Abstract
Free full text

In Silico Identification of an Aryl Hydrocarbon Receptor Antagonist with Biological Activity In Vitro and In Vivo
Abstract
The aryl hydrocarbon receptor (AHR) is critically involved in several physiologic processes, including cancer progression and multiple immune system activities. We, and others, have hypothesized that AHR modulators represent an important new class of targeted therapeutics. Here, ligand shape–based virtual modeling techniques were used to identify novel AHR ligands on the basis of previously identified chemotypes. Four structurally unique compounds were identified. One lead compound, 2-((2-(5-bromofuran-2-yl)-4-oxo-4H-chromen-3-yl)oxy)acetamide (CB7993113), was further tested for its ability to block three AHR-dependent biologic activities: triple-negative breast cancer cell invasion or migration in vitro and AHR ligand–induced bone marrow toxicity in vivo. CB7993113 directly bound both murine and human AHR and inhibited polycyclic aromatic hydrocarbon (PAH)– and TCDD-induced reporter activity by 75% and 90% respectively. A novel homology model, comprehensive agonist and inhibitor titration experiments, and AHR localization studies were consistent with competitive antagonism and blockade of nuclear translocation as the primary mechanism of action. CB7993113 (IC50 3.3 × 10−7 M) effectively reduced invasion of human breast cancer cells in three-dimensional cultures and blocked tumor cell migration in two-dimensional cultures without significantly affecting cell viability or proliferation. Finally, CB7993113 effectively inhibited the bone marrow ablative effects of 7,12-dimethylbenz[a]anthracene in vivo, demonstrating drug absorption and tissue distribution leading to pharmacological efficacy. These experiments suggest that AHR antagonists such as CB7993113 may represent a new class of targeted therapeutics for immunomodulation and/or cancer therapy.
Introduction
The aryl hydrocarbon receptor (AHR) field has undergone a dramatic paradigm shift in the last few years. Historically, the evolutionarily conserved AHR was studied for its ability, upon activation by environmental ligands, to regulate genes encoding a battery of cytochrome P450 enzymes that metabolize at least some of those ligands into metabolic intermediates, some of which are toxic (Hankinson et al., 1991; Nebert et al., 1991). Similarly, studies published over the last 20–30 years demonstrate the role of the AHR in the initiation of environmental chemical–induced cancers, to a large extent through P450-dependent generation of mutagenic intermediates. Consequently, studies on the effects of AHR activation, primarily by anthropogenic ligands, most frequently involved tissue toxicity or the initiation of malignancy.
However, several landmark studies now demonstrate that the AHR plays a critical role in several physiologic processes in the absence of environmental chemicals. These studies show that the AHR, presumably activated by endogenous ligands, plays a critical role in the manifestation or control of tissue inflammation and autoimmunity through enforcement of inflammatory cytokine production by monocytes (Wu et al., 2011) and synoviocytes (Lahoti et al., 2013), the production of inflammatory Th17 and immunosuppressive regulatory T cells (Veldhoen et al., 2008), and the regulation of apoptosis (Caruso et al., 2004, 2006). Indeed, the recent demonstration that AHR activation drives production of erythroid cells from pluripotent stem cell precursors (Smith et al., 2013), is required for development of gut-associated T cells (Kiss et al., 2011; Lee et al., 2012), and influences the differentiation of human hematopoietic stem cells (Boitano et al., 2010) suggests a critical role of the AHR in hematopoiesis in general.
Furthermore, several studies now implicate the AHR in cancer progression in the absence of environmental ligands. For example, AHR activated by an endogenous tryptophan–derived metabolite increases human glioblastoma cell survival and migration (Opitz et al., 2011). The AHR-repressor (AHRR) protein acts as a tumor suppressor gene in several human cancers (Zudaire et al., 2008). AHR expression and “constitutive” (endogenous ligand–driven) activity in breast cancer cells correlate with tumor aggressiveness (Schlezinger et al., 2006; Yang et al., 2008) and control expression of genes associated with tumor invasion (Yang et al., 2005). Surveys of cytochrome P450 protein expression identified the AHR gene targets CYP1A1, CYP1A2, and CYP1B1 in most types of primary human tumor samples examined, including solid and hematologic malignancies (Maecker et al., 2003), and found that expression of the AHR target gene CYP2S1 inversely correlates with patient survival (Murray GI et al., 2010). Finally, ectopic AHR expression in nonmalignant human mammary epithelial cells induces an epithelial-to-mesenchymal transition and a >50% increase in cell growth rates (Brooks and Eltom, 2011). Together, these studies strongly support the hypothesis that the AHR plays an important role in the later, more aggressive stages of cancer, even in the absence of environmental ligands.
Given the involvement of the AHR in blood cell development and multiple immune system phenomena, and its postulated role in cancer progression, we and others have hypothesized that AHR modulators, either agonists or antagonists, may represent an important new class of targeted therapeutics (Schlezinger et al., 2006; Zhang et al., 2009). We postulate that AHR antagonists in particular may be important for treatment of high AHR expressing, triple-negative breast cancers (TNBCs), malignancies which are particularly resistant to current chemotherapeutics and nonresponsive to hormone receptor–targeted therapeutics.
The identification of novel, potent AHR modulators has been hampered by the limited amount of data on the three-dimensional structure of the AHR protein, and specifically the structure of its ligand-binding domain (LBD). In its stead, researchers have developed structural homology models on the basis of ligand-binding domains of familial proteins (Motto et al., 2011; Xing et al., 2012). Although recent advancements in AHR-LBD models have improved our understanding of the requirements for AHR binding, the abilities of these programs to predict AHR ligands is only beginning to be realized.
Here, we used ligand shape–based virtual screening techniques to rapidly screen libraries of over 1 million commercially available small molecule compounds for potential AHR ligands. The focused library identified by this analysis was tested in a high-throughput in vitro bioassay for AHR-antagonist activity. Lead compounds chosen from the in vitro screening assays were characterized for their ability to directly bind the AHR and to block AHR nuclear translocation and transcriptional activity. One lead compound, CB7993113, was examined for its probable binding conformation to the AHR PAS-B domain. Finally, CB7993113 was tested for its ability to block three AHR-dependent biologic activities, triple-negative breast cancer cell invasion and migration in vitro, and AHR ligand–induced bone marrow toxicity in vivo.
Materials and Methods
Chemical Reagents
Commercial chemical libraries of test compounds were acquired from ChemBridge Corporation (San Diego, CA) and Enamine (Kiev, Ukraine). Dimethyl sulfoxide (DMSO), β-naphthoflavone (β-NF), 7,12-dimethylbenz[a]anthracene (DMBA), TCDD, and other chemical reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. CB7993113 and CH223191 were synthesized as below. All compounds submitted for biologic testing were deemed at least >98% pure by HPLC (with UV and mass spectral detection) and 1H NMR.
Chemical Synthesis of CB7993113.
1-(2-Hydroxyphenyl)ethanone (1.720 g, 14.29 mmol) and 5-bromofuran-2-carbaldehyde (2.5 g, 14.29 mmol) were dissolved in a round-bottom flask in 10 ml of ethanol. A solution of 17 M NaOH (1.5 ml) in water was then added under vigorous stirring. Precipitate formed under addition of base, and a thick paste was formed. The mixture was stirred for 24 hours at room temperature. Ethanol (50 ml) was added with 0.5 ml of 2.5 M NaOH. The mixture was cooled to 15°C and hydrogen peroxide was added (35% in water, 6.25 ml, 71.4 mmol). After 4 hours, dilute sulfuric acid was added to neutralize to pH 7.0, and the reaction was poured into 250 ml of water and stirred for 2 hours. The solid material was collected by filtration and dried under vacuum to give a yellow solid (1.01 g, 23% yield). 1 H NMR (399 MHz, DMSO-d6) δ ppm 4.60 (s, 2 H) 6.98 (d, J = 3.66 Hz, 1 H) 7.39 (br s, 1 H) 7.50 (t, J = 7.33 Hz, 1 H) 7.67–7.78 (m, 3 H) 7.81 (d, J = 7.33 Hz, 1 H) 8.08 (d, J = 8.06 Hz, 1 H).) A mixture of 2-(5-bromofuran-2-yl)-3-hydroxy-4H-chromen-4-one thus obtained (910 mg, 2.96 mmol), 2-bromoacetamide (409 mg, 2.96 mmol), potassium carbonate (1229 mg, 8.89 mmol), and dimethylformamide (30 ml) was stirred for 6 hours at 80°C. The solution was cooled and extracted with ethyl acetate. The combined organic layers were washed with water, dried with sodium sulfate, filtered and concentrated. Toluene was added and evaporated repeatedly until dry crystals of crude product were formed. The resulting material was purified by column chromatography, first eluting impurities with 100% ethyl acetate, followed by 10% methanol in methylene chloride to provide CB7993113 as a yellow solid (900 mg, 83% yield). 1H NMR (399 MHz, DMSO-d6) δ ppm 4.60 (s, 2 H) 6.98 (d, J = 3.66 Hz, 1 H) 7.39 (br. s, 1 H) 7.50 (t, J = 7.33 Hz, 1 H) 7.67–7.78 (m, 3 H) 7.81 (d, J = 7.33 Hz, 1 H) 8.08 (d, J = 8.06 Hz, 1 H). (ESI) found 363.9 [M + H]+.
Chemical Synthesis of CH223191 (Supplemental Fig. S1).
A solution containing 4-amino-2′,3-dimethylazobenzene (602 mg, 2.67 mM), 1-methyl-1H-pyrazole-5-carboxylic acid (372 mg, 2.95 mmol,), N,N-diisopropylethyl amine (1.4 ml, 8.04 mmol), and bromotripyrrolidinophosphonium hexafluorophosphate (1.857 g, 3.98 mmol, 1.5 Eq) in 10 ml of 1,2-dichloroethane was heated via microwave irradiation in two equal batches at 120°C for 22 minutes. The reaction mixture was reduced in vacuo, taken up in ethyl acetate (100 ml), and washed using 5% K2HPO4 (100 ml), saturated NaHCO3 (100 ml), and brine (100 ml). The isolated organic phase was dried (Na2SO4), filtered, and concentrated under reduced pressure to give a dark brown solid. The residue was purified twice by rapidly stirring in the minimal amount of warm CH2Cl2 and adding hexanes to precipitate the title compound (681 mg, 76%) as an amorphous tan solid. 1H NMR (300 MHz, CDCl3) δ ppm 8.21 (d, J = 8.7, 1 H), 7.86 (dd, J = 8.7, 2,1, 1 H), 7.82 (br s, 1 H), 7.64 (br s, 1 H), 7.61 (br s, 1 H), 7.54 (d, J = 2.1, 1 H), 7.39–7.32 (m, 2 H), 6.68 (d, J = 2.1, 1 H), 4.25 (s, 3 H), 2.73 (s, 3 H), 2.44 (s, 3 H). 13C NMR (100 MHz, CDCl3) δ ppm 157.9, 150.7, 149.9, 138.1, 137.8, 137.4, 135.2, 131.3, 130.9, 129.0, 126.4, 124.8, 122.6, 122.2, 115.4, 106.6, 39.5, 17.9, 17.6. LCMS (C18): tr(min) = 2.01, (ESI) found 334.2 [M + H]+.
Cell Culture
H1G1.1c3 cells were generously provided by M. S. Denison (University of California, Davis, CA) and maintained as previously described (Nagy et al., 2002). Cultures of H1G1.1c3 cells were maintained in selective medium consisting of DMEM (Mediatech, Manassas, VA) supplemented with 10% bovine growth serum (HyClone, Logan, UT), 2 mM l-glutamine (Mediatech), 5 μg/ml Plasmocin (InvivoGen, San Diego, CA), and 968 mg/l G-418 sulfate (American Bioanalytical, Natick, MA) in a 37°C humidified incubator in a 5% CO2 atmosphere. This murine hepatoma cell line contains a stable enhanced green fluorescent protein (EGFP) reporter construct regulated by AHR response elements (AHREs) derived from the CYP1A1 promoter.
ER–, PR–, HER– BP1 cells were generously provided by Dr. J. Russo (Fox Chase Cancer Center, Philadelphia, PA). BP1 cells were maintained in phenol red-free DMEM-F/12 medium (Mediatech) containing 5% equine serum (Sigma-Aldrich), 20 ng/ml of human recombinant epidermal growth factor (Life Technologies, Grand Island, NY), 0.5 μg/ml hydrocortisone (Sigma-Aldrich), 2 mM l-glutamine, 100 IU penicillin per 100 μg/ml streptomycin (Mediatech), 10 μg/ml insulin (Sigma-Aldrich), and 5 μg/ml Plasmocin. SUM149 cells were graciously provided by Dr. S. Ethier of Wayne State University (Detroit, MI), who isolated them from a primary inflammatory invasive ductal mammary carcinoma. Cells were maintained in Ham’s F-12 medium (Mediatech) containing 5% fetal bovine serum (Sigma-Aldrich), 0.5 μg/ml hydrocortisone, 2 μM l-glutamine, 100 IU penicillin per 100 μg/ml streptomycin, 10 μg/ml insulin, and 5 μg/ml Plasmocin. Hs578T-cell culture was described previously (Yang et al., 2008). BP1, Hs578T, and SUM149 cells were cultured and assayed at 37°C in a humidified incubator in a 5% CO2 atmosphere and grown as adherent monolayers at a maximum of 80% confluency.
Molecular Modeling/Predicting AHR Ligands
Commercial compound libraries were used for shape- and electrostatics-based comparisons. Databases of 445,418 compounds from ChemBridge Corporation (San Diego, CA) and 731,288 compounds from Enamine were used for this scaffold-hopping approach.
The structural file containing a representative flavonoid substructure, 4-oxo-2-phenylchroman-3-yl methylcarbamate, was expanded into a three-dimensional conformer database using OMEGA v.2.2.1 (OpenEye Scientific Software, Santa Fe, NM) (Hawkins and Nicholls, 2012), allowing an energy window of 8 kcal/mol above ground state, and an msd cutoff of 0.8 Å per the method described in Hawkins et al. (2007). The two lowest energy conformers were selected and used for subsequent studies.
In a similar way, three-dimensional conformer libraries of the Enamine and ChemBridge collections were generated. No limitation in terms of maximum number of conformers was set. To speed this computation, fragment libraries of each were pregenerated using the program makefragmentlib. The sample flavonoid conformers were compared against the Enamine and ChemBridge conformer databases using Rapid Overlay of Chemical Structures (ROCS; OpenEye). The highest scoring overlaps from ROCS were then subjected to electrostatic overlap comparison using EON (OpenEye). For hit-list ranking, the electrostatic Tanimoto combo (ET) score was used. This is the sum of the shape Tanimoto and the Poisson-Boltzmann electrostatic Tanimoto. The EON hit lists thus generated were merged into two separate structure files (Enamine and ChemBridge) using Pipeline Pilot (Accelrys, San Diego, CA) and each were sorted on the basis of the electrostatic Tanimoto combo score (ET_combo). An order list was thus generated consisting of the top 98 hits from ChemBridge and top 99 hits from Enamine. A summary of the computed properties of this library is presented in Supplemental Table S1.
High-Throughput AHR Reporter Assay
A previously described AHR reporter assay (Nagy et al., 2002) was adapted for high-throughput screening. To each well of a 384-well plate 105 H1G1.1c3 cells were added in selective medium and incubated at 37°C for 24 hours. Culture medium was replaced with nonselective medium consisting of MEMα (Life Technologies) supplemented with 10% BGS (HyClone/GE Healthcare Life Sciences, Logan, UT) and 2 mM l-glutamine (Mediatech) prior to application of the test compounds. β-NF was used as a positive control for induction of AHR reporter activity. A β-NF standard curve was generated by applying vehicle (0.1% DMSO) or final concentrations of 10−10 to 10−5 M β-NF to the cultures, with each concentration applied to 24 wells. To assess potential AHR agonist activity, vehicle or 2 μl of each of the 197 chemicals in DMSO diluted 1:10 in media (10−9 to 10−5 M, final concentration) chosen from commercial libraries were applied to triplicate wells. To screen for AHR antagonist activity, cells in 384-well plates were treated with β-NF (10−7 M) and 2 μl vehicle or test compounds at the indicated concentrations. The plates were incubated at 33°C for up to 72 hours. EGFP fluorescence was analyzed at 24, 48, and 72 hours using a fluorometric plate reader (SpectraFluor Plus; Tecan, Männedorf, Switzerland). Percent induction/inhibition of β-NF–induced AHR activity was calculated by subtracting the background fluorescence of untreated cells from all experimental values and dividing the background-adjusted fluorescence in the sample plus β-NF wells by the background-adjusted fluorescence in the β-NF–alone wells and then multiplying by 100.
Following the final fluorescence reading, the CellTitre-Blue cell viability assay was used to determine toxicity as per the manufacturer’s instructions (Promega, Madison, WI). Toxicity was calculated by dividing background-subtracted fluorescent readings of samples by those of untreated cells. Compounds consistently inducing a ≥10% increase in cell death, relative to baseline levels (generally 2–5%), were excluded from further studies.
Antagonism of TCDD-Induced AHR Activity
H1G1.1c3 cells (6 × 105) were added to each well of a 96-well plate in selective medium and incubated at 37°C for 24 hours. Culture medium was replaced with nonselective medium prior to application of the test compounds. Eight culture wells were treated with 0.5% DMSO (vehicle) or 10−7 to 5 × 10−11 M TCDD with or without 0.5% DMSO or 5 × 10−5 to 10−9 M CB7993113. Plates were incubated at 33°C for 24 hours, and EGFP fluorescence was analyzed using a Synergy 2 Multi-Mode Plate Reader (BioTek Inc., Winooski, VT). For each plate, specific fluorescence was calculated by subtracting the background fluorescence in untreated wells from the fluorescence in vehicle- or compound-treated wells. Percent induction was calculated by dividing by the specific fluorescence in the CB7993113-plus-TCDD wells by the specific fluorescence in cultures treated with vehicle-plus-TCDD for each given TCDD concentration and multiplied by 100. This experiment was repeated six times.
Efficacy and Potency Analyses
To determine 50% inhibitory concentration (IC50) values, 5 × 104 H1G1.1c3 cells were added to each well of a 96-well plate and cultured as above. For each experiment, a TCDD standard curve (to normalize fluorescence readings between experiments) was prepared by applying TCDD (10−10 to 10−6 M) or vehicle (DMSO, 0.5%), with each concentration applied to six wells. H1G1.1c3 cells were treated with vehicle (DMSO) or titered doses of CB7993113 or CH223191 (10−9 to 10−5 M) immediately following addition of vehicle (to assay potential agonist activity), 10−7 M 7,12-dimethylbenz[a]anthracene or 10−7 M β-NF (to assay antagonist activity) (six wells/condition). Plates were incubated at 33°C for 24 hours and EGFP fluorescence was analyzed using a Synergy 2 plate reader. Percent AHR induction was calculated as described above. Following the final fluorescence reading, the MTT cell viability assay was used to determine toxicity (Sigma-Aldrich). Briefly, after a 48-hour incubation, 10 μl MTT (5 mg/ml) was added to each well and incubated at 37°C for 4 hours. Formazan crystals were solubilized by the addition of 100 μl/well DMSO and incubation at 37°C for 2 hours. Absorbance at 570 nm was quantified using a Synergy 2 plate reader. Toxicity was calculated by subtracting the average absorbance in the untreated wells from the average absorbance in the experimental wells.
Homology Modeling/Theoretical AHR Binding
The X-ray structure of HIF-2α PAS-B domain cocrystallized with N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine was chosen as the template for model-building (Scheuermann et al., 2009). The structure was downloaded from the Protein Data Bank (PDB) (Berman et al., 2002). The PDB identifier (PDB ID) of the structure is 4GHI. MODELER (Fiser and Sali, 2003) was used to model the human AHR PAS-B domain. Only nonidentical residues and regions around the gaps were optimized. To assure that the binding site would not collapse in the process of model building, CB7993113 was aligned inside the homology model, on the basis of the position of the double ring of the ligand in the template and considering the polarity/hydrophobicity of the environment around it. The ligand-protein complex was then minimized using the CHARMM potential to avoid possible steric clashes (Brooks et al., 1983). The computational solvent mapping algorithm FTMap was used to predict the most probable CB7993113-AHR PAS-B domain–binding position. This method places small molecular probes of various sizes and shapes on a dense grid around the protein, finds favorable positions using empirical energy functions, clusters the conformations, and ranks the clusters on the basis of the average energy. All ligands and crystallographic water molecules were removed prior to mapping, and the probes were initially distributed over the entire protein surface (including all cavities) without any assumptions about the binding site. The regions that bind multiple low energy probe clusters [consensus cluster (CC) sites], identified the most important ligand-binding sites. The hot spots were ranked in terms of the number of overlapping probe clusters contained. The consensus cluster with the highest number of probe clusters was ranked first as CC1 and nearby consensus clusters within 7 Å were also joined with CC1 to form the predicted ligand-binding site.
Only nonidentical residues and regions around the gaps were optimized and the predicted ligand-protein complex was then minimized using the CHARMM potential (Brooks et al., 1983) to avoid potential steric clashes. The FTMap algorithm (http://ftmap.bu.edu)(Brenke et al., 2009) was used to predict the AHR PAS-B domain-binding hot spots.
[A model of the human AHR PAS-B domain in complex with CB7993113 is provided in Supplemental Data file CID1.pdb. A model of the human AHR PAS-B domain in complex with CB7993113, together with computational solvent mapping (http://ftmap.bu.edu) of the PAS-B domain, is provided in Supplemental Data file CID.pdb.]
In Vitro Protein Synthesis and Competitive-Binding Assay
Murine and human AHR proteins were synthesized from AHR expression constructs (pSportMAHR or pSportAHR2, respectively, gifts of Dr. C. Bradfield, University of Wisconsin, Madison, WI) (Burbach et al., 1992; Dolwick et al., 1993) using a TnT Quick Coupled reticulocyte lysate system (Promega). The ability of CB7993113 or CH223191 to compete with [3H]TCDD (35 Ci/mmol; Chemsyn Science Laboratories, Lenexa, KS) for binding to human or mouse AHR was measured by velocity sedimentation on sucrose gradients in a vertical tube rotor as described earlier (Karchner et al., 2006). Briefly, single TnT reactions (50 μl) were diluted 1:1 with MEEDMG buffer (25 mM MOPS, 1 mM EDTA, 5 mM EGTA, 0.02% NaN3, 1 mM DTT, 20 mM molybdate, 10% (v:v) glycerol, pH 7.5) and incubated overnight at 4°C with [3H]TCDD (2 nM) ± DMSO or competitor (10 μM final concentration, dissolved in DMSO). Incubations were applied to 10- to 30%-sucrose gradients and analyzed as described (Karchner et al., 2006). Nonspecific binding was determined by reactions containing an empty vector [unprogrammed lysate].
Human AHR–Driven Reporter Assay
BP1 cells (2 × 104/well) were plated in 24-well plates and allowed to adhere overnight. Cells were cotransfected with 0.1 μg of the pGudluc reporter plasmid, 0.1 μg CMV green, and 0.5 μg of either pcDNA or an Ahrr expression plasmid using Lipofectamine 2000 (Life Technologies), as we previously described (Yang et al., 2008). Cultures were incubated for 3 hours and then dosed with vehicle (DMSO, 0.1%), CH223191 (0.01–1 μM), or CB7993113 (1–20 μM). After 1 hour, the transfection medium was replaced, the cultures were redosed, and then incubated for 24 hours. Cells were harvested in Glo Lysis Buffer (Promega) and luciferase activity was determined with the Bright-Glo Luciferase System per the manufacturer’s instructions (Promega). Luminescence and fluorescence values were determined using a Synergy 2 plate reader. To calculate “fold change from naïve” the luminescence value was divided by the fluorescence value for each sample.
Human Peroxisome Proliferator–Activated Receptor Gamma– and Cytomegalovirus-Driven Reporter Assays
Cos-7 cells were transiently transfected with vectors containing human peroxisome proliferator–activated receptor gamma (PPARγ, PPARG1) (kindly provided by V. K. Chatterjee, University of Cambridge, Cambridge, UK) (Gurnell et al., 2000) and human RXRA (plasmid 8882; Addgene, Cambridge, MA) with PPRE ×3-TK-luc (plasmid 1015; Addgene) and cytomegalovirus (CMV)–eGFP-reporter constructs using Lipofectamine 2000 (Life Technologies). Transfected cultures were incubated for 3 hours. The medium was replaced with antibiotic-free DMEM with 5% fetal bovine serum and the cultures were incubated overnight. Cultures received no treatment (naïve) or were treated with vehicle (Vh; DMSO, 0.1%) or with rosiglitazone (1 μM) and Vh, CH223191 (10 μM), or CB7993113 (10 μM), and incubated for 24 hours. Cells were lysed in Glo Lysis Buffer (Promega). Lysates were transferred to a 96-well plate to which Bright Glo Reagent (Promega) was added. Luminescence and fluorescence were determined using a Synergy 2 plate reader. PPARγ-specific luminescence was normalized to the GFP fluorescence in the same well. For the CMV-driven reporter assay, human mammary tumor cells (BP1) were transfected with CMV-driven–eGFP-reporter plasmid (>80% transfection efficiency) and treated with CH223191 or CB7993113 as described above. GFP fluorescence was assayed 24 hours later. Constitutive CMV-reporter activity was calculated relative to levels seen in naïve cultures.
Mouse AHR Immunoblotting
H1G1.1c3 cells were plated at 2 × 106 cells in T75 flasks and allowed to adhere overnight. Cells were treated with CB7993113 (10 μM), vehicle (DMSO, 0.1%), or left untreated for 1 hour, followed by DMBA (10−7 M) treatment for 30 minutes. Nuclear and cytoplasmic cell extracts were prepared using the Nuclear Extract Kit (Active Motif, Carlsbad, CA) per the manufacturer’s instructions. Protein concentration was quantified using Protein Assay Reagent (Bio-Rad, Hercules, CA). Protein (30 μg) was resolved on 12% SDS-polyacrylamide gels and transferred to 0.2-μm nitrocellulose membranes (Bio-Rad). Membranes were probed with the following primary antibodies: mouse anti-AHR (Pierce, Rockford, IL), rabbit anti-Lamin-A/C (Cell Signaling Technologies, Danvers, MA), and mouse anti-α-tubulin (EMD Millipore, Billerica, MA). Immunoreactive bands were detected using horseradish peroxidase (HRP)–conjugated secondary antibodies [goat anti-rabbit (Bio-Rad), goat anti-mouse (Pierce)], and ECL substrate.
Human CYP1B1 mRNA Expression
BP1 cells (3 × 106) were plated in T225 flasks and allowed to adhere overnight. Cultures were dosed with CB7950998 (10 μM) or vehicle (DMSO, 0.1%) and incubated for 24 hours. mRNA was extracted using RNeasy Plus Mini Kit (Qiagen, Valencia, CA). cDNA was prepared from total RNA using the GoScript Reverse Transcription System (Promega), with a 1:1 mixture of random and oligo (dT)15 primers. All real-time qPCR reactions were performed using the GoTaq RT-qPCR Master Mix System (Promega). Validated primers were purchased from Qiagen: human CYP1B1–QT00209496 and human RRN18S–QT00199367. Real-time qPCR reactions were performed using a 7900HT Fast Real-Time PCR instrument (Applied Biosystems, Carlsbad, CA): Hot-Start activation at 95°C for 2 minutes, 40 cycles of denaturation (95°C for 15 seconds), and annealing/extension (55°C for 60 seconds). Relative gene expression was determined using the Pfaffl method (Pfaffl, 2001) with the threshold value for RRN18S for normalization. The Cq value from untreated cultures was used as the reference point.
Invasion and Migration Assays
For Matrigel assays, BP1 cells (3 × 105 cells/well) were plated in six-well plates and allowed to adhere overnight. Cells were pretreated with vehicle (DMSO), CH223191 (10 μM), CB7993113 (5 μM), or left untreated for 24 hours. Cells were harvested and prepared as a single cell suspension for addition to the Matrigel branching assay. Matrigel basement membrane matrix (BD Biosciences, Bedford, MA) was diluted to 6.3 mg/ml. Matrigel solution (200 μl) was added to a 24-well plate and solidified at 37°C for 30–45 minutes to form a base layer. Single-cell suspensions containing 2 × 104 pretreated BP1 cells in 10 μl serum-free media were mixed with 190 μl of Matrigel containing 0.1% vehicle or 5–10 μM CB7993113. Vehicle or CB7993113 was also added to the Matrigel top layer and the layer allowed to solidify at 37°C for 30–45 minutes. Complete medium (0.5 ml) containing vehicle (DMSO, 0.1%) or CB7993113 (5 or 10 μM) was added on top of solidified Matrigel and was replaced with fresh dosing solution every other day. Colony morphology was captured using a Zeiss Axiovert 200 M microscope. Images were captured using a Nikon Coolpix4300 digital camera.
For “scratch-wound” assays (Li et al., 2013), Hs578T or SUM149 cells (500,000 cells/well) were plated in six-well plates and allowed to grow until 100% confluent. Cultures were scratched with a pipette tip, left untreated, or treated with vehicle, 10 μM CH223191, or 10 μM CB7993113, and allowed to regrow for 48 hours. Cultures were photographed using a Nikon CoolPix 4300 attached to a Zeiss Telaval31 inverted microscope at 0, 24, and 48 hours.
In Vivo Studies
Male, 6–8 week old C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at Boston University. For pharmacokinetic studies, 13-week-old mice (four/group) were treated with 50 mg/kg CB7993113 by oral gavage or intraperitoneal injection. Serum was collected 4, 8, and 16 hours after treatment and from untreated mice. Serum samples were analyzed by liquid chromatography–tandem mass spectrometry for CB7993113 at Apredica/Cyprotex (Watertown, MA). For DMBA-induced bone marrow toxicity studies, mice (six per group) were injected intraperitoneally with vehicle (100 μl vegetable oil), CH223191 (50 mg/kg), or CB7993113 (50 mg/kg) 24 hours and 1 hour before dosing with 200 μl sesame oil or 50 mg/kg DMBA by oral gavage. Mice were euthanized 48 hours after DMBA treatment. Liver was snap frozen for mRNA analysis. Bone marrow was flushed from femurs and tibiae and red blood cells (RBCs) were removed using ACK Lysing Buffer (BioWhittaker, Lonza, Allendale, NJ).
For gene expression analyses, mRNA was extracted from liver tissues using the RNeasy Plus Mini Kit (Qiagen) and concentrated, if required, using the RNeasy MinElute Cleanup Kit (Qiagen). cDNA was prepared and real-time qPCR was carried out, as described above. Primer sequences for murine Cyp1a1 and murine Gapdh were previously described (Xu and Miller, 2004) and were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Relative gene expression was determined using the Pfaffl method (Pfaffl, 2001), with the threshold value for Gapdh used for normalization. The Cq value from untreated animals was used as the reference point.
For flow cytometry analyses, RBC-depleted bone marrow samples were treated with Fc-blocking solution (BD Biosciences, San Jose, CA) and suspended in phosphate-buffered saline (Life Technologies) supplemented with 2% fetal bovine serum (Gemini, West Sacramento, CA) for 15 minutes at 4°C. Samples were subsequently surface-stained with a cocktail of IgM-, CD24-, CD43-, and B220-specific antibodies, a cocktail of Gr-1-, CD3-, NK1.1-, CD11b-, and B220-specific antibodies, or their appropriate isotype-matched antibodies for 30 minutes at 4°C in the above staining buffer. All flow cytometry antibodies were obtained from BD Biosciences or eBiosciences (San Diego, CA). Samples were analyzed on a BD LSR II instrument. Post-acquisition analysis was performed using FlowJo analysis software (TreeStar).
Statistical Analysis
Statistical analyses were performed with GraphPad Prism (La Jolla, CA). Data are presented as means ± S.E. where applicable. One-tailed Student’s t tests and one-way analysis of variance with Dunnett, Tukey-Kramer, or Newman-Keuls post-hoc were used to determine significance.
Results
Virtual Screening and Generation of Focused Libraries.
Development of modulators that can alter AHR function is of growing importance given the established role that the AHR plays in mediating toxic effects of environmental chemicals and the emerging role of the AHR in inflammatory diseases, autoimmunity, hematopoiesis, and cancer. To this end, known AHR-ligand structures and shape-based modeling were used to predict new AHR ligands and to generate targeted libraries of small molecules with substructures similar to those of known natural and anthropogenic AHR ligands. Noting the prevalence of flavonoid-like molecules in known AHR ligands, a generic substructure, 4-oxo-2-phenylchroman-3-yl methylcarbamate (Supplemental Fig. S2), was selected as a basis to search for shape- and electrostatic-based similarities in commercial sets of compounds from two vendors, Enamine and ChemBridge, Inc.
Databases consisting of 445,418 and 731,288 small molecules from ChemBridge and Enamine, respectively, were scrutinized for three-dimensional conformers similar to the flavonoid substructure using the OMEGA algorithm. The three-dimensional conformer set generated from this analysis was then compared with generic flavonoid substructures by ROCS shape comparison. Subsequently, compounds that ranked highest in structural overlap from the ROCS comparison were subjected to an electrostatic analysis using EON electrostatic comparison and were reranked according to their electrostatic Tanimoto (ET) score. Focused libraries of the top 197 small-molecule hits from the Enamine and ChemBridge databases were generated (Supplemental Table S1) and tested for AHR modulating activity in vitro.
In Vitro Identification of Novel AHR Antagonists.
A murine reporter–based bioassay (Nagy et al., 2002) was adapted for semiautomated, high-throughput screening of the aforementioned focused library for AHR agonists and antagonists. This assay utilizes the murine hepatoma cell line H1G1.1c3 that is stably transfected with an AHR-responsive EGFP reporter to measure AHR transcriptional activity. To assess AHR agonist activity, each experimental compound at 1 μM or 5 μM concentration was added alone to H1G1.1c3 cells. For AHR-antagonist screening, each experimental compound was added immediately prior to addition of 10−7 M β-NF, a well-described AHR agonist. EGFP expression was assayed 24, 48, and 72 hours later.
Of the 197 compounds screened, 31 compounds exhibited agonist activity, as defined by a ≥25% increase in reporter activity at 1 and 5 μM and at all three time points when added alone to the reporter assay (data not shown). Four compounds demonstrated significant antagonist activity as defined by a ≥25% reduction in β-NF responses at 1 and 5 μM and at all three time points (Fig. 1, arrows). The structures of the putative antagonist compounds and, for comparison, those of other previously described AHR antagonists, CH223191 (Kim et al., 2006), StemRegenin (SR1) (Boitano et al., 2010), GNF351 (Smith et al., 2011), and 6, 2′,4′trimethoxyflavone (Zhao et al., 2010) are provided in Fig. 2. Note the relative structural dissimilarity of CB7993113 when compared with most of the other “pure” AHR antagonists.
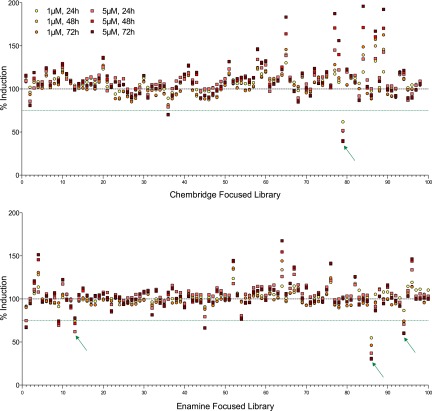
An in vitro AHR bioassay identifies novel AHR antagonists. H1G1.1c3 cells were left untreated or treated in triplicate wells with vehicle (0.1% DMSO), 1 or 5 μM concentrations of 197 compounds from the ChemBridge and Enamine focused libraries immediately prior to addition of 10−7 M β-NF. Cells were cultured at 33°C for 72 hours. EGFP fluorescence was analyzed at 24, 48, and 72 hours. Data are presented as the average of three wells, and are calculated as the percent induction, with 100% set at levels seen with 10−7 M β-NF alone (dashed line). Data points representing AHR antagonist hits, defined as those compounds with an average induction of ≤75% of β-NF treated cells (green dotted line) for all measured endpoints, are indicated with arrows. Data are representative of a single set of high-throughput screen experiments.
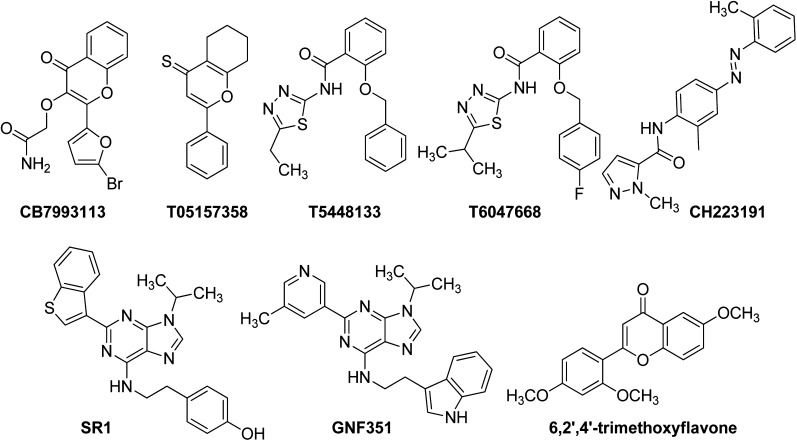
Structures of antagonist hit compounds. Structures of antagonist hits are presented along with their chemical identification numbers. The structure of the previously described AHR antagonists CH223191, SR1, GNF351, and trimethoxyflavone are presented for comparison.
Several compounds (e.g., α-naphthoflavone, galangin) exhibit partial AHR agonism (Santostefano et al., 1993; Zhang et al., 2003; Dvorak et al., 2008). Partial AHR agonists may appear to inhibit the AHR in bioassays since they compete with high-efficacy ligands for AHR binding but cannot fully activate the receptor. None of the four putative AHR antagonists discovered in this screen induced AHR-driven reporter activity in the high-throughput assay, even at the highest concentration tested (10 μM) (data not shown and Figs. 3 and and4),4), indicating that they are not partial agonists.
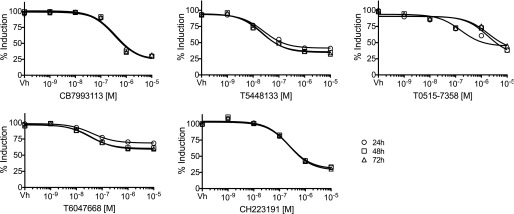
Hit compound characterization led to selection of lead compound, CB7993113. H1G1.1c3 cells were treated with vehicle (0.1% DMSO) or 10−9 to 10−5 M AHR antagonists immediately prior to stimulation with 10−7 M β-NF. Fluorescence (AHR-dependent reporter activity) was assayed 24, 48, and 72 hours later as in Fig. 1. Dose response curves were generated utilizing a three-parameter dose-response curve model in GraphPad Prism with a standard Hill slope of –1. Data are presented as the average of three wells and are calculated as the percent induction with 100% set at levels seen with 10−7 M β-NF alone.
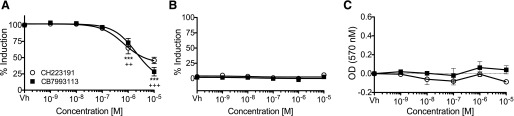
CB7993113 blocks DMBA-induced AHR-dependent reporter activity. (A) H1G1.1c3 cells were treated with vehicle (0.1% DMSO) or 10−9 to 10−5 M CB7993113 or CH223191 immediately prior to stimulation with 10−7 M DMBA and culture at 33°C. EGFP fluorescence (AHR reporter activity) was analyzed 24 hours later. (B) AHR agonist activity was measured by treating H1G1.1c3 cells with vehicle or 10−9 to 10−5 M CB7993113 or CH223191. Cells were cultured at 33°C and EGFP fluorescence was analyzed after 24 hours. Dose-response curves were generated utilizing a three-parameter dose-response curve model in GraphPad Prism with a standard Hill slope of –1 (A and B). (C) Cellular toxicity after the above treatment was measured by the MTT reduction assay 48 hours after addition of compounds. Data are presented as means ± S.E. of three independent experiments. (One-way analysis of variance with Dunnett’s post-test, ***P < 0.001 for CB7993113 compared with vehicle, ++P < 0.01 or +++P < 0.001 for CH223191 compared with vehicle.)
The potency and efficacy of the four discovered AHR antagonists were compared with CH223191 using the high-throughput H1G1.1c3 assay (Fig. 3). At maximal concentrations, the four discovered AHR antagonists inhibited between 40 and 70% of the β-NF–induced EGFP signal, with CB7993113 exhibiting the greatest efficacy (Table 1). IC50 values ranged from 0.023 to 1.85 μM (Table 1). None of the compounds were toxic even at the highest dose tested, as defined by a ≥10% increase in dead cells after 48 hours of exposure. All of the compounds were within Lipinski’s guidelines for drug-likeness (Lipinski et al., 2001). Interestingly, the low IC50 (i.e., high potency) exhibited by T0515-7358 (0.02 μM) was not reflected in a significantly higher efficacy (68% inhibition) than the other discovered compounds. This is not an unusual finding since the potency, in part a function of ligand-receptor affinity, does not necessarily correlate directly with efficacy, a function of the biologic response to the ligand. This result further highlights differences between AHR antagonists.
TABLE 1
AHR antagonist hit compounds
Vendor catalog numbers, chemical names, molecular weights, and Log P (partition coefficient) values are presented. The c Log P values were determined using Pipeline Pilot (Accelrys). Maximum percent inhibition values were determined from data presented in Fig. 3 using 50 μM concentrations of the respective compounds and assaying on day 3. IC50 values were determined from the dose-response curves generated in Fig. 3 using GraphPad Prism.
| Chemical ID | Chemical name | MW | Log P | Max % inhibition | IC50 |
|---|---|---|---|---|---|
| μM | |||||
| CB7993113 | 2-{[2-(5-Bromo-2-furyl)-4-oxo-4H-chromen-3-yl]oxy}acetamide | 364.1 | 1.03 | 70.3 | 0.33 |
| T0515-7358 | 2-Phenyl-5,6,7,8-tetrahydro-4H-chromene-4-thione | 242.3 | 3.88 | 67.7 | 0.02 |
| T5448133 | 2-(Benzyloxy)-N-(5-ethyl-1,3,4-thiadiazol-2-yl)benzamide | 339.4 | 3.91 | 61.6 | 1.85 |
| T6047668 | 2-((4-Fluorobenzyl)oxy)-N-(5-isopropyl-1,3,4-thiadiazol-2-yl)benzamide | 371.4 | 4.60 | 40.3 | 0.04 |
| CH223191 | (E)-1-Methyl-N-(2-methyl-4-(o-tolyldiazenyl)phenyl)-1H-pyrazole-5-carboxamide | 333.4 | 4.85 | 69.6 | 0.24 |
Compounds CB7993113 and T0515 exhibited the greatest efficacy (maximum percent inhibition). As anticipated on the basis of calculated lipophilicity (c Log P) values, CB7993113 was qualitatively observed to be more soluble in phosphate-buffered saline than the other compounds and therefore was prioritized for further study.
In a separate series of experiments, head-to-head comparisons were made between CH223191 and lead compound CB7993113, using a prototypic, toxic AHR ligand, DMBA (Teague et al., 2010), which also was used in in vivo bone marrow toxicity assays (see below). CB7993113 was slightly less potent than CH223191 as measured by reduction of DMBA-induced AHR reporter activity in H1G1.1C3 cells (IC50s 2.1 μM versus 0.7 μM, respectively; Fig. 4A). However, CB7993113 was more effective than CH223191 at reducing DMBA-induced reporter activity under these conditions (46 versus 28% respectively). Again, neither CB7993113 nor CH223191 acted as a partial agonist at doses ranging from 1 nM to 10 μM (Fig. 4B) and neither compound exhibited cytotoxicity as determined in an MTT assay (Fig. 4C). Furthermore, neither compound affected reporter activity mediated by an unrelated nuclear receptor, PPARγ, or reporter activity driven by a constitutive CMV promoter (Supplemental Fig. 3). Collectively, these data indicate that at least one predicted new compound, CB7993113, specifically antagonizes murine AHR transcriptional activity, does not exhibit partial AHR agonist activity, and is not cytotoxic, even at relatively high concentrations.
To begin to assess whether CB7993113 is probably a competitive or allosteric inhibitor, titered concentrations of β-NF (10−8 to 10−5 M) or TCDD (10−13 to 10−9 M) and CB7993113 (10−7 to 10−5 M) were added to H1G1.1c3 cells and the percent induction of TCDD-induced, AHR-driven reporter activity assayed 24 hours later. If CB7993113 is an allosteric inhibitor, then little or no change in the potency (EC50) of an AHR agonist should be seen after addition of titered concentrations of the inhibitor. In contrast, if CB7993113 is a competitive inhibitor, then agonist EC50 should increase as increasing concentrations of inhibitor are added. Indeed, the β-NF and TCDD titration curves shifted to the right (increasing EC50) as the concentration of CB7993113 was increased (Supplemental Fig. 4). These data are consistent with competitive inhibition.
A competitive AHR antagonist would be expected to bind directly to the AHR and, at a minimum, block agonist-induced nuclear translocation. Therefore, the capacity of CB7993113 to directly bind to in vitro translated murine AHR protein was determined in a [3H]TCDD competitive binding assay. CH223191 was included as a positive control. As predicted from the calculated IC50s (Figs. 3 and and4,4, Table 1), 10 μM CB7993113 and CH223191 were comparable in their ability to block [3H]TCDD-AHR binding (Fig. 5A).
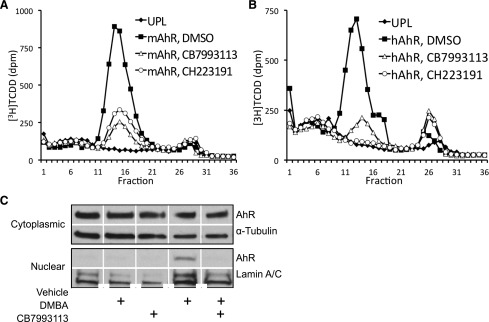
CB7993113 directly binds murine AHR protein and blocks AHR nuclear translocation. In vitro-expressed murine AHR (mAHR) (A) or human AHR (hAHR) (B) protein was incubated with 2 nM [3H]TCDD in the presence of vehicle (DMSO), 10 μM CH223191, or 10 μM CB7993113 for 12 hours. Quantification of [3H]TCDD-AHR binding was analyzed by velocity sedimentation on sucrose gradients. Data are representative of two independent experiments. (C) H1G1.1c3 cells were treated with vehicle (0.1% DMSO) or 10 μM CB7993113 for 1 hour, followed by treatment with 10−7 M DMBA for 30 minutes. Cytoplasmic and nuclear extracts were isolated and analyzed for AHR, α-tubulin, and lamin A/C content by immunoblotting. Data are representative of four independent experiments.
Several AHR ligands, including at least one AHR competitive antagonist (Boitano et al., 2010), exhibit species-specific AHR-binding characteristics. Therefore, the ability of CB7993113 to bind human AHR also was assessed using in vitro translated human AHR. The ability of CB7993113 to inhibit the binding of [3H]TCDD to human AHR was similar to that seen with murine AHR, whereas CH223191 was slightly more effective as an inhibitor of [3H]TCDD binding to human AHR compared with mouse AHR (Fig. 5B).
As would be expected from a competitive antagonist, 10 μM CB7993113 completely prevented DMBA-induced AHR nuclear translocation (Fig. 5C). Partial inhibition of AHR nuclear translocation was seen in two experiments with as little as 1 μM CB7993113 (data not shown). Consistent with our findings with the AHR-driven reporter assay, 10 μM CB7993113 alone did not induce nuclear translocation of the AHR protein and thus was not acting as a partial AHR agonist (Fig. 5C). These data collectively indicate that CB7993113 is probably a competitive AHR antagonist and that it blocks AHR nuclear translocation and, thereby, transcriptional activity.
Homology Modeling of CB7993113-AHR Binding.
To begin to visualize the orientation of this putative competitive inhibitor in the binding pocket of the human AHR PAS-B domain, the X-ray structure of the HIF-2α PAS-B domain, cocrystallized with the small antagonist N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine (4GH1) (Scheuermann et al., 2013) was selected as the template for model building. The recent crystal structure of the mouse PAS1 (PAS-A) domain (PDB ID 4M4X) (Wu et al., 2013) indicates that the human AHR PAS-B domain begins after residue Q273, and hence AHR residues 287–390 were aligned with HIF-2α residues 244–348 (Supplemental Fig. 5). The two sequences have 27% identity and 50% similarity in this range. Although this level of sequence identity is relatively low for homology modeling, the identical residues distribute almost equidistantly along the domain with only a two-percent (three amino acids) gap. This type of distribution makes it very probable that the backbones of the two proteins are very similar. Indeed, structures are available for a number of PAS domains with similar level of sequence identity and good three-dimensional overlap.
The MODELER mapping results were used in two different ways to approximate CB7993113 docking. First, a box with 4-Å padding was created around the predicted binding site. The docking was carried out restricting consideration to this box and using the standard settings of AutoDock Vina 1.1.0 (Scripps Institute) (Trott and Olson, 2010). The 10 lowest energy binding poses were retained for each ligand. Second, for the selection of the most probable pose, the AutoDock Vina energy score and the atom densities calculated from the mapping results were included in the calculus. Probe density was defined at each point binding site as the total number of probe atoms within a 1.25-Å radius. We considered each retained ligand pose and summed the atomic densities for all heavy atoms, resulting in a measure of overlap between the pose and the probe density. The poses with low energy scores were ranked on the basis of this overlap measure, and the pose with the best overlap was selected (Kozakov et al., 2011).
Figure 6A shows the template, the PAS-B domain of HIF-2α, with the bound antagonist. The backbone of the homology model of the human AHR is very similar (Fig. 6B). There is no template for the AHR loop 371–376 and thus it was constructed by MODELER. (Note that this loop is far from the binding site and its exact structure is not important for this model.) Figure 6B also shows the probe clusters obtained by the mapping program FTMap and indicating the energetically most important regions of the binding site. We note that the sequence differences between HIF-2α and AHR results in a larger binding cavity and greater hydrophobicity of the AHR. Consequently, the site can accommodate CB7993113 in a number of conformations and the 10 lowest energy docked poses of CB7993113 show substantial variation. Figure 6C shows one of the lowest energy structures (pose 2 from AutoDock Vina) that also overlaps well with the binding hot spots. The hydrophobic part of the chromen-3 moiety of the ligand is surrounded by the side chains of F295, M340, and A367, whereas H291 and Q383 donate hydrogen bonds to the more polar regions of CB7993113. For better visibility, Fig. 6D shows only a few of the side chains that are within 5 Å of the ligand. Interestingly, the differences in the amino acids shown in the AHR yield less bulky side chains, and thus increase the size of the ligand-binding cavity relative to HIF-2α.
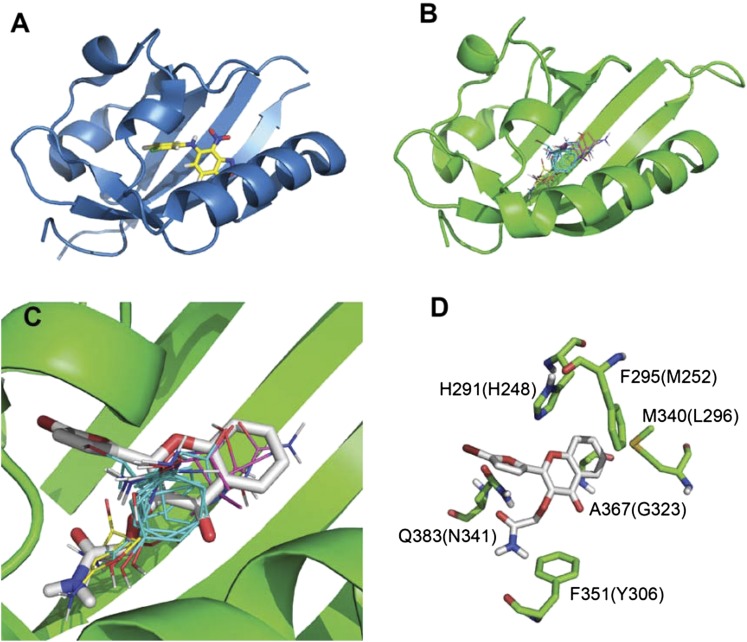
Homology modeling of CB7993113 binding to human AHR. Homology modeling on the basis of the crystal structure of ligand-bound HIF-2α was employed as described in the text. (A) Structure of ligand-bound HIF-2α. (B) A proposed model of CB7993113 docking to human AHR, including the probe clusters, obtained by the mapping program FTMap, and indicating the energetically most important regions of the binding site. (C) Hydrophobic part of the chromen-3 moiety of the ligand that is surrounded by the side chains of F295, M340, and A367. (D) A few side chains that are within 5 Å of the ligand. Residue numbers in parentheses represent the corresponding residues of HIF-2α (see Supplemental Fig. 5).
CB7993113-Mediated Inhibition of AHR-Dependent Biologic Responses.
AHR expression and activation in the absence of environmental ligands is associated with several human cancers, including breast cancer (Brooks and Eltom, 2011; Opitz et al., 2011; Schlezinger et al., 2006). It is presumed that this basal activity is mediated by endogenous AHR ligands. Therefore, it was predicted that CB7993113, like AHR repressor protein (Yang et al., 2008), would lower baseline levels of AHR-driven reporter activity and endogenous target gene expression in human breast cancer cells. Malignant triple-negative BP1 cells were used for these studies since we had previously established relatively high baseline levels of AHR (reporter) activity and AHR-dependent enforcement of high CYP1B1 levels in this invasive cell line (Yang et al., 2008). BP1 cells were transiently transfected with the pGudLuc AHR-driven reporter plasmid. As previously reported (Yang et al., 2008), cotransfection with a plasmid carrying the AHR repressor gene (Ahrr) significantly reduced baseline pGudLuc reporter activity (Fig. 7A). Similarly, treatment of pGudLuc-transfected cells with CB7993113 significantly decreased baseline AHR activity at concentrations as low as 5 μM (Fig. 7A). CB7993113 also tended to decrease endogenous CYP1B1 levels in this short-term experiment, although statistical significance was only reached at a concentration of 20 μM (Fig. 7B).
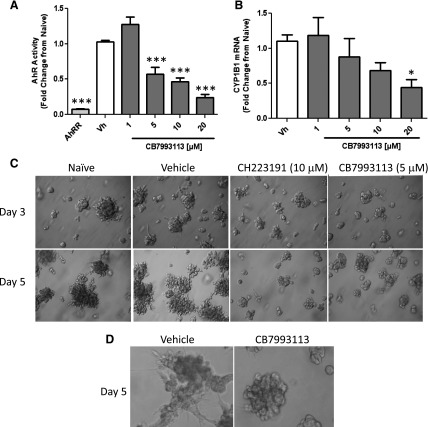
CB7993113 and CH223191 reduce the invasive phenotype of human breast cancer cell colonies in 3D Matrigel assays. (A) BP1 cells were cotransfected with AHRE-driven firefly luciferase reporter (pGudluc) and control CMV green (GFP) vectors and incubated for 3 hours. In some groups, Ahrr plasmid was cotransfected as a positive control. Cultures were left untreated or were treated with vehicle (0.1% DMSO) or 1–20 μM CB7993113. Cells were harvested 24 hours later and luciferase activity assayed. Luciferase (AHR-reporter) activity was first normalized to the GFP signal to control for transfection efficiency and then normalized to the luciferase signal obtained from untreated cells. Data are expressed as means ± S.E. obtained from three independent experiments. (***P < 0.001 compared with vehicle groups; one-way analysis of variance (ANOVA) with Dunnett’s post-test.) (B) BP1 cells were treated with vehicle (DMSO) or 1–20 μM CB7993113 for 24 hours. mRNA was extracted and analyzed by qPCR for CYP1B1 expression normalized to 18sRNA expression. qPCR was performed in duplicate. Data are expressed as means ± S.E. from three to five independent experiments. (*P < 0.05, compared with vehicle groups, ANOVA with Dunnett’s post-test.) (C) BP1 cells were treated with vehicle (0.1% DMSO), 10 μM CH223191, 5 μM CB7993113, or left untreated for 24 hours. Cells were harvested, counted, and plated in Matrigel. Representative images from three independent experiments were taken on days 3 and 5. (D) BP1 cells were treated as in (C) with vehicle or CB7993113 (10 μM). High magnification images were captured on day 5 to illustrate cell morphology. CB7993113-treated cells extracted from Matrigels were ~95% viable. No significant differences in the numbers of cells in cultures after extraction from three-dimensional cultures were seen.
Our laboratory (Trombino et al., 2000; Schlezinger et al., 2006) and others (Brooks and Eltom, 2011; Opitz et al., 2011) have postulated that AHR hyperexpression and activity facilitate malignant transformation. It would then be predicted that AHR antagonists would reverse at least some component of the malignant phenotype. The growth of immortalized cells in irregular, branching colonies in Matrigel is widely seen as a marker for invasiveness (Hughes et al., 2008). Indeed, we have demonstrated that inhibition of AHR activity with AHRR or AHR knockdown with AHR-specific small-interfering RNA blocks the formation of branching colonies in human breast cancer cell lines (S. Narasimhan et al., manuscript in preparation). Therefore, the ability of CB7993113 to alter the morphology of BP1 cell colonies in Matrigel was assessed. Untreated or vehicle-treated BP1 cell colonies exhibited the branched, irregular morphology typical of invasive cells as early as day 3 of culture (Fig. 7C). However, treatment with either 10 μM CH223191 or 5 μM CB7993113 clearly reduced the size of the colonies and their degree of branching. Higher magnification images revealed that CB7993113 tended to decrease formation of invasive cell processes (Fig. 7D).
It is possible that the morphologic changes seen in the presence of CB7993113 are, in part, a function of altered cell growth or toxicity. However, [3H]thymidine incorporation experiments in two-dimensional BP1 cultures failed to demonstrate a significant change in cell growth or viability (more than five experiments, data not shown). Furthermore, the number of viable cells recovered from 7- to 10-day-old three-dimensional Matrigel cultures, as determined by Trypan blue exclusion and visual inspection or by propidium iodide exclusion and flow cytometry, was not affected by CB7993113 treatment (three experiments, data not shown). Therefore, the changes in morphology observed with CB7993113 were not attributable to decreased cell growth or increased toxicity.
To assess whether CB7993113 inhibits another marker of tumor invasiveness, i.e., migration across a “scratch wound,” and to extend studies to two other cell lines, confluent cultures of AHRhigh (Yang et al., 2008) Hs578T (triple-negative) and SUM149 (inflammatory breast cancer–derived) cells were “wounded” in a scratch assay (Li et al., 2013), treated with vehicle (DMSO), 10 μM CH223191, or 10 μM CB7993113, and photographed 24 and 48 hours later. Naïve or vehicle-treated Hs578T and SUM149 cultures closed the scratch wound completely by approximately 56 and 72 hours, respectively (not shown). Whereas vehicle had no effect on the rate of wound closure, treatment with either CH223191 or CB7993113 consistently inhibited wound closure in both cell lines at 24 and 48 hours (Fig. 8).
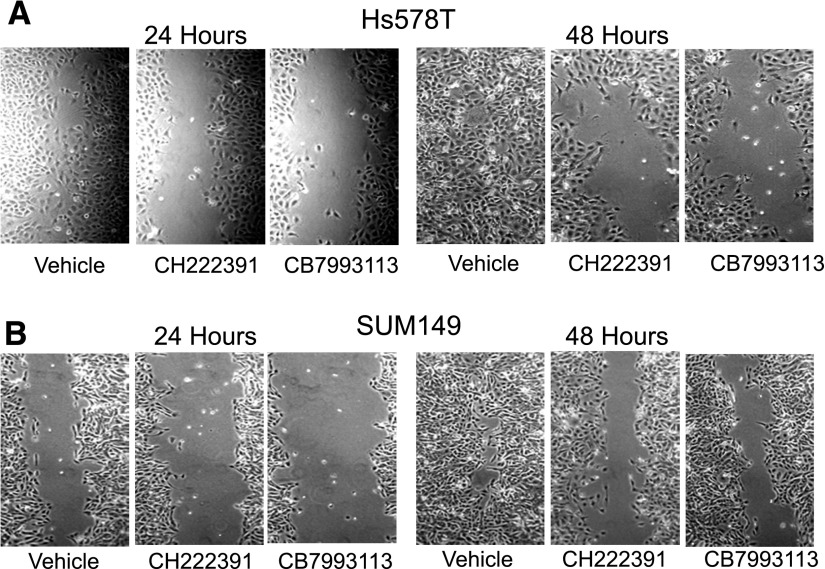
CB7993113 blocks mammary tumor cell migration in a scratch-wound assay. Confluent monolayers of Hs578T (triple-negative breast cancer) and SUM149 (inflammatory breast cancer) cells, scratched with a pipette tip, were treated with vehicle, 10 μM CH223191, or 10 μM CB7993113, and allowed to regrow for 48 hours. Cultures were photographed 24 and 48 hours after wounding. Images taken at 24 and 48 hours are representative of results obtained in six experiments.
Results in both the Matrigel and scratch-wound assays are consistent with a role for the AHR in breast cancer cell invasiveness and suggest the use of AHR antagonists to pharmacologically alter constitutive AHR activity in and potential invasiveness of cancer cells.
In Vivo Efficacy of the AHR Antagonist CB7993113.
Drugs to be used as therapeutics must exhibit several properties in vivo, including sufficient solubility, absorption, stability, and accumulation in target organs at biologically relevant concentrations. Although in silico computation predicted that CB7993113 would satisfy these criteria, in vivo studies were required to confirm this prediction. In the absence of an animal model system that faithfully and consistently recapitulates breast cancer cell invasion in humans, we chose to evaluate CB7993113 in vivo efficacy using surrogate endpoints of bioactivity, i.e., the ability to block acute AHR-mediated CYP1A1 induction in liver and AHR-regulated bone marrow toxicity (Teague et al., 2010; N’jai et al., 2011).
In pharmacokinetics experiments, 50 mg/kg CB7993113 was readily absorbed in vivo following either intraperitoneal or oral administration. Thus, 823 ± 263 nM and 395 ± 162 nM CB7993113 were detected in sera 1 hour after intraperitoneal injection or oral gavage, respectively. The compound exhibited a serum half-life of 4.0 hours (data not shown). Therefore, a dose of 50 mg/kg was used for further in vivo studies.
As expected from in vitro studies, intraperitoneal injection of 50 mg/kg CB7993113 did not induce CYP1A1 mRNA expression in liver as assessed by real-time qPCR (Fig. 9A), indicating that neither this compound nor its metabolites are partial AHR agonists in vivo. In contrast, an equal concentration of DMBA induced a significant, 200- to 400-fold, increase in CYP1A1 at this 48-hour time point. DMBA-induced CYP1A1 induction was inhibited by both CH223191 and CB7993113, although inhibition of DMBA-induced CYP1A1 expression was less consistent with CH223191 than with CB7993113.
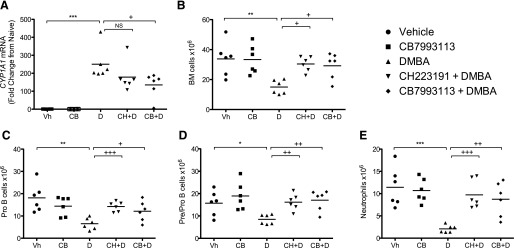
CB7993113 prevents DMBA-induced toxicity in vivo. Eight-week-old C57BL/6J mice (six per group) were treated by intraperitoneal injection with vehicle (vegetable oil), 50 mg/kg CH223191, or 50 mg/kg CB7993113 24 hours and 1 hour before DMBA treatment. Vehicle (sesame oil) or 50 mg/kg DMBA was then administered by oral gavage. Mice were sacrificed 48 hours after DMBA treatment. (A) Liver mRNA was extracted and analyzed by qPCR for CYP1A1 expression normalized to GAPDH expression. qPCR was performed in duplicate. Data bars represent the mean values from six mice. (B) Bone marrow was harvested from the right tibia and femur of each mouse and viable cells were counted by Trypan blue exclusion. Data bars represent the mean values from six mice. (C–E) Bone marrow was harvested and analyzed by flow cytometry. (C) Pre/Pro-B cells are defined as IgM–/B220+/CD43+/HSA–. (D) Pro-B cells are defined IgM–/B220+/CD43+/HSA+. (E) Neutrophils are defined as CD11bhi/GR-1hi. Data bars represent the mean values from six mice. Data were analyzed by one-way analysis of variance with Tukey-Kramer post-test (A and B) or Newman-Keuls post-test (C–E). (*P < 0.05, **P < 0.01, ***P < 0.001 compared with vehicle; +P < 0.05, ++P < 0.01, or +++P < 0.001 compared with DMBA treatment.)
In previous studies, we and others demonstrated that DMBA exposure induces a dramatic loss of bone marrow pro- and pre-B cells and other hematopoietic cell types, probably through induction of apoptosis (Mann et al., 1999; Teague et al., 2010; N’jai A et al., 2011). To determine if CB7993113 could reach sufficient concentrations in situ to block this AHR-dependent bone marrow toxicity, C57Bl/6 mice were treated with vehicle, 50 mg/kg CB7993113, or, as a positive control, 50 mg/kg CH223191, by intraperitoneal injection 24 hours and 1 hour before oral gavage with 50 mg/kg DMBA. Mice were euthanized 48 hours later, bone marrow cells were collected, and hematopoietic cells were phenotyped by flow cytometry. As expected from in vitro studies in which CB7993113 failed to exhibit toxicity, it also failed to affect the number of bone marrow cells recovered 48 hours after intraperitoneal injection (Fig. 9B). In contrast, a significant reduction in the total number of bone marrow cells was observed after DMBA treatment. This acute bone marrow cell ablation was inhibited by treatment with either CB7993113 or CH223191 (Fig. 9B).
Phenotypes of bone marrow subpopulations were analyzed to determine which hematopoietic cell subsets were affected and to determine if CB7993113 could protect all subsets from DMBA-induced toxicity. No significant changes were seen in the resident bone marrow T cell (CD3+) or natural killer cell (NK1.1+) populations following treatment with DMBA, CH223191, CB7993113, or combinations of DMBA with either antagonist (data not shown). In contrast, DMBA treatment significantly reduced the number of pre/pro-B cells (IgM–/B220+/CD43+/HSA–), pro-B cells (IgM–/B220+/CD43+/HSA+) (Hardy and Hayakawa, 2001), and neutrophils (CD11bhi/GR-1hi) (Sukhumavasi et al., 2007) (Fig. 9, C–E). Notably, pretreatment of mice with either antagonist significantly inhibited DMBA-induced toxicity in all three cell populations. These data confirm the ability of a prototypic AHR ligand to adversely affect bone marrow cells destined to contribute to the adaptive immune response, i.e., pre/pro- and pro-B cells, and to the innate immune response, i.e., neutrophils. Importantly, CB7993113 prevented a significant DMBA-induced loss of these three bone marrow cell subsets. These data demonstrate the achievement of physiologically relevant doses of CB7993113 in vivo, and suggest that this nontoxic antagonist could be used to block AHR activity either induced acutely by environmental ligands or chronically during a variety of pathologic conditions.
Discussion
The AHR has been recognized for many years as a key regulator of environmental chemical toxicity and carcinogenicity. Indeed, it mediates the biologic effects of some of the most potent toxicants known, including TCDD. Of equal concern, from an environmental point of view, is the ability of the AHR to bind and respond to a variety of structurally disparate chemicals, including planar polychlorinated biphenyls, dioxins, polycyclic aromatic hydrocarbons, plant-derived flavonoids and tryptophan-derived metabolites. Whereas this receptor’s promiscuity and the resulting pathologic consequences are reason enough to study AHR signaling pathways, in the end, the AHR’s role in regulating normal and pathologic biologic activities in the absence of environmental ligands may prove to be the more compelling story. In this vein, the AHR promotes the production of IL-17–secreting T cells critical to inflammation-based diseases and some forms of autoimmunity (Quintana et al., 2012), and affects production of regulatory T cells that oppose inflammatory and autoimmune responses (Apetoh et al., 2010; Gandhi et al., 2010). AHR signaling also plays a key role in development of gut-associated leukocytes that mediate both the innate and adaptive immune responses required to prevent microbial infiltration and the resulting inflammatory colitis (Kiss et al., 2011; Li et al., 2011). In various model systems, the AHR also contributes to hematopoietic stem cell development (Boitano et al., 2010; Casado et al., 2010; Smith et al., 2013). Consequently, the ability to regulate AHR activity may be important for treatment of hematologic diseases. Accordingly, studies have shown that AHR modulation attenuates disease in models of multiple sclerosis and type 1 diabetes (Quintana et al., 2008; Kerkvliet et al., 2009). Perhaps most strikingly, AHR modulators have a promising application in stem cell biology as demonstrated by the expansion of human CD34+ progenitor cells from cord blood by culture with the purine-derived AHR antagonist SR1 (Boitano et al., 2010) and the development of bipotential hematopoietic stem cells, megakaryocytes, and erythroid cells from AHR-activated, induced pluripotent stem cells (Smith et al., 2013).
Here, we present a combined in silico and high-throughput in vitro screening platform for the identification of AHR modulators, both agonists and antagonists. Several bioflavonoids, among other phytochemicals, have been identified as AHR ligands. The activity of most of these previously identified compounds ranges from weakly agonistic to weakly antagonistic (Lu et al., 1996; Zhang et al., 2003). However, select compounds induce significant AHR-dependent biologic effects, including several flavones, such as β-NF, used here as a prototypic AHR agonist. Therefore, we chose a generic substituted flavone backbone as our pharmacophore upon which to perform shape and electrostatic comparisons. In a novel approach to generating a template molecule that could be used to screen over a million small molecules in silico, the flavone-based model pharmacophore was expanded into a three-dimensional conformer database and the molecular shapes were compared with a similar database containing conformers of 1,176,756 commercially available, drug-like molecules. A focused library of 197 compounds was selected on the basis of three-dimensional electrostatic similarities.
The validity of this approach was confirmed by a relatively high “hit” rate. Of the 197 compounds assayed in the bioassay, 31 consistently induced AHR reporter activity with 27 of those exhibiting an EC50 of <10 μM. Four compounds exhibited AHR antagonist activity, all with IC50s <5 μM. This represents a hit rate for probable AHR ligands of 17.8%. Cell-free AHR binding studies performed on one antagonist (CB7993113, Fig. 5) and one agonist (data not shown) confirmed that each molecule was in fact an AHR ligand. Since both AHR agonists and antagonists may be useful in various therapeutic settings, this new approach to rapidly screening large libraries in silico for AHR ligands has great utility.
Our AHR ligand screen identified a disproportionate number of agonists. This result is consistent with the literature in which identification of AHR agonists is far more common than the identification of pure antagonists, i.e., competitive AHR antagonists that do not exhibit partial agonism at higher doses. To date, relatively few “pure” AHR antagonists have been discovered. These include CH223191 (Kim et al., 2006), 6,2′,4′-trimethoxyflavone (TMF) (Murray IA et al., 2009), SR1 (Boitano et al., 2010), and GNF351 (Smith et al., 2011). Identification of additional AHR antagonists, such as CB7993113, is important because each antagonist appears to exhibit unique properties in terms of affinity for AHRs from different species, “ligand selectivity,” or ability to block AHR response element–dependent signaling (Smith et al., 2011). For example, SR1 is a potent antagonist of the human AHR but has little or no effect on ligand binding to murine AHR. CH223191 exhibits ligand-selective inhibition, e.g., a propensity to block halogenated hydrocarbon–induced but not flavone-induced AHR activation (Zhao et al., 2010). In contrast, CB7993113 appears to be a more generally applicable AHR antagonist in that it inhibits both human and murine AHR, blocks DMBA-, TCDD-, flavone (β-NF)–, and tryptophan metabolite (e.g., FICZ; data not shown)–induced AHR activity and reduces baseline AHR activity (e.g., baseline pGudLuc activity). It shows no agonist activity either in vitro at doses at least as high as 20 μM or in vivo at doses at least up to 50 mg/kg. Given the number and diversity of possible therapeutic applications for AHR antagonists, including but probably not limited to human hematopoietic stem cell expansion (Boitano et al., 2010), T-cell expansion (Carlin et al., 2013), inhibition of inflammatory Th17 development (Veldhoen et al., 2009), reduction in regulatory T-cell development (Apetoh et al., 2010), expansion of erythroid and megakaryocyte lineage cells (Smith et al., 2013), and inhibition of breast cancer cell invasion (Figs. 7 and and8),8), identification of new, nontoxic AHR antagonists is of considerable import. Furthermore, expanding the database of AHR antagonists, which tend to be dissimilar in structure, will facilitate the definition of critical structural characteristics that confer the ability to inhibit, as opposed to induce, AHR activity. Again, this knowledge is critical to the rational design of more potent, nontoxic, pure AHR antagonists for therapeutic use.
Paramount to the identification of a novel antagonist is the confirmation of its drug-likeness and lack of toxicity. In our in vitro screen, compounds exhibiting any level of toxicity in murine H1G1 hepatoma cells were discarded. CB7993113 exhibited no toxicity when added at concentrations at least up to 20 μM to several human cells including HepG2 hepatoma cells, BP1, D3, Hs578T, or MDA-MB-231 breast cancer cells, and primary human-induced pluripotent stem cells (data not shown). Furthermore, CB7993113 is predicted to conform to Lipinski’s rules and thereby is expected to demonstrate oral bioavailability. Pharmacokinetics studies demonstrating significant serum bioavailability after either oral or intraperitoneal administration were consistent with this conclusion. Most importantly, CB7993113 effectively blocked acute, 50 mg/kg DMBA–induced hepatic CYP1A1 induction and bone marrow toxicity in vivo, demonstrating not only sufficient drug absorption but adequate tissue distribution and persistence for inhibiting a strong AHR-dependent (Mann et al., 1999; N’jai A et al., 2011) biologic signal.
Finally, pure AHR antagonists like CB7993113 may be useful as cancer therapeutics. Earlier studies implicated the AHR in the control of important cell functions dysregulated during malignant transformation, including cell growth (Barhoover et al., 2010) and cell migration (Dietrich and Kaina, 2010). A more recent, high-profile study demonstrated that AHR, constitutively activated by an endogenous ligand, drives human glioblastoma invasion (Opitz et al., 2011). Similarly, accumulating evidence suggests that hyper-expressed, “constitutively active” AHR plays a role in the malignant transformation of breast epithelial cells (Hall et al., 2010; Brooks and Eltom, 2011; Goode et al., 2013), in the expression of epithelial-to-mesenchymal transition markers (Schlezinger et al., 2006; Shin et al., 2006), and, most recently, in breast cancer stem cell homeostasis (Dubrovska et al., 2012; Zhao et al., 2012). As shown here, CB7993113 significantly reduces the invasive phenotype of ER–/PR–/HER2– breast cancer cells in vitro (Figs. 7 and and8).8). Although further in vivo experiments are required, these experiments collectively suggest that this, or similar AHR antagonists may represent new targeted therapeutics for some kinds of cancers, including triple-negative breast cancers.
Acknowledgments
The authors thank Jessalyn Ubellacker and Faye Andrews for their outstanding technical contributions. MPP acknowledges a free academic license to the OpenEye Suite of Software.
Abbreviations
| AHR | Aryl hydrocarbon receptor |
| AHRE | aryl hydrocarbon receptor response element |
| AHRR | aryl hydrocarbon receptor repressor |
| β-NF | beta-naphthoflavone |
| CC | consensus cluster |
| CB7993113 | 2-((2-(5-bromofuran-2-yl)-4-oxo-4H-chromen-3-yl)oxy)acetamide |
| CH223191 | (E)-1-methyl-N-(2-methyl-4-(o-tolyldiazenyl)phenyl)-1H-pyrazole-5-carboxamide |
| CMV | cytomegalovirus |
| DMBA | 7,12-dimethylbenz[a]anthracene |
| DMSO | dimethyl sulfoxide |
| EGFP | enhanced green fluorescent protein |
| IC50 | inhibitory concentration 50% |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| P450 | cytochrome P450 |
| PDB | Protein Database |
| PPARγ | peroxisome proliferator–activated receptor gamma |
| ROCS | rapid overlay of chemical structures |
| SR1 | StemRegenin 1 |
| TCDD | 2,3,7,8-tetrachlorodibenzo-p-dioxin |
| TMF | 6,2′,4′-trimethoxyflavone |
Authorship Contributions
Participated in research design: Parks, Sherr, Schlezinger, Pollastri, Hahn, Haigh, Vajda, Kozakov, Beglov.
Conducted experiments: Parks, Stanford, Novikov, Franks, Hahn, Narasimhan, Kozakov, Beglov, Schlezinger.
Contributed new reagents or analytic tools: Ashton, Hopper, Pollastri.
Performed data analysis: Parks, Sherr, Schlezinger, Pollastri, Hahn, Vajda, Kozakov, Beglov.
Wrote or contributed to the writing of the manuscript: Parks, Pollastri, Hahn, Kozakov, Beglov, Vajda, Schlezinger, Sherr.
Footnotes
This work was supported by the National Institutes of Health National Institute of Environmental Health Sciences [R01-ES11624;, P42-ES007381;, R01-ES006272;] and National Institute of General Medical Sciences [R01-GM064700]; the Art beCAUSE Breast Cancer Foundation; The Mary Kay Foundation; and a Boston University Ignition Award.
dx.doi.org/10.1124/mol.114.093369.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, Burns EJ, Sherr DH, Weiner HL, Kuchroo VK. (2010) The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol 11:854–861 [Europe PMC free article] [Abstract] [Google Scholar]
- Barhoover MA, Hall JM, Greenlee WF, Thomas RS. (2010) Aryl hydrocarbon receptor regulates cell cycle progression in human breast cancer cells via a functional interaction with cyclin-dependent kinase 4. Mol Pharmacol 77:195–201 [Abstract] [Google Scholar]
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S et al. (2002) The Protein Data Bank. Acta Crystallogr D Biol Crystallogr 58:899–907 [Abstract] [Google Scholar]
- Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, et al. (2010) Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329:1345–1348 [Europe PMC free article] [Abstract] [Google Scholar]
- Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S. (2009) Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics 25:621–627 [Europe PMC free article] [Abstract] [Google Scholar]
- Brooks B, Bruccoleri R, Olafson B, States D, Swaminathan S, Karplus M. (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217 [Google Scholar]
- Brooks J, Eltom SE. (2011) Malignant transformation of mammary epithelial cells by ectopic overexpression of the aryl hydrocarbon receptor. Curr Cancer Drug Targets 11:654–669 [Europe PMC free article] [Abstract] [Google Scholar]
- Burbach KM, Poland A, Bradfield CA. (1992) Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci USA 89:8185–8189 [Europe PMC free article] [Abstract] [Google Scholar]
- Carlin SM, Ma DD, Moore JJ. (2013) T-cell potential of human adult and cord blood hemopoietic stem cells expanded with the use of aryl hydrocarbon receptor antagonists. Cytotherapy 15:224–230 [Abstract] [Google Scholar]
- Caruso JA, Mathieu PA, Joiakim A, Leeson B, Kessel D, Sloane BF, Reiners JJ., Jr (2004) Differential susceptibilities of murine hepatoma 1c1c7 and Tao cells to the lysosomal photosensitizer NPe6: influence of aryl hydrocarbon receptor on lysosomal fragility and protease contents. Mol Pharmacol 65:1016–1028 [Abstract] [Google Scholar]
- Caruso JA, Mathieu PA, Joiakim A, Zhang H, Reiners JJ., Jr (2006) Aryl hydrocarbon receptor modulation of TNFalpha -induced apoptosis and lysosomal disruption in a hepatoma model that is caspase-8 independent. J Biol Chem 281:10954–10967 [Abstract] [Google Scholar]
- Casado FL, Singh KP, Gasiewicz TA. (2010) The aryl hydrocarbon receptor: regulation of hematopoiesis and involvement in the progression of blood diseases. Blood Cells Mol Dis 44:199–206 [Europe PMC free article] [Abstract] [Google Scholar]
- Dietrich C, Kaina B. (2010) The aryl hydrocarbon receptor (AhR) in the regulation of cell-cell contact and tumor growth. Carcinogenesis 31:1319–1328 [Europe PMC free article] [Abstract] [Google Scholar]
- Dolwick KM, Schmidt JV, Carver LA, Swanson HI, Bradfield CA. (1993) Cloning and expression of a human Ah receptor cDNA. Mol Pharmacol 44:911–917 [Abstract] [Google Scholar]
- Dubrovska A, Hartung A, Bouchez LC, Walker JR, Reddy VA, Cho CY, Schultz PG. (2012) CXCR4 activation maintains a stem cell population in tamoxifen-resistant breast cancer cells through AhR signalling. Br J Cancer 107:43–52 [Europe PMC free article] [Abstract] [Google Scholar]
- Dvorak Z, Vrzal R, Henklova P, Jancova P, Anzenbacherova E, Maurel P, Svecova L, Pavek P, Ehrmann J, Havlik R, et al. (2008) JNK inhibitor SP600125 is a partial agonist of human aryl hydrocarbon receptor and induces CYP1A1 and CYP1A2 genes in primary human hepatocytes. Biochem Pharmacol 75:580–588 [Abstract] [Google Scholar]
- Fiser A, Sali A. (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374:461–491 [Abstract] [Google Scholar]
- Gandhi R, Kumar D, Burns EJ, Nadeau M, Dake B, Laroni A, Kozoriz D, Weiner HL, Quintana FJ. (2010) Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat Immunol 11:846–853 [Europe PMC free article] [Abstract] [Google Scholar]
- Goode GD, Ballard BR, Manning HC, Freeman ML, Kang Y, Eltom SE. (2013) Knockdown of aberrantly upregulated aryl hydrocarbon receptor reduces tumor growth and metastasis of MDA-MB-231 human breast cancer cell line. Int J Cancer 133:2769–2780 [Europe PMC free article] [Abstract] [Google Scholar]
- Gurnell M, Wentworth JM, Agostini M, Adams M, Collingwood TN, Provenzano C, Browne PO, Rajanayagam O, Burris TP, Schwabe JW, et al. (2000) A dominant-negative peroxisome proliferator-activated receptor gamma (PPARgamma) mutant is a constitutive repressor and inhibits PPARgamma-mediated adipogenesis. J Biol Chem 275:5754–5759 [Abstract] [Google Scholar]
- Hall JM, Barhoover MA, Kazmin D, McDonnell DP, Greenlee WF, Thomas RS. (2010) Activation of the aryl-hydrocarbon receptor inhibits invasive and metastatic features of human breast cancer cells and promotes breast cancer cell differentiation. Mol Endocrinol 24:359–369 [Europe PMC free article] [Abstract] [Google Scholar]
- Hankinson O, Brooks BA, Weir-Brown KI, Hoffman EC, Johnson BS, Nanthur J, Reyes H, Watson AJ. (1991) Genetic and molecular analysis of the Ah receptor and of Cyp1a1 gene expression. Biochimie 73:61–66 [Abstract] [Google Scholar]
- Hardy RR, Hayakawa K. (2001) B cell development pathways. Annu Rev Immunol 19:595–621 [Abstract] [Google Scholar]
- Hawkins PC, Nicholls A. (2012) Conformer generation with OMEGA: learning from the data set and the analysis of failures. J Chem Inf Model 52:2919–2936 [Abstract] [Google Scholar]
- Hawkins PCD, Skillman AG, Nicholls A. (2007) Comparison of shape-matching and docking as virtual screening tools. J Med Chem 50:74–82 [Abstract] [Google Scholar]
- Hughes L, Malone C, Chumsri S, Burger AM, McDonnell S. (2008) Characterisation of breast cancer cell lines and establishment of a novel isogenic subclone to study migration, invasion and tumourigenicity. Clin Exp Metastasis 25:549–557 [Abstract] [Google Scholar]
- Karchner SI, Franks DG, Kennedy SW, Hahn ME. (2006) The molecular basis for differential dioxin sensitivity in birds: role of the aryl hydrocarbon receptor. Proc Natl Acad Sci USA 103:6252–6257 [Europe PMC free article] [Abstract] [Google Scholar]
- Kerkvliet NI, Steppan LB, Vorachek W, Oda S, Farrer D, Wong CP, Pham D, Mourich DV. (2009) Activation of aryl hydrocarbon receptor by TCDD prevents diabetes in NOD mice and increases Foxp3+ T cells in pancreatic lymph nodes. Immunotherapy 1:539–547 [Europe PMC free article] [Abstract] [Google Scholar]
- Kim SH, Henry EC, Kim DK, Kim YH, Shin KJ, Han MS, Lee TG, Kang JK, Gasiewicz TA, Ryu SH, et al. (2006) Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) prevents 2,3,7,8-TCDD-induced toxicity by antagonizing the aryl hydrocarbon receptor. Mol Pharmacol 69:1871–1878 [Abstract] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. (2011) Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334:1561–1565 [Abstract] [Google Scholar]
- Kozakov D, Hall DR, Chuang GY, Cencic R, Brenke R, Grove LE, Beglov D, Pelletier J, Whitty A, Vajda S. (2011) Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci USA 108:13528–13533 [Europe PMC free article] [Abstract] [Google Scholar]
- Lahoti TS, John K, Hughes JM, Kusnadi A, Murray IA, Krishnegowda G, Amin S, Perdew GH. (2013) Aryl hydrocarbon receptor antagonism mitigates cytokine-mediated inflammatory signalling in primary human fibroblast-like synoviocytes. Ann Rheum Dis 72:1708–1716 [Europe PMC free article] [Abstract] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, et al. (2012) AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol 13:144–151 [Europe PMC free article] [Abstract] [Google Scholar]
- Li NY, Weber CE, Wai PY, Cuevas BD, Zhang J, Kuo PC, Mi Z. (2013) An MAPK-dependent pathway induces epithelial-mesenchymal transition via Twist activation in human breast cancer cell lines. Surgery 154:404–410 [Abstract] [Google Scholar]
- Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. (2011) Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147:629–640 [Abstract] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26 [Abstract] [Google Scholar]
- Lu YF, Santostefano M, Cunningham BD, Threadgill MD, Safe S. (1996) Substituted flavones as aryl hydrocarbon (Ah) receptor agonists and antagonists. Biochem Pharmacol 51:1077–1087 [Abstract] [Google Scholar]
- Maecker B, Sherr DH, Vonderheide RH, von Bergwelt-Baildon MS, Hirano N, Anderson KS, Xia Z, Butler MO, Wucherpfennig KW, O’Hara C, et al. (2003) The shared tumor-associated antigen cytochrome P450 1B1 is recognized by specific cytotoxic T cells. Blood 102:3287–3294 [Abstract] [Google Scholar]
- Mann KK, Matulka RA, Hahn ME, Trombino AF, Lawrence BP, Kerkvliet NI, Sherr DH. (1999) The role of polycyclic aromatic hydrocarbon metabolism in dimethylbenz[a]anthracene-induced pre-B lymphocyte apoptosis. Toxicol Appl Pharmacol 161:10–22 [Abstract] [Google Scholar]
- Motto I, Bordogna A, Soshilov AA, Denison MS, Bonati L. (2011) New aryl hydrocarbon receptor homology model targeted to improve docking reliability. J Chem Inf Model 51:2868–2881 [Europe PMC free article] [Abstract] [Google Scholar]
- Murray GI, Patimalla S, Stewart KN, Miller ID, Heys SD. (2010) Profiling the expression of cytochrome P450 in breast cancer. Histopathology 57:202–211 [Abstract] [Google Scholar]
- Murray IA, Flaveny CA, DiNatale BC, Chairo CR, Schroeder JC, Kusnadi A, Perdew GH. (2009) Antagonism of aryl hydrocarbon receptor signaling by 6,2′,4′-trimethoxyflavone. J Pharmacol Exp Ther 332:135–144 [Europe PMC free article] [Abstract] [Google Scholar]
- N’jai AU, Larsen MC, Bushkofsky JR, Czuprynski CJ, Jefcoate CR. (2011) Acute disruption of bone marrow hematopoiesis by benzo(a)pyrene is selectively reversed by aryl hydrocarbon receptor-mediated processes. Mol Pharmacol 79:724–734 [Europe PMC free article] [Abstract] [Google Scholar]
- Nagy SR, Sanborn JR, Hammock BD, Denison MS. (2002) Development of a green fluorescent protein-based cell bioassay for the rapid and inexpensive detection and characterization of ah receptor agonists. Toxicol Sci 65:200–210 [Abstract] [Google Scholar]
- Nebert DW, Petersen DD, Puga A. (1991) Human AH locus polymorphism and cancer: inducibility of CYP1A1 and other genes by combustion products and dioxin. Pharmacogenetics 1:68–78 [Abstract] [Google Scholar]
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, et al. (2011) An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478:197–203 [Abstract] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [Europe PMC free article] [Abstract] [Google Scholar]
- Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. (2008) Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453:65–71 [Abstract] [Google Scholar]
- Quintana FJ, Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D, Rangachari M, Zhu C, Xiao S, Seavitt J, et al. (2012) Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol 13:770–777 [Europe PMC free article] [Abstract] [Google Scholar]
- Santostefano M, Merchant M, Arellano L, Morrison V, Denison MS, Safe S. (1993) alpha-Naphthoflavone-induced CYP1A1 gene expression and cytosolic aryl hydrocarbon receptor transformation. Mol Pharmacol 43:200–206 [Abstract] [Google Scholar]
- Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. (2009) Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci USA 106:450–455 [Europe PMC free article] [Abstract] [Google Scholar]
- Scheuermann TH, Li Q, Ma HW, Key J, Zhang L, Chen R, Garcia JA, Naidoo J, Longgood J, Frantz DE, et al. (2013) Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol 9:271–276 [Europe PMC free article] [Abstract] [Google Scholar]
- Schlezinger JJ, Liu D, Farago M, Seldin DC, Belguise K, Sonenshein GE, Sherr DH. (2006) A role for the aryl hydrocarbon receptor in mammary gland tumorigenesis. Biol Chem 387:1175–1187 [Abstract] [Google Scholar]
- Shin SR, Sánchez-Velar N, Sherr DH, Sonenshein GE. (2006) 7,12-dimethylbenz(a)anthracene treatment of a c-rel mouse mammary tumor cell line induces epithelial to mesenchymal transition via activation of nuclear factor-kappaB. Cancer Res 66:2570–2575 [Abstract] [Google Scholar]
- Smith BW, Rozelle SS, Leung A, Ubellacker J, Parks A, Nah SK, French D, Gadue P, Monti S, Chui DH, et al. (2013) The aryl hydrocarbon receptor directs hematopoietic progenitor cell expansion and differentiation. Blood 122:376–385 [Europe PMC free article] [Abstract] [Google Scholar]
- Smith KJ, Murray IA, Tanos R, Tellew J, Boitano AE, Bisson WH, Kolluri SK, Cooke MP, Perdew GH. (2011) Identification of a high-affinity ligand that exhibits complete aryl hydrocarbon receptor antagonism. J Pharmacol Exp Ther 338:318–327 [Europe PMC free article] [Abstract] [Google Scholar]
- Sukhumavasi W, Egan CE, Denkers EY. (2007) Mouse neutrophils require JNK2 MAPK for Toxoplasma gondii-induced IL-12p40 and CCL2/MCP-1 release. J Immunol 179:3570–3577 [Abstract] [Google Scholar]
- Teague JE, Ryu H-Y, Kirber M, Sherr DH, Schlezinger JJ. (2010) Proximal events in 7,12-dimethylbenz[a]anthracene-induced, stromal cell-dependent bone marrow B cell apoptosis: stromal cell-B cell communication and apoptosis signaling. J Immunol 185:3369–3378 [Europe PMC free article] [Abstract] [Google Scholar]
- Trombino AF, Near RI, Matulka RA, Yang S, Hafer LJ, Toselli PA, Kim DW, Rogers AE, Sonenshein GE, Sherr DH. (2000) Expression of the aryl hydrocarbon receptor/transcription factor (AhR) and AhR-regulated CYP1 gene transcripts in a rat model of mammary tumorigenesis. Breast Cancer Res Treat 63:117–131 [Abstract] [Google Scholar]
- Trott O, Olson AJ. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461 [Europe PMC free article] [Abstract] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. (2008) The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453:106–109 [Abstract] [Google Scholar]
- Veldhoen M, Hirota K, Christensen J, O’Garra A, Stockinger B. (2009) Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med 206:43–49 [Europe PMC free article] [Abstract] [Google Scholar]
- Wu D, Nishimura N, Kuo V, Fiehn O, Shahbaz S, Van Winkle L, Matsumura F, Vogel CF. (2011) Activation of aryl hydrocarbon receptor induces vascular inflammation and promotes atherosclerosis in apolipoprotein E-/- mice. Arterioscler Thromb Vasc Biol 31:1260–1267 [Europe PMC free article] [Abstract] [Google Scholar]
- Wu D, Potluri N, Kim Y, Rastinejad F. (2013) Structure and dimerization properties of the aryl hydrocarbon receptor PAS-A domain. Mol Cell Biol 33:4346–4356 [Europe PMC free article] [Abstract] [Google Scholar]
- Xing Y, Nukaya M, Satyshur KA, Jiang L, Stanevich V, Korkmaz EN, Burdette L, Kennedy GD, Cui Q, Bradfield CA. (2012) Identification of the Ah-receptor structural determinants for ligand preferences. Toxicol Sci 129:86–97 [Europe PMC free article] [Abstract] [Google Scholar]
- Xu M, Miller MS. (2004) Determination of murine fetal Cyp1a1 and 1b1 expression by real-time fluorescence reverse transcription-polymerase chain reaction. Toxicol Appl Pharmacol 201:295–302 [Abstract] [Google Scholar]
- Yang X, Liu D, Murray TJ, Mitchell GC, Hesterman EV, Karchner SI, Merson RR, Hahn ME, Sherr DH. (2005) The aryl hydrocarbon receptor constitutively represses c-myc transcription in human mammary tumor cells. Oncogene 24:7869–7881 [Abstract] [Google Scholar]
- Yang X, Solomon S, Fraser LR, Trombino AF, Liu D, Sonenshein GE, Hestermann EV, Sherr DH. (2008) Constitutive regulation of CYP1B1 by the aryl hydrocarbon receptor (AhR) in pre-malignant and malignant mammary tissue. J Cell Biochem 104:402–417 [Abstract] [Google Scholar]
- Zhang S, Lei P, Liu X, Li X, Walker K, Kotha L, Rowlands C, Safe S. (2009) The aryl hydrocarbon receptor as a target for estrogen receptor-negative breast cancer chemotherapy. Endocr Relat Cancer 16:835–844 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang S, Qin C, Safe SH. (2003) Flavonoids as aryl hydrocarbon receptor agonists/antagonists: effects of structure and cell context. Environ Health Perspect 111:1877–1882 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao B, Degroot DE, Hayashi A, He G, Denison MS. (2010) CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci 117:393–403 [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao S, Kanno Y, Nakayama M, Makimura M, Ohara S, Inouye Y. (2012) Activation of the aryl hydrocarbon receptor represses mammosphere formation in MCF-7 cells. Cancer Lett 317:192–198 [Abstract] [Google Scholar]
- Zudaire E, Cuesta N, Murty V, Woodson K, Adams L, Gonzalez N, Martínez A, Narayan G, Kirsch I, Franklin W, et al. (2008) The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. J Clin Invest 118:640–650 [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Molecular Pharmacology are provided here courtesy of American Society for Pharmacology and Experimental Therapeutics
Full text links
Read article at publisher's site: https://doi.org/10.1124/mol.114.093369
Read article for free, from open access legal sources, via Unpaywall:
http://molpharm.aspetjournals.org/content/molpharm/86/5/593.full.pdf
Citations & impact
Impact metrics
Article citations
In silico approaches for drug repurposing in oncology: a scoping review.
Front Pharmacol, 15:1400029, 11 Jun 2024
Cited by: 0 articles | PMID: 38919258 | PMCID: PMC11196849
Review Free full text in Europe PMC
Aryl hydrocarbon receptor: An emerging player in breast cancer pathogenesis and its potential as a drug target (Review).
Mol Med Rep, 29(1):11, 24 Nov 2023
Cited by: 3 articles | PMID: 37997818 | PMCID: PMC10704547
Review Free full text in Europe PMC
Aryl Hydrocarbon Receptor as an Anticancer Target: An Overview of Ten Years Odyssey.
Molecules, 28(10):3978, 09 May 2023
Cited by: 1 article | PMID: 37241719 | PMCID: PMC10221042
Review Free full text in Europe PMC
Identification of environmental factors that promote intestinal inflammation.
Nature, 611(7937):801-809, 20 Oct 2022
Cited by: 44 articles | PMID: 36266581 | PMCID: PMC9898826
The Role of the Aryl Hydrocarbon Receptor (AhR) and Its Ligands in Breast Cancer.
Cancers (Basel), 14(22):5574, 14 Nov 2022
Cited by: 20 articles | PMID: 36428667 | PMCID: PMC9688153
Review Free full text in Europe PMC
Go to all (29) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (2)
-
(1 citation)
PDBe - 4M4XView structure
-
(1 citation)
PDBe - 4GHIView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The bioflavonoid galangin blocks aryl hydrocarbon receptor activation and polycyclic aromatic hydrocarbon-induced pre-B cell apoptosis.
Mol Pharmacol, 58(3):515-525, 01 Sep 2000
Cited by: 32 articles | PMID: 10953044
Activation of the aryl hydrocarbon receptor/transcription factor and bone marrow stromal cell-dependent preB cell apoptosis.
J Immunol, 158(5):2165-2173, 01 Mar 1997
Cited by: 33 articles | PMID: 9036962
Expression of functional aromatic hydrocarbon receptor and aromatic hydrocarbon nuclear translocator proteins in murine bone marrow stromal cells.
Arch Biochem Biophys, 352(1):9-18, 01 Apr 1998
Cited by: 9 articles | PMID: 9521805
Aryl Hydrocarbon Receptor (AHR) Ligands as Selective AHR Modulators (SAhRMs).
Int J Mol Sci, 21(18):E6654, 11 Sep 2020
Cited by: 63 articles | PMID: 32932962 | PMCID: PMC7555580
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NHLBI NIH HHS (1)
Grant ID: T32 HL007501
NIEHS NIH HHS (6)
Grant ID: P01 ES011624
Grant ID: P42 ES007381
Grant ID: R01-ES006272
Grant ID: P42-ES007381
Grant ID: R01 ES006272
Grant ID: R01-ES11624
NIGMS NIH HHS (2)
Grant ID: R01 GM064700
Grant ID: R01-GM064700






