPSS (Chicago, USA) software. To statistically compare histograms, the parameter-free Kolmogorov-Smirnov test (44) was employed, and to compare means, statistical significance was calculated using the t-test.
Stereotactic cell transplantation
Rats (male Wistars, 250-350 g) were the subjects of these experiments. Animal husbandry and all aspects of experimental manipulation of our animals were in strict accord with guidelines from the National Institute of Health and approved by members of the Stanford Institutional Animal Care and Use Committee. Rats were anaesthetized by i.p. injection (90 mg ketamine and 5 mg xylazine per kg of rat body weight). For cell transplantation, a 1 mm craniotomy was drilled over motor cortex. 1 μl of ESCs expressing ChR2-EYFP fusion protein at a density of 50 × 103 cells/μl, suspended in PBS was injected (26g Hamilton Syringe) into rat motor cortex (AP +1.5 mm, ML +1.5 mm, DV +1.5 mm). The injection duration was 10 min; an additional 10 min delay followed before syringe withdrawal, and electrophysiology was conducted after 1 week.
Electrophysiology
For acute slice electrophysiological experiments, 1 week post cell transplantation, 250 μm cortical slices were prepared in ice-cold cutting buffer (64 mM NaCl, 25 mM NaHCO3, 10 mM glucose, 120 mM sucrose, 2.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2 and 7 mM MgCl2, equilibrated with 95 % O2/5 % CO2) using a vibratome (VT 1000 S; Leica). After a recovery period of 30 min in cutting buffer at 32-35 °C, slices were gently removed to a recording chamber mounted on an upright microscope (DM LFSA, Leica) and continuously perfused at a rate of 3-5 mL/min with carbonated ACSF (124 mM NaCl, 3 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 2.4 mM CaCl2, 1.3 mM MgCl2, 10 mM Glucose), ventilated with 95% O2/5% CO2.ChR2-YFP-ESCs were identified on an upright fluorescence microscope (DM LFSA, Leica) with a 20X, 0.5 NA water immersion objective and a YFP filter set. Images were recorded with a CCD camera (Retiga Exi, Qimaging) by Qimaging software. Electrophysiological recordings in cultured ChR2-YFP ESCs were performed as previously described (35, 39), in Tyrode solution containing (in mM): NaCl 125, KCl 2, CaCl2 3, MgCl2 1, glucose 30 and HEPES 25 (pH 7.3 with NaOH). Membrane currents were measured with the patch-clamp technique in whole-cell mode (45) using Axon Multiclamp 700B (Axon Instruments) amplifiers. Pipette solution consisted of (in mM): 97 potassium gluconate, 38 KCl, 6 NaCl, 0.35 sodium ATP, 4 magnesium ATP, 0.35 EGTA, 7 phosphocreatine and 20 HEPES (pH 7.25 with KOH). Pipette resistance was 4-8 MΩ. Membrane potential was noted at the time of establishing the whole cell configuration. We employed pClamp 9 acquisition software (Axon Instruments), a DG-4 high-speed optical switch with 300 W xenon lamp (Sutter Instruments) and a GFP filter set (excitation filter HQ470/40x, dichroic Q495LP; Chroma) to deliver blue light for ChR2 activation. Through a 20x objective lens, power density of the blue light was 8−12 mW/mm2, measured by power meter (Newport). All experiments were performed at room temperature (22−24 °C).
Results
Functional expression of ChR2 in embryonic stem cells
To assess the potential of optogenetics in stem cells, mouse ESCs were transduced with a lentiviral ChR2-YFP-construct under the control of the EF1α promoter (46); after sorting for the top 5 % based on YFP fluorescence intensity (Fig. 1a), we found that the population doubling time and vitality of the resulting ChR2-YFP-ESCs did not differ significantly compared to non-transduced ESCs (Suppl. Fig. 1), and confocal microscopy demonstrated membrane localization of ChR2-YFP with high, uniform expression levels in the ESC population (Fig. 1b). ChR2-ESCs continued to express the embryonic stem cell markers Oct4 and Sox2 as measured by RT-PCR (Fig. 1c) and SSEA1 (Suppl. Fig. 1), maintaining the undifferentiated state as did non-transduced control cells. Electrophysiologically, the ChR2-ESCs displayed typical outwardly-rectifying and passive currents, while illumination with blue light (470 nm, 500 ms pulse duration) evoked inward photocurrents (Fig. 1d,e); steady-state photocurrents showed little inactivation while peak photocurrents showed inactivation and recovery with kinetics similar to that previously shown in neurons(35) (Fig. 1f).

(a) FACS analysis of a sample of 105 ChR2-ESCs before sorting. 2D dot plot, sideward scattering (SSC) versus green fluorescence (FITC). A significant variation of YFP-expression in cell population becomes apparent. A gate enclosing 5 % of cells with the strongest fluorescence was applied for sorting. (b) Confocal micrograph of ChR2-ESCs revealing membrane localized expression of ChR2-YFP. Vitality (assessed by trypan blue exclusion), morphology, and population doubling times of ChR2-YFP ESC were indistinguishable from native cells. Scale bar: 50 μm. (c) RT-PCR analysis of pluripotency marker expression of both ChR2-ESCs as well as native ESCs revealed strong expression of Oct4 and Sox2, compared to mouse hippocampal cells HT22 (negative control). The housekeeping gene GAPDH was used as loading control. (d) Pulsed light (473 nm) evoked inward currents in a ChR2–YFP ESC in voltage clamp (light pulse indicated by blue bar). (e) Summary data on evoked photocurrents (mean +/- s.e.m., n=15 cells). (f) Kinetics of de-inactivation of the peak ChR2 current in ESCs. Ten overlaid photocurrent traces obtained in voltage clamp are shown; pairs of 0.5 s light pulses (indicated by blue bars) were separated by increasing intervals from 1 to 10 seconds; traces are aligned to the initiation of the first pulse in each sweep. Note that steady-state current is constant while the peak current shows inactivating behavior. (g-i) Light evoked currents in transplanted ChR2-YFP ESCs in rat cortical brain tissue. (g) YFP fluorescence of ChR2-YFP-ESCs 7 days after transplantation, observed in an acute cortical slice. Scale bar: 500μm. (h) Higher magnification reveals cellular morphology and membrane localization of ChR2-YFP. Scale bar: 80 μm. (i) Electrophysiological recordings in transplanted ChR2-ESC in acute brain slice revealed blue light evoked inward photocurrents. As in culture, the steady-state current is stable and the peak current displays inactivation.
To test whether transplanted ChR2-ESCs could still respond to optical stimulation, 5×105 ChR2-YFP expressing ESCs were stereotaxically injected into the cortex of healthy rats. One week after transplantation, animals were sacrificed and in acute slices, transplanted cells could be identified by YFP fluorescence (Fig. 1g,h). Patch clamp recordings were conducted, revealing inward currents upon illumination with blue light (Fig. 1i) that displayed typical inactivation of the peak current and stability of the steady-state current.
ChR2-ESCs express voltage gated Ca2+-channels
Intracellular Ca2+ is a major mediator of differentiation and survival in stem cells and their progeny, especially in neural lineages (29). ChR2 itself is a nonselective cation channel that directly allows Ca2+ entry into cells (47, 48). Additional routes of photo-evoked Ca2+ entry could include activation of voltage-gated Ca2+ channels (VGCCs) by virtue of ChR2-induced membrane voltage changes. Notably, we find that mouse ES cells express four major VGCCs assessed by RT-PCR and immunoreactivity (Fig. 2a), and this supplementary mechanism for photoactivated Ca2+ entry could become increasingly potent as cells proceed down the neuronal lineage and develop hyperpolarized membrane potentials. Moreover, the known Ca2+ flux of ChR2 itself suggested the potential for optical control of stem cell processes.

(a) In addition to the known Ca2+ permeability of ChR2 itself, RT-PCR analysis revealed multiple voltage-gated Ca2+-channels in ESCs and ChR2-ESCs. The transcript for the L-type voltage-gated Ca2+ channel α1C subunit is present in hippocampal RNA (HPC), ESCs, and ChR2-ESCs, but not in HEK 293 cells (left). This transcriptional pattern was also observed for the L-type voltage-gated Ca2+-channel α1D subunit, and the T-type voltage-gated Ca2+ channel subunits α1G and α1H (right). All cells show strong expression of housekeeping gene β-actin. Right: Immunocytochemical detection of membrane-localized Ca2+-channel subunit expression in ChR2-ESCs (L-type Ca2+-channel subunit α1C and T-type Ca2+-channel subunit α1G). Cellular nuclei were visualized by DAPI staining. Scale bars: 25 μm. (b) Transcriptional analysis of neural lineage markers of differentiated ChR2-ESCs at day 22. ChR2-ESCs were compared to cells re-infected with EF1α-ChR2 lentivirus at neural stage (day 15) and mouse hippocampal HT22 cells. Transcripts of neural markers nestin were present in ChR2-ESCs as well as in re-infected ChR2-ESCs, in contrast to HT 22 cells. Neuronal cell adhesion molecule 1 (NCAM1) transcript could be detected in all three cell populations, microtubule-associated protein 2 is only strongly transcribed in both ESC populations. GAPDH was used as loading control. Right: Immunocytochemistry for stage specific markers. After 20 days cells adapt bipolar morphology, a dense network of β-3-tubulin-expressing cells can be observed indicating progression down the neuronal lineage; scale bar 50 μm. After 30 days, cells express the mature neuronal marker MAP2; scale bar: 100 μm. Re-infected ChR2-ESC derived neurons display strong expression of ChR2, scale bar: 10 μm.
ChR2-ESCs differentiate into functional excitatory neurons
We first verified that ChR2-ESCs were capable of neural lineage differentiation, using a retinoic acid-(RA) based neural differentiation protocol. RT-PCR revealed gene expression of neural as well as neuronal markers, complemented with immunofluorescent stainings for neuronal cytoskeletal proteins β-3 tubulin and MAP2 (Fig. 2b). Reinfection with EF1α-ChR2 lentivirus did not alter gene expression profile. By day 28 the resulting ChR2-ESC-derived neurons displayed strong ChR2-expression (Fig. 2b) and mature neuronal morphology. Generated neurons were expressing the vesicular glutamate transporter II (Fig. 3a). Electrophysiologically, cells displayed sodium currents, action potentials, and excitatory postsynaptic currents which could be blocked by excitatory synaptic transmission glutamate receptor antagonists CNQX and D-AP5 (Fig. 3b-d).
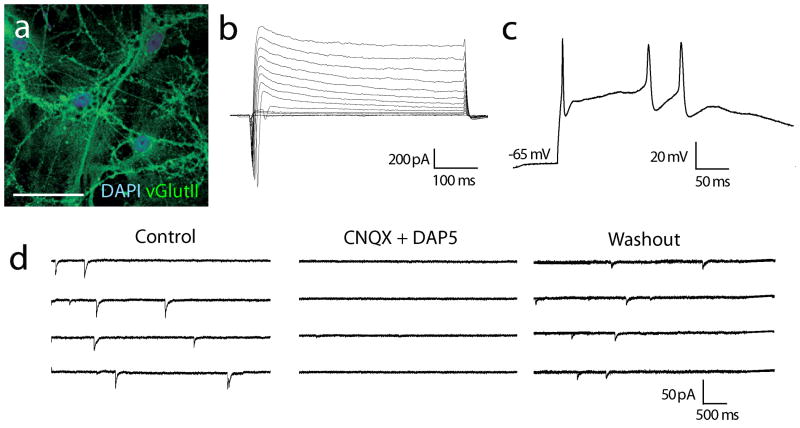
(a) ChR2-ESC-derived neurons after 36 days of differentiation, displaying mature dendritic morphology and vGlutII expression. Scale bar: 30 μm. (b) Active membrane currents were observed in ChR2-ESC-derived neurons. After establishing the whole-cell configuration, voltage steps were applied from a holding potential of -80mV (in 10mV steps up to +20mV); fast voltage-activated inward currents were evoked. (c) Corresponding to the fast active inward currents, action potentials could be recorded in current clamp in response to current injection (150 pA). (d) Spontaneous excitatory synaptic activity was recorded (left panel), abolished with co-application of the excitatory glutamate receptor antagonists CNQX (10 μM) and D-AP5 (25 μM) (middle panel) and restored after drug washout (right panel).
Automated optical stimulation
One challenge in deriving replacement tissues from ES cells is that differentiation takes place over many days, and therefore also the patterning and differentiation stimuli. Hence, to be applicable, optogenetic stimulation must be deliverable in chronic fashion. In designing the system to meet this challenge, it is also important to consider that since knowledge of the precise combinations and timing of signaling events required for stem cell differentiation is limited, a multiwell configuration would in principle be desireable, to allow for fast optical mapping of cell lines, conditions, and “differentiation space” in the laboratory. We therefore devised an automated multiwell optogenetic stimulation approach designed to precisely revisit and optically stimulate multiple regions of interest (ROIs) in defined patterns over extended periods of time (Fig. 4a).
ROIs in multiwell plates were user-defined in a custom GUI and their locations saved for rapid and reproducible access by a robotic stage. Stimulation parameters (excitation filter wavelength, optical switch pulse duration, and frequency/duty cycle of excitation) were set in the custom software, which also controls the microscopic stage in all three spatial dimensions, and controls operation of the DG-4 optical switch which employs spinning galvanometers to deliver light with submillisecond precision. The microscope itself is surrounded by a climate controlled Plexiglas chamber wherein both temperature and CO2-level are tightly regulated and temporally precise imaging can proceed in parallel with optical stimulation (Fig. 4b). Embryonic stem cells can be cultured and photostimulated in this environment rather than in a standard incubator for many weeks, allowing us to investigate the effect of optogenetic stimulation on the differentiation of embryonic stem cells in a controlled, reproducible manner.
Optogenetic modulation of ESC neural and neuronal differentiation
Using a multi-well setup, differentiation space could be efficiently mapped while controlling for nonspecific effects related to the rig or to illumination. Cells were stimulated for 5 days with blue light (470 nm at 15 Hz for 10s) delivered every 60 min using a 10x objective. The survival and morphology of the cells was monitored using time-lapse imaging every 8 hours. Cells incubated without RA and 2.5 μM RA showed no decrease in cellular viability, however incubation with 5 μM RA resulted in significant reduction of cell viability of both ChR2-ESCs as well as native ESCs. Consequently, cells incubated with 5 μM RA were not used for subsequent analysis.
To identify rapidly-acting effects of optical stimulation on ESC differentiation, cells were simultaneously assayed following the conclusion of stimulation. Immunostaining for the neural marker nestin as well as for the neuronal marker β-3-tubulin followed by analysis of fluorescence histograms and direct quanitification of neuronal cells obtained by confocal microscopy was used to quantify neural and neuronal lineage differentiation, along with imaging of cellular nuclei using DAPI. Figure 5 a,b shows a 3D projection of two typical confocal z-stacks of single ROIs, displaying both DAPI (blue) and nestin (red) fluorescence. Optically stimulated cells consistently showed higher nestin immunoreactivity (Fig. 5b) compared to non-stimulated cells (Fig. 5a), while optical stimulation interestingly was ineffective in the absence of retinoic acid (RA) (Fig. 5c). To quantify this effect, we generated fluorescence intensity histograms of all ROIs across all wells in each condition (resulting in more than 150 confocal images per condition). These intensity histograms revealed considerable differences between stimulated and non-stimulated ChR2-ESCs (Fig. 5f; p<0.01, Kolmogorov-Smirnov test (44)). We next conducted an experiment to test the possibility that the nestin distributions of unmodified (“native”) optically stimulated ESCs (Fig. 5d) and ChR2-YFP optically stimulated ESCs (Fig. 5f) could represent samples from the same distribution; after automated optical stimulation, repeated as in the above experiment and subsequent blinded analysis, we found that this hypothesis could be rejected (p<0.001; two-tailed K-S Z=5.43); Fig. 5f,g shows the observed increase in high levels of optically-induced nestin expression in the ChR2-YFP cells. We calculated the mean nestin fluorescence intensity in each condition, and comparing optically stimulated with non-optically stimulated cells across all conditions revealed that only ChR2-YFP ESCs incubated with 2.5 μM RA showed a significant optogenetically-induced increase in mean nestin expression (p<0.01, two-tailed t-test; Fig. 5g). In the presence of 1 μM RA, a nonsignificant trend toward higher nestin expression in the setting of optical stimulation was observed, while in 0 μM RA no effect of optical stimulation was observed (e.g. Fig. 5c). To exclude the hypothesis, that optogenetic stimulation only modulates neural, but not neuronal differentiation, we conducted an independent quantification of the early neuronal marker β-3-tubulin. Blinded analysis of confocal images upon staining with β-3-tubulin allowed for a direct quantification of β-3-tubulin positive cells (Fig. 5 h,i). Neuronal, β-3-tubulin expressing cells could be easily distinguished from non-neuronal cells. For normalization, the number of β-3-tubulin positive cells of the experimental condition native ESCs, no stimulation, 2.5 μM RA was defined as 100%. Across all stimulation conditions, no or only single β-3-tubulin expressing cells could be detected without the presence of RA. Incubation of native ESCs with 1 μM RA lead to about 30% β-3-tubulin positive cells, with or without optical stimulation. A non-significant increase of neuronal differentiation became apparent when comparing optically stimulated and non-stimulated ChR2-ESCs. At 2.5 μM RA, again, no increase in neuronal cells in response to optical stimulation could be observed (Fig. 5j) in native ESCs. In contrast, optically stimulating ChR2 ESCs in presence of 2.5 μm RA led to a significant increase of neuronal cells (Fig. 5k).
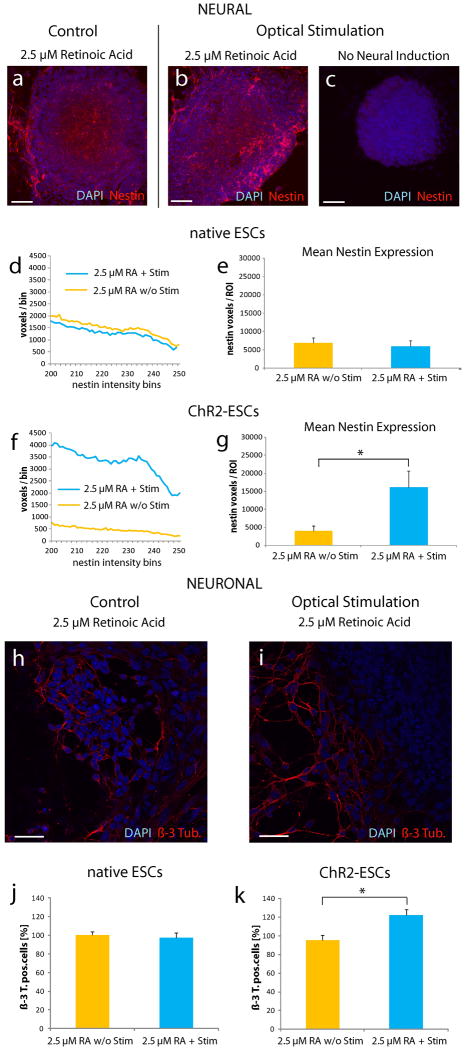
(a-c) 3D image stacks of ChR2-ESCs after 5 days with (b, c) or without (a) optical stimulation. (a, b) Cells were incubated with 2.5 μM retinoic acid; as a negative control, in (c) no neural lineage inducing agents were added. Cells were stained for cellular nuclei (DAPI; blue fluorescence) and the neural marker nestin (red fluorescence). 3D reconstruction consists of 10 x-y sections, z-step size is 0.98 μm. Scale bars: 50 μm. (d - g) Summary of blinded experiments comparing response to optical stimulation of native ESCs (d, e) and ChR2-ESCs (f, g). (d, f) Histograms of nestin fluorescence intensities of optically-stimulated versus non-stimulated cells. All individual x-y sections of all embryoid body-containing ROIs in each condition were used to calculate the intensity histograms (8-bit images gave rise to 256 intensity bins). (d) Optical stimulation of native ESCs did not significantly affect nestin expression. (f) Increased nestin expression in the optically-driven ES cells; histograms were normalized revealing an optically-driven increased fraction of nestin-positive voxels, as evaluated by non-parametric statistical testing. DAPI histograms reflecting the number of cells did not reveal any significant changes upon light stimulation, no proliferative effect could be observed. (e, g) Summary data of mean nestin-positive voxels per embryoid body-containing ROI (+/- s.e.m., n = 10 ROIs, two independent experiments). (e) Optical stimulation of native ESCs did not significantly affect nestin expression. (g) Optically-stimulated ChR2-ESCs gave rise to significantly increased nestin expression in the presence of 2.5 μM RA (two tailed t-test, p=0.03). (h-i) Confocal images of ChR2-ESCs after 5 days with (i) or without (h) optical stimulation. Cells were incubated with 2.5 μM retinoic acid. Cells were stained for cellular nuclei (DAPI; blue fluorescence) and the neuronal marker β-3-tubulin (red fluorescence). Scale bars: 30 μm. (j-k) Normalized percentage of β-3-tubulin expressing cells incubated with 2.5 μM RA, two independent experiments per condition, mean +/- s.e.m. (j) No significant change in cell numbers upon optically stimulated native ESCs can be detected. (k) In contrast, stimulation of ChR2-ESCs results in a significant increase (P=0.01, two tailed t-test) of the number of β-3-tubulin expressing cells of about 25%.
Discussion
Depolarization has been reported in other studies to modulate neural differentiation processes in dividing cells (49-52), and depolarization and calcium waves have both been observed in proliferating CNS progenitors in situ (30, 53). The specific signal transduction cascades mediating the influence of membrane depolarization events in early development remains unclear, but Ca2+ and Ca2+ channels may play a key role and ChR2 is well suited to recruit these mechanisms; indeed, emerging evidence points to the expression of VGCCs during early stages of embryonic development (54, 55), which may allow ChR2 to recruit Ca2+ -dependent cellular processes not only via its own light-activated Ca2+ flux but also by activating native VGCCs as differentiating cells mature. Krtolica et al. (56) reported enhancement of hematoendothelial differentiation upon chronic depolarization of human ESCs. In all of these cases, as we observed with the RA gating of optogenetic modulation, depolarization or Ca2+ influx will likely depend on other patterning and lineage-specific differentiation factors, and clearly more studies are needed to investigate the precise temporal patterns and combinatorial interactions of Ca2+ with other signalling messengers.
Studies have shown the induction of pluripotent stem cells (iPSCs) from somatic cells (57-61), significantly expanding the possible sources of stem cells in regenerative medicine but further highlighting the ongoing need for selective and highly sensitive stem cell differentiation and control tools. Globally applied stimuli such as growth factors and organic compounds will affect all cells present, including non-dividing constituents of the stem cell niche as well as the stem cells and their progeny, but it is unlikely that these growth factors will have the same desired effect in all of the very different cells present in the typical differentiation milieu. By targeting optical control to either the proliferating cells or to niche constituents like astrocytes, optogenetic control of intracellular signaling will allow selective control of the desired cell type (41). Future studies may further capitalize on the genetic targeting of optical control to selectively drive differentiating cells within complex multiple cell-type environments.
Indeed, this optical specificity principle extends to the selective control of fully differentiated stem cell progeny in situ. Minimally invasive fiberoptic strategies (33, 34, 37) have brought optogenetics to the fully intact, behaving mammal. Transplanted cells may require electrical activity to drive the final stages of phenotype consolidation and to fully integrate into host neural circuitry, representing the central goal of stem cell based regeneration medicine. Yet the lack of precise tools to drive only the transplanted cells has hindered progress on this front. Compared to conventional electric stimulation or drugs, the genetic targeting of ChR2 makes it possible to specifically and reversibly drive precise amounts of activity in the transplanted ESCs and their progeny, which moreover do not require addition of chemical cofactors in vivo for ChR2 function. Finally, optically driving only the transplanted cells, with behavioral readouts or noninvasive imaging readout modalities like fMRI (and without the serious problem of signal interference from metal electrodes), opens the door to imaging and tuning the specific contribution of transplanted cells in the restoration of network activity and circuit dynamics, for example in Parkinson's disease (62). With these approaches and others, optogenetic technologies have the potential to become valuable tools in stem cell biology and regenerative medicine.
Supplementary Material
Supp Fig S1
Supplementary Figure 1: Characterization of ChR2-ESCs. (a) Expression of pluripotency marker SSEA-1 in ChR2-ESCs. Scale bar: 50 μm (b) Vitality of native ESCs and ChR2-ESCs determined by trypan-blue exclusion, mean +/- s.e.m of four independent experiments. Vitality of both populations ranges above 95%, no significant differences in vitally can be observed. (c) FACS analysis of ChR2-ESC population. 2-D dot plot of 105 ChR2 ESCs, forward and sideward scattering.
Acknowledgments
This work was supported by CIRM, NSF, NIDA, NIMH, and the NIH Director's Pioneer Award, as well as by the Snyder, Kinetics, Culpeper, Coulter, Klingenstein, Whitehall, McKnight, and Albert Yu and Mary Bechmann Foundations (KD), CIRM (LPW), the Bavarian State Ministry of Sciences, Research and the Arts (“ForNeuroCell”) (AS and JK) and the Stanford Graduate Fellowship (HCT). The authors wish to thank B. Cord and K. Schrenk-Siemens for assistance with ESC culture, K. Lee for assistance with confocal imaging, W. Beisker for assistance with FACS analysis and C. Zimmer for support.
Footnotes
Albrecht Stroh: Collection of Data, Conception and Design, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Hsing-Chen Tsai: Collection of Data, Data analysis and interpretation, Manuscript writingLi Ping Wang: Collection of Data, Conception and Design, Data analysis and interpretation
Feng Zhang: Collection of Data
Jenny Kressel: Collection of Data
Alexander Aravanis: Collection of Data, Data analysis and interpretation
Nandhini Santhanam: Collection of Data
Karl Deisseroth: Conception and Design, Data analysis and interpretation, Manuscript writing
Arthur Konnerth: Conception and Design
M. Bret Schneider: Conception and Design, Data analysis and interpretation, Manuscript writing
.Competing Interests Statement: The authors declare that they have no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1002/stem.558
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4182948?pdf=render
Free after 12 months at intl-stemcells.alphamedpress.org
http://intl-stemcells.alphamedpress.org/cgi/reprint/29/1/78.pdf
Free after 12 months at intl-stemcells.alphamedpress.org
http://intl-stemcells.alphamedpress.org/cgi/content/full/29/1/78
Free to read at intl-stemcells.alphamedpress.org
http://intl-stemcells.alphamedpress.org/cgi/content/abstract/29/1/78
Citations & impact
Impact metrics
Citations of article over time
Article citations
Optical mapping and optogenetics in cardiac electrophysiology research and therapy: a state-of-the-art review.
Europace, 26(2):euae017, 01 Feb 2024
Cited by: 4 articles | PMID: 38227822 | PMCID: PMC10847904
Review Free full text in Europe PMC
Harnessing the Stem Cell Niche in Regenerative Medicine: Innovative Avenue to Combat Neurodegenerative Diseases.
Int J Mol Sci, 25(2):993, 12 Jan 2024
Cited by: 3 articles | PMID: 38256066 | PMCID: PMC10816024
Review Free full text in Europe PMC
On-Demand Chemomagnetic Modulation of Striatal Neurons Facilitated by Hybrid Magnetic Nanoparticles.
Adv Funct Mater, 32(35):2204732, 26 Jun 2022
Cited by: 3 articles | PMID: 36339020 | PMCID: PMC9635318
Tracking connectivity maps in human stem cell-derived neuronal networks by holographic optogenetics.
Life Sci Alliance, 5(7):e202101268, 13 Apr 2022
Cited by: 4 articles | PMID: 35418473 | PMCID: PMC9008225
Current Topics of Optogenetics for Medical Applications Toward Therapy.
Adv Exp Med Biol, 1293:513-521, 01 Jan 2021
Cited by: 2 articles | PMID: 33398838
Go to all (56) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Optogenetic control of iPS cell-derived neurons in 2D and 3D culture systems using channelrhodopsin-2 expression driven by the synapsin-1 and calcium-calmodulin kinase II promoters.
J Tissue Eng Regen Med, 13(3):369-384, 30 Jan 2019
Cited by: 14 articles | PMID: 30550638 | PMCID: PMC6492196
Optogenetics reveal delayed afferent synaptogenesis on grafted human-induced pluripotent stem cell-derived neural progenitors.
Stem Cells, 32(12):3088-3098, 01 Dec 2014
Cited by: 36 articles | PMID: 25183299
Functional control of transplantable human ESC-derived neurons via optogenetic targeting.
Stem Cells, 28(11):2008-2016, 01 Nov 2010
Cited by: 59 articles | PMID: 20827747 | PMCID: PMC2988875
Neural tissue engineering using embryonic and induced pluripotent stem cells.
Stem Cell Res Ther, 2(2):17, 15 Apr 2011
Cited by: 27 articles | PMID: 21539726 | PMCID: PMC3226288
Review Free full text in Europe PMC




