Abstract
Summary
Clustered regularly interspaced short palindromic repeats (CRISPR)-based technologies have revolutionized human genome engineering and opened countless possibilities to basic science, synthetic biology and gene therapy. Albeit the enormous potential of these tools, their performance is far from perfect. It is essential to perform a posterior careful analysis of the gene editing experiment. However, there are no computational tools for genome editing assessment yet, and current experimental tools lack sensitivity and flexibility. We present a platform to assess the quality of a genome editing experiment only with three mouse clicks. The method evaluates next-generation data to quantify and characterize insertions, deletions and homologous recombination. CRISPR Genome Analyzer provides a report for the locus selected, which includes a quantification of the edited site and the analysis of the different alterations detected. The platform maps the reads, estimates and locates insertions and deletions, computes the allele replacement efficiency and provides a report integrating all the information.Availability and implementation
CRISPR-GA Web is available at http://crispr-ga.net. Documentation on CRISPR-GA instructions can be found at http://crispr-ga.net/documentation.htmlContact
[email protected].Free full text

Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA)
Abstract
Summary: Clustered regularly interspaced short palindromic repeats (CRISPR)-based technologies have revolutionized human genome engineering and opened countless possibilities to basic science, synthetic biology and gene therapy. Albeit the enormous potential of these tools, their performance is far from perfect. It is essential to perform a posterior careful analysis of the gene editing experiment. However, there are no computational tools for genome editing assessment yet, and current experimental tools lack sensitivity and flexibility.
We present a platform to assess the quality of a genome editing experiment only with three mouse clicks. The method evaluates next-generation data to quantify and characterize insertions, deletions and homologous recombination. CRISPR Genome Analyzer provides a report for the locus selected, which includes a quantification of the edited site and the analysis of the different alterations detected. The platform maps the reads, estimates and locates insertions and deletions, computes the allele replacement efficiency and provides a report integrating all the information.
Availability and implementation: CRISPR-GA Web is available at http://crispr-ga.net. Documentation on CRISPR-GA instructions can be found at http://crispr-ga.net/documentation.html
Contact: ude.dravrah.dem.sciteneg@lleugm
1 INTRODUCTION
The journal Science selected CRISPR (clustered regularly interspaced short palindromic repeats) as one of the top 10 breakthroughs of 2013 (Clear, 2013). CRISPR-based technologies (Cho et al., 2013; Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013) have revolutionized human genome engineering and opened unlimited possibilities to basic science, synthetic biology and gene therapy. Its design simplicity and efficiency makes it an extremely versatile tool (Jinek et al., 2012).
Most of the genome engineering methods are based on the combined effort of a nuclease, and an exogenous DNA, which encodes the desired edits. The nuclease specifically cleaves the genome in a region close to the targeting site, and the cellular machinery repairs the genome using the exogenous DNA as template. CRISPR gene editing is based on Cas9. This protein can be programmed easily to cleave specific DNA sequences and target à la carte the desired genome region (Jinek et al., 2012).
Although these technologies have been widely implemented in many fields and organisms [human, cynomolgus monkeys, mouse, rat, zebrafish, drosophila, yeast, Caenorhabditis elegans, Arabidopsis, bacteria and others (DiCarlo et al., 2013; Esvelt et al., 2013; Feng et al., 2013; Hwang et al., 2013; Li et al., 2013; Niu et al., 2014; Yu et al., 2013)], we do not have any computational tools to analyze the outcome of these experiments yet. Several tools have proliferated for off-target prediction (http://bioanalysis.otago.ac.nz/CRISPRTarget) and experiment design (http://www.e-crisp.org, http://zifit.partners.org/ZiFiT/, http://www.crispr-cas.org/, http://wormcas9hr.weebly.com/), but none of them supports experimental analysis.
We present CRISPR Genome Analyzer (CRISPR-GA), a platform to assess the quality of a genome editing experiment only with three mouse clicks. It can be used in any genomic locus of any organism that the sequence is available. The method uses next-generation data to quantify and characterize insertions, deletions and homologous recombination (HR) at the intended targeting sites. Current methods to assess genome engineering are based on enzymatic mutation detection techniques (Kim et al., 2009; Qiu et al., 2004) or reporter assays. Both methods suffer from important limitations. In optimal conditions, an enzymatic method such as Surveyor’s assay, it reaches a detection limit of 3%, but often editing efficiencies are well below this value (Yang et al., 2013) (Fig. 1C). Another enzymatic method, the T7 endonuclease I (T7E) may achieve detection limits around 1%. Both methods have biases on the detection limit. Surveyor’s assay is more sensitive to transition and transversion mutations, whereas T7E to indels (Huang et al., 2012). Reporter assays are more sensitive, as they detect single cells, but they require additional steps to generate exogenous DNA reporter constructs. More importantly, reporters incorporate other by-products (i.e. fluorescent proteins), which may not be desired for certain applications. In addition, both methods deliver a binary output, and none provides with a complete description of the genome editing experiment.
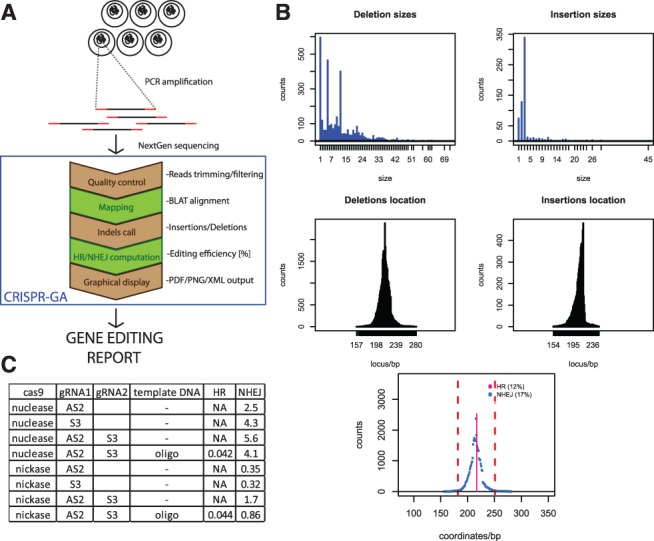
CRISPR-GA pipeline. (A) From experiment to report. Schematic pipeline of gene editing assessment. (B) Output of CRISPR-GA: different information is estimated. Deletions, insertions, HR and corresponding efficiencies. Upper panels estimate the number of insertions and deletions and each corresponding size. Middle panels estimate the number of insertions and deletions, and their corresponding location within the genomic locus of interest. The bottom panel shows the number of deletions and HR at each corresponding locations and outputs the HR and NHEJ efficiency. (C) Experimental results assessed by CRISPR-GA from testing several mutants of cas9, gRNAs and a DNA template. HR and NHEJ values are presented
CRISPR-GA requires little experimental labor, and it provides a complete report of the genome editing results with a fast turnover. The user is only required to use polymerase chain reaction (PCR) to amplify the area of the genome to analyze (the specific instructions to carry out this PCR can be found in supplementary). This amplified library is sequenced with an Illumina Miseq Machine (other platforms may also be used; see online documentation). The user provides to the pipeline CRISPR-GA, the next-generation sequencing obtained reads, the intended editing sequence and the original sequence. CRISPR-GA will estimate the HR (normally associated with intended edits), non-homologous end-joining (NHEJ; normally associated with non-desired edits) and a complete report of the location and characteristics of the indels.
Next-generation sequencing is becoming available for most research institutions. The possibility of bar coding hundreds of samples in a single Illumina Miseq lane makes it competitive from a time and economic perspective. Hundred samples can be analyzed for a bit more than $1000 with cost of $10 per sample, in one overnight Illumina Miseq Run.
2 IMPLEMENTATION
CRISPR-GA pipeline consists of five different steps: reads quality control, mapping, indel calling, HR and NHEJ estimation and graphical representation (Fig. 1A). Initially, the reads are uploaded, the 3′ end is trimmed of nucleotides with a Phred score <20 and reads shorter than 80 bp are discarded [fastx toolkit, (Pearson et al., 1997)]. This filtering and trimming step eliminates reads with sequencing errors. Then, they are mapped to the user-incorporated reference sequence using BLAT [(Kent, 2002)], which has a good support for indels. If the user inputs paired-end reads, the first two steps will be done independently, and the results intersected. Most of the users will supply paired-end reads, as all new Illumina kits only support paired ends. However, CRISPR-GA single end compatibility will be maintained to support all possible experimental setups. In the third step, R statistical language (http://www.R-project.org/) is used to process the mapped BLAT results and call the insertions and deletions. Fourth, pattern matching is used to compute the number of reads matching the expected sequence and other variants generated. NHEJ and HR are estimated computing the Equations (1) and (2), respectively. Fifth, R statistical language is used to produce a report that integrates the results (Fig. 1B). It consists of three parts: (i) analysis of indels sizes, (ii) analysis of indels locations within the edited genomic locus and (iii) an integrated plot representing indels, NHEJ and HR. Additionally, a FASTA file is generated with all reads containing indels, as well as an XML text output with HR, NHEJ values and indels relative location in genomic locus.
We provide an example of the potential of CRISPR-GA (Fig. 1C). We made an experiment where we analyze the efficiency of different variants of cas9 (nuclease and nickases), and different guide RNAs (gRNAs) (AS2 + S3) targeting two adjacent sites on the AAVS1 locus (Yang et al., 2013). We have also used a single-stranded oligonucleotide DNA template to introduce specific mutations via HR. CRISPR-GA is sensitive to detect the differences of editing efficiency with the Human Induced Pluripotent Stem Cells (hiPSC). We see that nickase NHEJ activity is significantly lower than that of nuclease. However, HR efficiency shows similar results. These are expected results, as the nickase only excises one genomic strand, whereas the nuclease both, stimulating NHEJ pathway.
3 CONCLUSIONS
CRISPR-GA is a tool that provides easy (three mouse clicks), economic (~$10), sensitive (detection limit <0.1%) and comprehensive analysis of gene editing results.
ACKNOWLEDGEMENTS
The authors would like to thank John Aach and Susan Byrne for their comments; Suzanne Clewley for her help with infrastructure and Dan Goodman for interesting hints for the Amazon EC2.
Funding: CEGS grant (Centers of Excellence in Genomic Science, grant number HG005550, National Institutes of Health). M.G. is funded by a Human Frontiers Science Program Long Term Fellowship.
Conflict of interest: none declared.
REFERENCES
- Cho SW, et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. [Abstract] [Google Scholar]
- Clear P. 2013 runners-up. Genetic microsurgery for the masses. Science. 2013;342:1434–1435. [Abstract] [Google Scholar]
- Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. [Europe PMC free article] [Abstract] [Google Scholar]
- DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. [Europe PMC free article] [Abstract] [Google Scholar]
- Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1121. [Europe PMC free article] [Abstract] [Google Scholar]
- Feng Z, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–1232. [Europe PMC free article] [Abstract] [Google Scholar]
- Huang MC, et al. A simple, high sensitivity mutation screening using Ampligase mediated T7 endonuclease I and Surveyor nuclease with microfluidic capillary electrophoresis. Electrophoresis. 2012;33:788–796. [Abstract] [Google Scholar]
- Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. [Europe PMC free article] [Abstract] [Google Scholar]
- Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. [Abstract] [Google Scholar]
- Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. [Europe PMC free article] [Abstract] [Google Scholar]
- Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. [Europe PMC free article] [Abstract] [Google Scholar]
- Kim HJ, et al. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. [Europe PMC free article] [Abstract] [Google Scholar]
- Li D, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:681–6. [Abstract] [Google Scholar]
- Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. [Europe PMC free article] [Abstract] [Google Scholar]
- Niu Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. [Abstract] [Google Scholar]
- Pearson WR, et al. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. [Abstract] [Google Scholar]
- Qiu P, et al. Mutation detection using Surveyor nuclease. Biotechniques. 2004;36:702–707. [Abstract] [Google Scholar]
- Yang L, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. [Europe PMC free article] [Abstract] [Google Scholar]
- Yu Z, et al. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195:289–291. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Bioinformatics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/bioinformatics/btu427
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/bioinformatics/article-pdf/30/20/2968/48929935/bioinformatics_30_20_2968.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/bioinformatics/btu427
Article citations
Quantifying allele-specific CRISPR editing activity with CRISPECTOR2.0.
Nucleic Acids Res, 52(16):e78, 01 Sep 2024
Cited by: 0 articles | PMID: 39077930 | PMCID: PMC11381363
CRISPR-Cas guide RNA indel analysis using CRISPResso2 with Nanopore sequencing data.
BMC Res Notes, 17(1):205, 26 Jul 2024
Cited by: 0 articles | PMID: 39061110 | PMCID: PMC11282726
The impact and future of artificial intelligence in medical genetics and molecular medicine: an ongoing revolution.
Funct Integr Genomics, 24(4):138, 16 Aug 2024
Cited by: 0 articles | PMID: 39147901
Review
Protocol for the electroporation of CRISPR-Cas for DNA and RNA targeting in Bos taurus zygotes.
STAR Protoc, 5(1):102940, 08 Mar 2024
Cited by: 2 articles | PMID: 38460133 | PMCID: PMC10941008
Systematic Comparison of Computational Tools for Sanger Sequencing-Based Genome Editing Analysis.
Cells, 13(3):261, 30 Jan 2024
Cited by: 0 articles | PMID: 38334653 | PMCID: PMC10854981
Go to all (85) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Advances and perspectives on the use of CRISPR/Cas9 systems in plant genomics research.
Curr Opin Plant Biol, 30:70-77, 18 Feb 2016
Cited by: 47 articles | PMID: 26896588
Review
Web-based design and analysis tools for CRISPR base editing.
BMC Bioinformatics, 19(1):542, 27 Dec 2018
Cited by: 71 articles | PMID: 30587106 | PMCID: PMC6307267
CRISPR-Cas9: A revolution in genome editing in rheumatic diseases.
Joint Bone Spine, 84(1):1-4, 04 Nov 2016
Cited by: 2 articles | PMID: 27825565
Review
CRISPR-Cpf1: A New Tool for Plant Genome Editing.
Trends Plant Sci, 22(7):550-553, 19 May 2017
Cited by: 52 articles | PMID: 28532598
Funding
Funders who supported this work.
NHGRI NIH HHS (2)
Grant ID: P50 HG005550
Grant ID: HG005550





