Abstract
Free full text

The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs
Abstract
hStaufen is the human homolog of dmStaufen, a double-stranded (ds)RNA-binding protein involved in early development of the fly. hStaufen-containing complexes were purified by affinity chromatography from human cells transfected with a TAP-tagged hStaufen gene. These complexes showed a size >10 MDa. Untagged complexes with similar size were identified from differentiated human neuroblasts. The identity of proteins present in purified hStaufen complexes was determined by mass spectrometry and the presence of these proteins and other functionally related ones was verified by western blot. Ribosomes and proteins involved in the control of protein synthesis (PABP1 and FMRP) were present in purified hStaufen complexes, as well as elements of the cytoskeleton (tubulins, tau, actin and internexin), cytoskeleton control proteins (IQGAP1, cdc42 and rac1) and motor proteins (dynein, kinesin and myosin). In addition, proteins normally found in the nucleus, like nucleolin and RNA helicase A, were also found associated with cytosolic hStaufen complexes. The co-localization of these components with hStaufen granules in the dendrites of differentiated neuroblasts, determined by confocal immunofluorescence, validated their association in living cells. These results support the notion that the hStaufen-containing granules are structures essential in the localization and regulated translation of human mRNAs in vivo.
INTRODUCTION
Although sorting of proteins to specific points within the cell can be a post-translational process, their intracellular accumulation at specific sites is often accomplished by the previous localization of the corresponding mRNAs (reviewed in 1–5). The specific intracellular localization of certain proteins is a key event in many biological phenomena. One of the best known examples is determination of the antero-posterior axis during the early development of Drosophila melanogaster. Thus, the localization of oskar mRNA in the fly oocyte defines the location of the pole plasm at the posterior pole (6) and later the specific posterior expression of nanos in the embryo (7). Likewise, the localization of bicoid mRNA at the anterior pole determines the position of the head in the body plan (7). The specific localization of proteins is also essential in the definition of cell asymmetries in cell division and differentiation. prospero mRNA is localized in the basal area of fly mitotic neuroblasts and determines the fate of one daughter cell as the ganglion mother cell (8,9). Similarly, mating type switching of the daughter cell in yeast is determined by the specific accumulation of Ash1 mRNA at the budding tip. Ash1p inhibits the expression of HO endonuclease and stops the shift of mating type in the daughter cell (10,11). In addition to these biological processes, cell comunication events are also affected by the localization of specific mRNAs. Consequently, mRNA localization at the synapse has been suggested as a means to increase neuronal plasticity by local translation of specific proteins (reviewed in 4,12,13) and cell motility in a specific direction depends on the asymmetric localization of β-actin mRNA (14).
For it to be effective, mRNA localization has to be tightly coupled with translation at the site. Thus, several types of trans factors, some of which act in conjunction with the cis signals present in the messengers, are involved in confined protein expression via mRNA localization. (i) RNA-binding proteins that associate with specific mRNAs. Among them, there are proteins containing double-stranded (ds)RNA-binding domains, of which dmStaufen is the prototype (7); Zipcode-binding proteins like ZBP-1 (15) or Vera (16); some hnRNP components (17); a new type of RNA-binding protein that recognizes Ash1 mRNA (18,19). (ii) Motor proteins that associate with specific RNPs to transport them, like dynein (20) or kinesin (21). (iii) Adaptor proteins that interact with other elements in the mRNA RNPs and are essential for their transport, like barentsz and miranda (22–24). (iv) Cytoskeletal structures along which the specific RNPs move, like microtubules (25) or actin fibers (26,27). (v) Specific repressors of protein synthesis that prevent translation before the target mRNA is properly localized, like bruno activity on oskar mRNA translation (28).
dmStaufen is an RNA-binding protein crucial for the localization of specific mRNAs in fly early development. It is a member of a family of dsRNA-binding proteins that include PKR, RNase III, HIV TAR-binding protein and vaccinia E3L protein (7,29). These proteins contain several dsRNA-binding domains (dsRBDs) whose structure has been determined (30,31). The interaction of dmStaufen with its cognate mRNAs in vivo involves the formation of large RNA granules in which both RNA–RNA and protein–RNA interactions are important (25,32). We and others have identified the human homolog of dmStaufen (hStaufen) (33,34). The various isoforms of hStaufen contain four dsRBDs homologous to dmStaufen but lack the sequences corresponding to the N-terminal half of the fly protein. hStaufen protein binds dsRNA in vitro without any sequence specificity and is localized in human cells in culture in the rough endoplasmic reticulum (ER), in association with polysomes (33). In contrast to dmStaufen, it contains a tubulin-binding domain (34) that may be involved in the transport of specific mRNAs along microtubules. In fact, the rat homolog of hStaufen localizes at the dendrites of neurons in culture and associates with RNA-containing granules that move in a microtubule-dependent manner (35–37). More recently, a second hStaufen homolog has been described that also localizes to the somato-dendritic domains in neurons and is associated with RNA-containing granules (38–40).
In this report we describe the purification of hStaufen-containing complexes isolated from cells expressing a tagged hStaufen gene and the characterization of untagged complexes in differentiated human neuroblasts in culture. The hStaufen-containing complexes contain ribosomes, proteins from the cytoskeleton, motor proteins and regulatory proteins, as well as other RNA-binding proteins normally localized to the nucleus. Altogether, the composition and size of these complexes is consistent with a role in the regulated transport and translation of mRNAs.
MATERIALS AND METHODS
Plasmids and cells
The 293T cell line (41) and the neuroblast SHSY5Y cell line were provided by A.Portela and J.R.Naranjo, respectively. Cell culture was carried out as described (42). Differentiation of neuroblasts was performed by incubation with 10 µM retinoic acid for 6–7 days. The vaccinia recombinant virus vTF7-3 (43) was a gift of B.Moss. Plasmid pBS 1479 (44), containing the TAP tag, was provided by B.Seraphin. The TAP sequence was amplified by PCR and cloned in pCDNA3 (Invitrogen) to generate plasmid pC-TAP. Likewise, it was cloned downstream of the hStaufen cDNA, previously cloned in the same vector, to generate pChStaufen-TAP. Plasmid pBS1456, which expresses the TEV protease with a histidine tag, was kindly provided by G.Stier. Standard conditions were used for DNA restriction, DNA isolation, ligation and Escherichia coli transformation (45).
Antisera
Rabbit antiserum specific for hStaufen protein has been described previously (33). Rat anti-hStaufen serum was prepared by hyperimmunization with purified His-hStaufen protein. Antibodies specific for CLIP115, CLIP170, CLASP-1 and CLASP-2 were obtained from N.Galjart. Those specific for CLIP170 and IQGAP-1 were provided by K.Kaibuchi. Antibodies directed to PABP1, eIF4G and PDI were obtained from A.Nieto. Antibodies specific for nucleolin and hnRNPU were provided by S.Piñol-Roma. Antibodies specific for influenza NP, Tau, FMRP and kinesin KMT3x were obtained from I.Salanueva, J.Avila, C.Bagni and I.Vernós, respectively. Antibodies directed to Rho and Rac were purchased from Upstate Biotechnology. Those specific for L28, S6, internexin and cdc42 were purchased from Santa Cruz Biotechnology and the antibody against calnexin was from Stressgene. The syto-14 dye was purchased from Molecular Probes.
Transfection
For infection–transfection, cultures of 293T cells were infected with vTF7-3 virus at a multiplicity of infection of 5–10 p.f.u./cell. After virus adsorption for 1 h at 37°C, the cultures were washed with DMEM and transfected with pChStaufen-TAP (10–20 µg/100 mm dish) as described (33). The same amount of pC-TAP plasmid was transfected as a control culture. From the transfection step on all media contained 50 µg/ml cytosine arabinoside (AraC) to inhibit viral DNA replication and late gene expression. Some transfections were carried out with the same protocol but in the absence of virus infection. Cationic liposomes were prepared as described (46).
Purification of tagged Staufen protein
For purification of complexes containing the tagged hStaufen protein, extracts of infected–transfected cultures were prepared in lysis buffer [100 mM NaCl, 50 mM Tris–HCl, 0.5% NP40, 1 mM dithiothreitol, pH 7.5, 170 µg/ml phenylmethylsulfonyl fluoride, 80 µg/ml Nα-p-tosyl-L-lysine chloromethyl ketone (TLCK), 80 µg/ml N-p-tosyl-L-phenylalanine chloromethyl ketone (TPCK), 150 µg/ml benzamidine]. After centrifugation for 3 min at 2500 r.p.m. and 4°C, the supernatant was further centrifuged for 10 min at 10 000 r.p.m. and 4°C and was incubated with IgG–Sepharose (100 µl bed volume, ~5 µl bed volume/mg protein) for 12 h at 4°C. The resin was washed 10 times with 1 ml of a buffer containing 150 mM NaCl, 10 mM Tris–HCl, 0.1% NP40, pH 8.0, and subsequently five times with 1 ml of 150 mM NaCl, 10 mM Tris–HCl, 0.1% NP40, 0.5 mM EDTA, pH 8.0. The material bound to the resin was digested with 80 µg TEV protease for 2 h at room temperature in the same buffer. The supernatant was mixed with six successive washes of the resin in a buffer containing 150 mM NaCl, 10 mM Tris–HCl, 0.1% NP40, 10 mM 2-mercaptoethanol, 1 mM Mg(AcO)2, 1 mM imidazole, 2 mM CaCl2, pH 8.0, and incubated for 12 h at 4°C with calmodulin–agarose resin. The resin was washed 10 times with the same buffer and eluted with a buffer containing 10 mM Tris–HCl, 0.1% NP40, 10 mM 2-mercaptoethanol, 1 mM imidazole, 2 mM EGTA, pH 8.0.
For fractionation of endogenous hStaufen complexes, cell extracts prepared as indicated above were applied to a Sephacryl S400 resin equilibrated in the same buffer, at a sample to bed volume ratio of 1:100 and the hStaufen complexes were revealed by western blot. The column was calibrated with thyroglobulin, ferritin, catalase and influenza virus ribonucleoproteins purified from virions. The latter were revealed by western blot with anti-NP antibodies.
Protein analyses
Isolation of polysomes was carried out as described previously (33). In brief, the cytoplasmic fraction was obtained by cell lysis in isotonic buffer (150 mM NaCl, 1.5 mM MgCl2, 10 mM Tris–HCl, pH 8.5) containing 0.5% NP40 and centrifuged for 10 min at 10 000 g and 4°C. The supernatant was centrifuged on a 7–47% sucrose gradient in isotonic buffer, for 2 h at 40 000 r.p.m. and 2°C in a SW41 rotor.
000 r.p.m. and 2°C in a SW41 rotor.
Western blotting was carried out as described (33). For silver staining the gels were fixed for 30 min each in 25% ethanol, 10% acetic acid and 25% ethanol, 0.5% acetic acid. After soaking twice for 20 min in silver nitrate (1.8 g/l), the gel was washed three times with water. Reduction was in 10 mM NaBH4, 0.75 M NaOH, 0.75% formaldehyde until the protein bands were apparent and the reaction was stopped in 10% acetic acid.
For confocal microscopy, cultures of differentiated human neuroblasts were fixed for 20 min at room temperature in phosphate-buffered saline (PBS) containing 3% paraformaldehyde and then permeabilized for 5 min with 0.5% Triton X-100 in PBS. The preparations were blocked with 2% bovine serum albumin (BSA) in PBS for 30 min and incubated for 45 min with the appropriate dilutions of primary antibodies in PBS containing 0.1% BSA. After washing with PBS, the preparations were incubated with secondary antibodies using the same conditions. Appropriate combinations of goat anti-rabbit, goat anti-mouse or donkey anti-goat IgG labelled with either alexa488, alexa594 or FITC were used for double labelling experiments. Finally, the preparations were mounted in Mowiol and observed in a Zeiss Axiophot fluorescence microscope equipped with a Bio-Rad confocal unit. Optical sections of 0.3 µm were acquired sequentially with the LaserSharp software and processed with Laserpix.
For identification of proteins by mass spectrometry, tryptic peptide mass fingerprints were obtained and compared with the database (http://www.matrixscience.com/). Identifications were considered positive for scores >73 (P < 0.05).
RESULTS
Expression and purification of TAP-tagged human Staufen protein
To characterize the hStaufen complexes present in human cells we attempted their purification by the strategy developed by Rigaut et al. for yeast (44). The sequence for the TAP tag was cloned downstream of the cDNA for hStaufen and the tagged gene was expressed in human cells by transfection. The TAP tag comprises two affinity sequences, an IgG-binding domain from protein A and the calmodulin-binding peptide, separated by a cleavage signal for the TEV protease (44) (Fig. (Fig.1A).1A). This allows purification of hStaufen-containing complexes under very mild conditions, as described in Materials and Methods. A critical issue when using a tagged protein as a molecular probe is whether it behaves biologically as the native protein. To check that point, we compared the relative association with polysomes of hStaufen-TAP, expressed by transfection of plasmid pChStaufen-TAP into 293T cells, and the endogenous hStaufen isoforms. The results are presented in Figure Figure1B.1B. The upper band represents hStaufen-TAP recombinant protein (677 amino acids), which could also be revealed by an irrelevant rabbit serum (data not shown), while the lower bands correspond to endogenous hStaufen isoforms. The results clearly indicate that the tagged protein is indistinguishable from the wild-type protein in its sedimentation pattern. The intracellular localization of both tagged and endogenous hStaufen proteins was studied by confocal immunofluorescence. The localization of tagged protein in transfected cells was determined using an irrelevant rabbit serum and is shown in Figure Figure1C1C (right panel), while the endogenous hStaufen protein was revealed with a specific antiserum in mock-transfected cells (Fig. (Fig.1C,1C, left panel). The appearance of similar granular structures in both cases indicates that the tagged and endogenous proteins have similar behaviours and that the tagged protein can be used as a marker of the normal one.

Generation of TAP-tagged hStaufen-containing complexes. The TAP tag was inserted downstream of hStaufen cDNA and the tagged gene was expressed by transfection into 293T human cells. (A) The diagram shows the general structure of hStaufen protein including its four dsRBDs (2–5), as well as the fusion protein hStaufen-TAP with its tags for affinity purification. (B) Soluble extracts derived from transfected cells were centrifuged on a 7–47% sucrose gradient and the endogenous and recombinant hStaufen proteins were revealed by western blotting with anti-hStaufen- specific serum. The numbers at the bottom indicate the fractions analyzed. The mobility of molecular weight markers is indicated on the left. (C) Cultures of 293T cells were transfected with pC-TAP or pChStaufen-TAP, fixed and analyzed by confocal immunofluorescence, using anti-hStaufen (for mock-transfected cells, left panel) or irrelevant rabbit serum (for hStaufen-TAP transfected cells, right panel).
To improve the yield of tagged protein, we purified hStaufen-containing complexes from 293T cells infected with vaccinia vTF7-3 virus, which expresses T7 RNA polymerase, and transfected with pChStaufen-TAP plasmid. AraC was included to avoid the replication and late gene expression of vaccinia virus. Under these conditions, accumulation of the tagged protein was 2- to 4-fold higher than endogenous hStaufen (data not shown). As a control, a parallel purification was carried out from cells infected with vTF7-3 virus and transfected with pC-TAP empty vector. The various purification steps were examined by western blot using a hStaufen-specific or an irrelevant rabbit serum to reveal the presence of the IgG-binding domain in the TAP tag, when appropriate. After purification, no endogenous hStaufen protein was detectable by western blot in the control preparation, while a clear band of hStaufen was obtained from the pChStaufen-TAP-transfected cells (Fig. (Fig.2A).2A). The presence of hStaufen as well as potentially associated proteins was analyzed by silver staining of purified preparations obtained from pC-TAP- or pChStaufen-TAP-transfected cells. Essentially no protein could be detected in the material obtained from pC-TAP-transfected cells, while recombinant and endogenous hStaufen proteins (see below) were observed in the complexes purified from pChStaufen-TAP-transfected cells (Fig. (Fig.2B,2B, stars). To further characterize the purified hStaufen complexes, their size was determined by gel filtration on Sephacryl S400, along with molecular weight standards. Only complexes with a size >10 MDa were observed (Fig. (Fig.22C).

Purification and characterization of hStaufen-containing complexes. Cultures of 293T cells were transfected with pChStaufen-TAP (hStau) and soluble extracts were used for TAP purification. (A) Western blot of the purified products revealed with anti-hStaufen serum. Purified His–hStaufen was used as an internal marker. (B) Silver staining of the purified material. The positions of hStaufen proteins (stars) are indicated on the right. The mobility of molecular weight markers is indicated to the left of each panel. (C) Purified hStaufen complexes were filtered on a Sephacryl S400 column and their elution was monitored by western blot with anti-hStaufen serum. The elution of molecular weight markers is indicated at the top. Blue dextran exclusion marker eluted in fraction 17. (D) Soluble extracts derived from differentiated neuroblast SHSY5Y cells were filtered on a Sephacryl S400 column and their elution was monitored by western blot with anti-hStaufen serum, as indicated above. Similar results were found when using Sephacryl S500 for filtration (data not shown).
Protein composition of purified, tagged hStaufen complexes
The pattern of proteins associated with purified hStaufen was very complex, but reproducibly obtained in several transfection and purification experiments. The various bands were excised from the gel, digested with trypsin and their tryptic fingerprints were obtained by MALDI-TOF mass spectrometry. The most abundant bands (Fig. (Fig.2B,2B, stars) were identified as hStaufen and corresponded to the recombinant (upper band) and endogenous (lower band) proteins, as their relative abundances varied from experiment to experiment. The presence of endogenous hStaufen in the purified complexes is not unexpected, as Staufen proteins can interact and multiple copies can be present in the hStaufen granules. Many other associated proteins were identified by mass spectrometry and the results are presented in Table Table1.1. The presence in these complexes of proteins already known to interact with hStaufen, like tubulin (34) and myosin (47), validated the purification approach taken. Many of the proteins found were ribosomal components, but cytoskeleton and motor proteins, as well as proteins involved in protein synthesis and regulatory proteins, like Rho-associated protein kinase II, IQGAP1 and Ras-GAP, were also present. Surprisingly, typical nuclear proteins, like nucleolin, RNA-dependent RNA helicase A and hnRNP U, were also found associated with the hStaufen complexes.
Table 1.
| Protein | Accession no. | Identification technique | ||
|---|---|---|---|---|
| | | Mass spectrometry | Western blot | Confocal immunofluorescence |
| β-5 Tubulin | gi|18088719 | + (187a) | + | + |
| α-Tubulin | | – | + | + |
| Tau | | – | + | + |
| hStaufen isoform 2 | gi|10944134 | + (273) | + | + |
| β-Actin | | – | + | + |
| Myosin heavy chain | gi|7669506 | + (103) | NT | NT |
| RNA-dependent RNA helicase A | gi|3915658 | + (139) | + | + |
| Nucleolin | gi|21750187 | + (86) | + | + |
| hnRNP U (SAF A) | gi|14044052 | + (91) | – | – |
| Poly(A)-binding protein | gi|4505575 | + (107) | + | + |
| α-Internexin (neuronal intermediate filament) | gi|14249342 | + (77) | + | + |
| Dynein intermediate chain | – | + | + | |
| Kinesin | | – | + | + |
| ρ-Associated protein kinase II | gi|4759044 | + (74) | NT | NT |
| Ras GAP | gi|627594 | + (74) | NT | NT |
| Rac1 | | – | + | + |
| Cdc42 | | – | + | + |
| IQGAP1 protein | gi|4506787 | + (87) | + | + |
| FMRP | | – | + | + |
| Ribosomal protein P0 | gi|12654583 | + (120) | NT | NT |
| Ribosomal protein S4b | gi|539681 | + (150) | NT | NT |
| Ribosomal protein S6 | gi|20381196 | + (120) | + | + |
| Ribosomal protein L6b | gi|18088374 | + (103) | NT | NT |
| Ribosomal protein L28 | gi|13904866 | + (116) | + | + |
aIdentification score (see Materials and Methods).
bMany other small and large ribosomal proteins were identified that are not included in this table (data not shown).
To verify the initial identification, complexes purified as indicated above, as well as the corresponding parallel purifications from cells expressing only the TAP tag but not recombinant hStaufen, were analyzed by western blot using antisera specific for many of the proteins identified and others known to interact with them. The results are presented in Figure Figure3.3. Most of the proteins detected by mass spectrometry were also identified immunologically in purified hStaufen complexes, the exception being hnRNP U, which could not be detected with the available reagents. In addition, β-actin, α-tubulin, kinesin, dynein, FMRP, Tau, Rac1 and cdc42 were also detected. None of the proteins tested for were present in the control purifications obtained from TAP-transfected complexes (Fig. (Fig.3,3, Table Table11 and data not shown). As the hStaufen complexes contain RNA (see below), we analyzed whether the presence of RNA was required for the association with hStaufen complexes of the identified proteins. Extracts from pChStaufen-TAP-transfected cultures were treated with RNase A and hStaufen complexes were purified as described. As expected, the yield of hStaufen was reduced (by ~50%), since RNA–RNA interactions as well as protein–RNA interactions are involved in the formation of Staufen granules (25,32). Some of the associated proteins were no longer detectable after RNase treatment, like ribosomal proteins, PABP and nucleolin (data not shown), suggesting that their presence in the complexes depends mainly on their interaction with RNA. Others, like actin, tubulin, tau, dynein and FMRP, were still present in purified complexes after RNase treatment (data not shown), an indication that protein–protein interactions are more important in these associations.

Identification of proteins associated with hStaufen-containing complexes. Cultures of 293T cells were transfected with pChStaufen-TAP (hStau) or pC-TAP (CTRL) and soluble extracts were used for TAP purification. Aliquots from either purified complexes (Stau) or control purifications (CTRL) were analyzed by western blot using the antisera indicated. The signal obtained from whole cell extracts (WE) by western blot with each antibody is presented in the panel on the right. Numbers to the left of each panel indicate the position of molecular weight markers (in kDa).
It should be stressed that, in spite of the complexity of the protein composition of purified hStaufen complexes, these appear to be specific, as no protein could be detected in control purifications (Fig. (Fig.1D)1D) and several abundant proteins, like the Golgi marker p115 and the ER markers calnexin and PDI, were not co-purified (Fig. (Fig.33 and data not shown). It is noteworthy that some proteins typically expressed in neural tissues, like internexin and FMRP, could be co-purified with hStaufen from HEK293T cells, a transformed cell line derived from kidney. This fact supports the contention that the hStaufen complexes purified are similar to those detected in neurons (37,40,48) (see below).
Characterization of endogenous, untagged hStaufen complexes from human neuroblasts
The experiments described so far were performed with complexes derived from hStaufen-TAP-transfected HEK293T cells. To verify the relevance of the observations made in a more physiological system, we characterized the endogenous hStaufen complexes derived from differentiated human neuroblasts. Cultures of the human SHSY5Y neuroblast cell line were differentiated by treatment with retinoic acid and total cytoplasmic extracts were analyzed by gel filtration as indicated above. The mobility of hStaufen complexes was determined by western blotting and is shown in Figure Figure2D.2D. The size obtained was indistinguishable from that of purified complexes presented in Figure Figure2C,2C, i.e. >10 MDa. In contrast to a recent report (40), we could not detect small hStaufen complexes (~0.65 MDa), in spite of using detergent-solubilized total cell extracts as input for purification of tagged complexes (Fig. (Fig.2C)2C) or for size fractionation of endogenous complexes (Fig. (Fig.2D).2D). The purified, tagged hStaufen complexes, as well as the endogenous, untagged ones, contained rRNAs as well as a number of mRNAs (R.M.Marión, manuscript in preparation). The presence of RNA in the hStaufen complexes was also verified in vivo. SHSY5Y differentiated neuroblasts were incubated in culture with syto14, fixed and analyzed by confocal immunofluorescence to detect both hStaufen and RNA (Fig. (Fig.4).4). hStaufen granules were detected both in the soma and the dendrites, although their morphology was affected by the dye incorporation. Most of the RNA was apparent in the nucleus, but co-localization with hStaufen granules was apparent in the cell processes (Fig. (Fig.4,4, inserts).
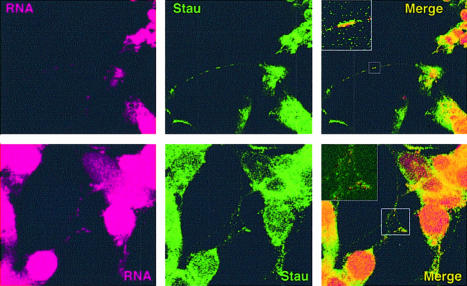
Co-localization of hStaufen granules and RNA. Cultures of differentiated neuroblasts were incubated with syto-14. The cultures were washed, fixed and analyzed by confocal immunomicroscopy using anti-hStaufen serum. The panels to the right represent merging of the left and center images. The inserts in the merged images are blow-ups of the regions of the images indicated.
The association with endogenous, untagged hStaufen complexes of the proteins already detected in purified, tagged hStaufen complexes was ascertained by confocal microscopy of human differentiated neuroblasts. The potential co-localization of these hStaufen granules with the proteins identified in purified complexes was studied by double labelling confocal immunofluorescence and the results are presented in Figures Figures55–9. The association of hStaufen with ribosomal proteins was assayed using S6 and L28 as representative markers. A good co-localization in the cell soma was apparent and, importantly, perfect co-localization was observed in hStaufen granules within dendrites (see Fig. Fig.5).5). Similar results were obtained for L28 (data not shown). The ribosomes included in the hStaufen granules were most probably loaded with mRNA, as they co-localized with PABP1 (see Fig. Fig.55 for a representative example). Interestingly, the hStaufen granules also co-localized with FMRP, a neuronal protein involved in the repression of translation of specific neural mRNAs (49). This is in agreement with the described association of these proteins by co-immunoprecipitation assays (47). We next studied the co-localization of hStaufen granules with various cytoskeletal components. Localization of these hStaufen complexes along microtubules, neurofilaments and actin fibers was observed, as presented in Figure Figure66 by staining with anti-tubulin, anti-tau, anti-internexin and phaloidin, respectively. Moreover, some of the hStaufen granules could be stained with anti-dynein or anti-kinesin antibodies (Fig. (Fig.7),7), indicating that they could move along microtubules in either direction. The presence of IQGAP1 protein in purified hStaufen complexes was also verified by immunofluorescence. Good co-localization of IQGAP1 with hStaufen granules was observed (Fig. (Fig.8).8). As IQGAP1 is a scaffolding protein involved in the activities of the cytoskeleton and cdc42 and rac1 signaling (50), the presence of the latter in hStaufen granules was also analyzed and the results indicate that these proteins are also associated with hStaufen granules (Fig. (Fig.8).8). On the other hand, the association of myosin with the purified hStaufen complexes (Table (Table1),1), together with the presence of β-actin in the complexes (Figs (Figs33 and and6),6), suggests that the hStaufen granules can also move on actin fibers.

Co-localization of translation-related proteins with hStaufen granules in differentiated neuroblasts. Cultures of human neuroblasts were differentiated, fixed and processed for confocal double immunofluorescence using anti-hStaufen serum and the sera indicated in each panel. The panels to the right represent merging of the left and center images. The inserts in the merged images are blow-ups of the regions of the images indicated. The bar indicates 10 µm.

Co-localization of cytoskeleton proteins with hStaufen granules in differentiated neuroblasts. Cultures of human neuroblasts were differentiated, fixed and processed for confocal double immunofluorescence using anti-hStaufen serum and the sera indicated in each panel. The panels to the right represent merging of the left and center images. The inserts in the merged images are blow-ups of the regions of the images indicated. The bar indicates 10 µm.
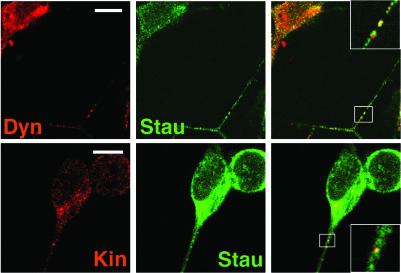
Co-localization of motor proteins with hStaufen granules in differentiated neuroblasts. Cultures of human neuroblasts were differentiated, fixed and processed for confocal double immunofluorescence using anti-hStaufen serum and the sera indicated in each panel. The panels to the right represent merging of the left and center images. The inserts in the merged images are blow-ups of the regions of the images indicated. The bar indicates 10 µm.

Co-localization of cytoskeleton-regulatory proteins with hStaufen granules in differentiated neuroblasts. Cultures of human neuroblasts were differentiated, fixed and processed for confocal double immunofluorescence using anti-hStaufen serum and the sera indicated in each panel. The panels to the right represent merging of the left and center images. The inserts in the merged images are blow-ups of the regions of the images indicated. The bar indicates 10 µm.
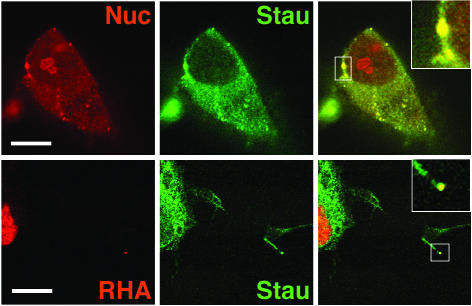
Co-localization of nuclear proteins with hStaufen granules in differentiated neuroblasts. Cultures of human neuroblasts were differentiated, fixed and processed for confocal double immunofluorescence using anti-hStaufen serum and the sera indicated in each panel. The panels to the right represent merging of the left and center images. The inserts in the merged images are blow-ups of the regions of the images indicated. The bar indicates 10 µm.
Finally two of the typically nuclear proteins identified in purified hStaufen complexes were also found associated with hStaufen granules in the cytoplasm of neuroblasts. Although nucleolin stained mainly the nucleoli, it was also found in cytoplasmic spots perfectly co-localized with hStaufen (Fig. (Fig.9).9). Likewise, RNA helicase A, although mainly localized in the nuclei of neuroblasts, was also found associated with some hStaufen granules in the dendrites (Fig. (Fig.99).
DISCUSSION
To investigate the possible roles of hStaufen protein in somatic human cells we set out to recover and characterize intracellular hStaufen-containing complexes. As a first step for this information-driven strategy we chose to express a TAP-tagged hStaufen protein in HEK293T human cells and use the tag as a handle to purify intracellular hStaufen complexes. The results presented in this report show that these complexes can be purified extensively, are very large in size and contain a recurrent set of associated proteins (Figs (Figs11 and and2).2). They could also be purified from cells in which protein synthesis had been inhibited by puromycin treatment (data not shown). The relevance of these complexes containing tagged hStaufen for the normal situation in unaltered cells was verified in human differentiated neuroblasts. Endogenous complexes containing hStaufen with a size similar to that of complexes purified by affinity chromatography were detected in these cells (Fig. (Fig.2)2) and, moreover, most of the proteins identified in the latter (Fig. (Fig.3)3) were also present in large hStaufen-containing granules observed in dendrites of differentiated neuroblasts in vitro (Figs (Figs55–9).
Biological implications of the structure of hStaufen complexes
The presence of many ribosomal components, as well as proteins involved in protein synthesis, like PABP1, suggests that the purified hStaufen-containing complexes are actively involved in protein synthesis or set for expression of the enclosed mRNAs. Of special relevance may be the finding of FMRP associated with the purified hStaufen complexes, even from HEK293T cells, and their co-localization in neuroblast processes. FMRP is involved in the translational control of a variety of mRNAs in the dendrites and its mutation leads to structural and functional defects in dendritic spines (for a recent review see 51). The repression of translation of specific mRNAs takes place by the association of FMRP with BC1 RNA, a small RNA abundantly expressed in neurones, by sequence-specific interaction with the target mRNAs (49). Interestingly, its human counterpart BC200 was found in purified hStaufen complexes (R.M.Marión, manuscript in preparation).
In addition, the elements required for intracellular transport of these complexes, like cytoskeleton and motor proteins (tubulins, actin, internexin, myosin, dynein and kinesin) are also constituents of the hStaufen complexes, suggesting that they may serve as transport vehicles for the localization of specific mRNAs at precise sites in the cell. In fact, the presence of actin and myosin in complexes in which Staufen is present have already been reported (47) and it has been shown that the localization of prospero mRNA in fly neuroblasts is dependent on actin filaments by interaction with miranda protein (52). It is worth mentioning that both plus-end and minus-end directed motors have been detected, implying that the hStaufen complexes can move along microtubules using anterograde and retrograde modes. This is consistent with the fact that the localization of dmStaufen complexes containing oskar mRNA to the posterior pole in D.melanogaster embryos is dependent on a kinesin motor (21) and the corresponding localization of bicoid mRNA complexes to the anterior pole relies on a dynein motor (20). In this context, the identification of IQGAP-1 protein in purified hStaufen complexes and its co-localization with hStaufen granules in dendrites is especially noteworthy. Mammalian IQGAP-1, like its homologs in other species, including yeast, is a central protein that integrates multiple signaling pathways in connection with the regulation of the cytoskeleton. By means of a series of interaction domains, it binds actin (53), myosin light chain and calmodulin, as well as the Rho-like GTPases cdc42 and Rac1, but not Ras (54). In addition, it binds the CLIP170 adaptor protein to associate with microtubules (55). By means of this complex set of interactions, IQGAP-1 modulates cytoskeletal structure. It enhances the growth of actin fibers directly and also by increasing the concentration of GTP-bound cdc42 (54). Thus, the identification of IQGAP-1 and several of its interactors in purified hStaufen complexes and in hStaufen RNA granules in vivo strongly supports a role for these complexes in the regulated transport of mRNA included in translation complexes.
Surprisingly, some typical nuclear proteins, like nucleolin, RNA helicase A and hnRNP U, were also found associated with the complexes in the cytoplasm. Although hnRNP A2 protein is associated with MBP mRNA in oligodendrocyte processes (17), there is no precedent for the presence of hnRNP U protein in RNA granules in the cytoplasm. As the presence of hnRNP U protein in hStaufen granules could not be detected by western blot or immunofluorescence, it is unclear at this time whether it plays a role in mRNA localization. Likewise, nucleolin, a specific component of the nucleolus, has not been found before in cytoplasmic mRNA-containing complexes, although it was detected as a component of the small Ro RNP particles (56). Although Staufen has been detected in association with telomerase in the cell nucleolus (57), the presence of nucleolin in purified hStaufen complexes and its co-localization with hStaufen in neuroblast dendrites adds to the growing evidence that some nuclear proteins remain as tags in cytoplasmic mRNA-containing complexes or behave as shuttling proteins (15,16,58). On the other hand, the presence of RNA helicase A in hStaufen granules may indicate a requirement for remodeling of the complex upon translation activation (37).
The complex set of proteins associated with hStaufen in vivo may indicate that we are dealing with a series of different complexes that represent various functional or positional variants of hStaufen RNA-containing granules that may participate in mRNA storage, transport or translation, depending on general or position-specific stimuli in the cell.
ACKNOWLEDGEMENTS
We are indebted to A.Nieto for critical comments on the manuscript. We thank J.R.Naranjo, B.Moss, B.Seraphin, A.Portela, G.Stier, N.Galjart, K.Kaibuchi, A.Nieto, S.Piñol-Roma, J.Ávila, C.Bagni, I.Salanueva and I.Vernós for providing biological materials. The technical assistance of S.Gutiérrez, Y.Fernández and J.Fernández is gratefully acknowledged. P.V. was a fellow from Gobierno Vasco. R.M.M. was a postdoctoral fellow from Comunidad de Madrid. This work was supported by Programa Sectorial de Promoción General del Conocimiento (grants PB97-1160 and BMC2001-1223).
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkh552
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/32/8/2411/9490051/gkh552.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Posttranscriptional regulation of neurofilament proteins and tau in health and disease.
Brain Res Bull, 192:115-127, 29 Oct 2022
Cited by: 4 articles | PMID: 36441047 | PMCID: PMC9907725
Review Free full text in Europe PMC
The double-stranded RNA-binding protein, Staufen1, is an IRES-transacting factor regulating HIV-1 cap-independent translation initiation.
Nucleic Acids Res, 50(1):411-429, 01 Jan 2022
Cited by: 15 articles | PMID: 34893869 | PMCID: PMC8754648
Unveiling mRNP composition by fluorescence correlation and cross-correlation spectroscopy using cell lysates.
Nucleic Acids Res, 49(20):e119, 01 Nov 2021
Cited by: 3 articles | PMID: 34478550 | PMCID: PMC8599746
The multifunctional RNA-binding protein Staufen1: an emerging regulator of oncogenesis through its various roles in key cellular events.
Cell Mol Life Sci, 78(23):7145-7160, 11 Oct 2021
Cited by: 13 articles | PMID: 34633481 | PMCID: PMC8629789
Review Free full text in Europe PMC
Walking the line: mechanisms underlying directional mRNA transport and localisation in neurons and beyond.
Cell Mol Life Sci, 78(6):2665-2681, 20 Dec 2020
Cited by: 15 articles | PMID: 33341920 | PMCID: PMC8004493
Review Free full text in Europe PMC
Go to all (102) article citations
Other citations
Wikipedia (Showing 7 of 7)
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro.
Nucleic Acids Res, 27(11):2241-2247, 01 Jun 1999
Cited by: 66 articles | PMID: 10325410 | PMCID: PMC148787
A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum.
Mol Cell Biol, 19(3):2212-2219, 01 Mar 1999
Cited by: 122 articles | PMID: 10022908 | PMCID: PMC84014
Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons.
Genes Brain Behav, 4(6):350-359, 01 Aug 2005
Cited by: 153 articles | PMID: 16098134
Cytoskeleton-dependent transport and localization of mRNA.
Int Rev Cytol, 211:1-31, 01 Jan 2001
Cited by: 15 articles | PMID: 11597002
Review





