Abstract
Context
Alcohol dependence is a serious and common public health problem. It is well established that genetic factors play a major role in the development of this disorder. Identification of genes that contribute to alcohol dependence will improve our understanding of the mechanisms that underlie this disorder.Objective
To identify susceptibility genes for alcohol dependence through a genome-wide association study (GWAS) and a follow-up study in a population of German male inpatients with an early age at onset.Design
The GWAS tested 524,396 single-nucleotide polymorphisms (SNPs). All SNPs with P < 10(-4) were subjected to the follow-up study. In addition, nominally significant SNPs from genes that had also shown expression changes in rat brains after long-term alcohol consumption were selected for the follow-up step.Setting
Five university hospitals in southern and central Germany.Participants
The GWAS included 487 male inpatients with alcohol dependence as defined by the DSM-IV and an age at onset younger than 28 years and 1358 population-based control individuals. The follow-up study included 1024 male inpatients and 996 age-matched male controls. All the participants were of German descent.Main outcome measures
Significant association findings in the GWAS and follow-up study with the same alleles.Results
The GWAS produced 121 SNPs with nominal P < 10(-4). These, together with 19 additional SNPs from homologues of rat genes showing differential expression, were genotyped in the follow-up sample. Fifteen SNPs showed significant association with the same allele as in the GWAS. In the combined analysis, 2 closely linked intergenic SNPs met genome-wide significance (rs7590720, P = 9.72 x 10(-9); rs1344694, P = 1.69 x 10(-8)). They are located on chromosome region 2q35, which has been implicated in linkage studies for alcohol phenotypes. Nine SNPs were located in genes, including the CDH13 and ADH1C genes, that have been reported to be associated with alcohol dependence.Conclusions
This is the first GWAS and follow-up study to identify a genome-wide significant association in alcohol dependence. Further independent studies are required to confirm these findings.Free full text

Genome-wide association study of alcohol dependence
Abstract
Context
Identification of genes contributing to alcohol dependence will improve our understanding of the mechanisms underlying this disorder.
Objective
To identify susceptibility genes for alcohol dependence through a genome-wide association study (GWAS) and follow-up study in a population of German male inpatients with an early age at onset.
Design
The GWAS included 487 male inpatients with DSM-IV alcohol dependence with an age at onset below 28 years and 1,358 population based control individuals. The follow-up study included 1,024 male inpatients and 996 age-matched male controls. All subjects were of German descent. The GWAS tested 524,396 single nucleotide polymorphisms (SNPs). All SNPs with p<10-4 were subjected to the follow-up study. In addition, nominally significant SNPs from those genes that had also shown expression changes in rat brains after chronic alcohol consumption were selected for the follow-up step.
Results
The GWAS produced 121 SNPs with nominal p<10-4. These, together with 19 additional SNPs from homologs of rat genes showing differential expression, were genotyped in the follow-up sample. Fifteen SNPs showed significant association with the same allele as in the GWAS. In the combined analysis, two closely linked intergenic SNPs met genome-wide significance (rs7590720 p=9.72×10-9; rs1344694 p=1.69×10-8). They are located on chromosome 2q35, a region which has been implicated in linkage studies for alcohol phenotypes. Nine SNPs were located in genes, including CDH13 and ADH1C genes which have been reported to be associated with alcohol dependence.
Conclusion
This is the first GWAS and follow-up study to identify a genome-wide significant association in alcohol dependence. Further independent studies are required to confirm these findings.
Introduction
Alcohol dependence is characterised by a cluster of cognitive, behavioural and physiological symptoms, with an affected individual continuing to drink despite significant alcohol-induced impairment or distress. Since alcohol affects most human organs, its abuse is associated with a wide range of physical, mental and social harm. According to the World Health Organisation,1 alcohol abuse constitutes a serious public health problem worldwide, accounting for 4% of the global burden, a burden comparable to the death and disability attributable to tobacco and hypertension.1,2
Alcohol dependence is a phenotypically heterogeneous disorder which runs in families and which has a high genetic loading. Twin and adoption studies have shown that 40-60% of the inter-individual phenotypic variance is accounted for by genetic factors.3-5 In congruence with the observed phenotypic heterogeneity of alcohol dependence, vulnerability to alcohol dependence on the molecular level is thought to be mediated by many genetic loci of small to modest effects in Europeans.6-14 Despite strenuous efforts and numerous linkage and candidate gene studies, identification of the underlying susceptibility genes has proven difficult. Genome-wide association studies (GWAS) conducted in other complex disorders have been shown to be a successful tool in identifying underlying susceptibility genes (for all published GWAS see: http://www.genome.gov/26525384) and have already resulted in the identification of genetic susceptibility variants for psychiatric disorders such as nicotine addiction, schizophrenia and bipolar disorder15-24 which were mostly not detected through other diagnosis based discovery approaches. For alcohol dependence, only one GWAS using pooled DNA samples has been published to date, and it has proposed several new susceptibility loci for alcohol dependence. Their products are implicated in cellular signalling, gene regulation, development and cell adhesion.25 Convergent translational approaches integrate genetic findings from animal models with a candidate gene or GWAS approach in humans and have proven to be very successful.26,27 The goal of the present study was to conduct a GWAS and follow-up study for alcohol dependence using individual genotyping in samples stratified for homogeneity with respect to sex, ethnicity, age at onset, and recruitment procedures. In order to increase the explanatory power of our findings, we applied a Convergent Functional Genomics28 approach to integrate findings from gene expression data in alcohol dependent rats with our GWAS findings.
Methods
Human Studies
Patients
Patients were recruited from consecutive admissions to the psychiatric and addiction medicine departments of 5 different study centers at University hospitals across Southern and Central Germany: Regensburg, Mannheim, Munich, Bonn/Essen/Düsseldorf/Homburg and Mainz. These centres are members of the German Addiction Research Network (GARN; http://www.bw-suchtweb.de) for which one of the co-authors (KM) is spokesman. All patients included in the study suffered from alcohol dependence of such severity that hospitalisation for the treatment or prevention of severe withdrawal symptoms was warranted. All patients received a diagnosis of alcohol dependence (DSM-IV) by consensus of two clinical psychiatrists. Within the GARN study, DSM-IV criteria were ascertained systematically through use of semi-structured and independently rated interviews conducted by trained staff members.
In Munich and Bonn, patients were assessed using the Semi Structured Assessment for Genetics of Alcoholism (SSAGA),29 in Mainz the Composite International Diagnostic Interview (CIDI)30 was used, in Regensburg, Essen/Düsseldorf/Homburg and Mannheim the Structured Clinical Interview for DSM31 was used. The latter tool was also applied in Munich. All patients and controls were of self reported German ancestry. The study was approved by all relevant local Ethics Committees and all subjects provided written informed consent. In order to increase the power of the study, we increased the homogeneity of the patient sample by including male patients only. Furthermore, only subjects with an early age at onset of alcohol dependence were included, a feature which has a higher heritability in males.32,33Patients included in the GWAS (Mannheim N=98; Bonn/Essen/Düsseldorf/Homburg N=30; Mainz N=30; Munich N=53; and Regensburg N=276) had an age at onset <28 (median=20, mean±std=21±4.1) years, defined as the age at which DSM-IV criteria for alcohol dependence were fulfilled for the first time.
The follow-up sample consisted of 1,024 German males recruited at the same sites and under the same protocol as the patients who were entered in the GWAS (Mannheim N=256; Bonn/Essen/Düsseldorf/Homburg N=149; Mainz N=63; Munich N=136; Regensburg N=420). Since most of the patients with an earlier age at onset had already been included in the GWAS, age at onset for the follow-up study was necessarily less stringent and defined as < 45 (median=30, mean±std=29.3±7) years. Phenotypes and genotypes resulting from GWAS are stored in a comprehensive database at the Institute for Medical Biometry, Informatics and Epidemiology in Bonn (IMBIE).
Control subjects
A total of 1,358 control subjects were included in the GWAS. Controls were taken from three population-based epidemiological studies: 1) N=487 from PopGen34 N=488 from KORA35 and 3) N=383 from Heinz Nixdorf Recall Study (HNR)36. These three recruitment areas are located in Schleswig-Holstein (North Germany), Augsburg (South Germany) and Essen, Bochum, Mülheim (Ruhr-Area, West Germany). The PopGen (Population-based Recruitment of Patients and Controls for the Analysis) project (http://www.science.ngfn.de/10_233.htm; www.popgen.de;) was initiated to provide all locally prevalent cases with the diseases in question for the disease-orientated projects from the NGFN as well as population-based control samples which will ultimately comprise 7200 persons. The KORA project (Cooperative Health Research in the Region of Augsburg http://www.science.ngfn.de/10_234.htm; www.gsf.de/KORA), which has evolved from the WHO MONICA study (Monitoring of Trends and Determinants of Cardiovascular Disease), has the biosamples, phenotypic characteristics and environmental parameters of 18,000 adults from Augsburg and the surrounding counties. The biological specimen bank was established in order to enable researchers to perform epidemiological research into molecular and genetic factors. The Heinz Nixdorf Recall (Risk Factors, Evaluation of Coronary Calcification, and Lifestyle) study (http://www.recall-studie.uni-essen.de) was started in 2000 as a prospective cohort study to determine predictors of coronary heart disease. It has the biosamples, phenotypic characteristics and environmental parameters of 4500 adults. PopGen and Kora are the two major German biobanking resources available to participants of the National Genome Research Project (NGFN) (http://www.ngfn.de/) for use as universal controls,37 and they have already been used as samples in other GWAS.37-38
Male controls for the follow-up study were drawn from the KORA study (N=382) and from a population based sample (N=614) which had been collected in the area of Bonn by one of the authors (MR) for use as a control sample for association studies within the framework of the NGFN. Controls were not stratified for alcohol abuse/dependence, which may introduce a conservative bias. Tests for genetic differentiation between German populations have shown low levels of population substructure, thus demonstrating that the German population is an appropriate source for use in association studies of complex diseases.40
DNA preparation, genotyping and statistical analysis
Genomic DNA was prepared from whole blood according to standard procedures. For all samples, DNA concentrations were quantified in triplicate measurements by Picogreends DNA Quantitation Kit (www.invitrogen.com) and normalized to 50 ng/ul at the CIMH, Molecular Genetics Laboratory. For the GWAS, all samples were genotyped individually using Human Hap 550 BeadChips (Illumina, Inc., San Diego, CA, USA). The patient and the PopGen and KORA samples were genotyped at Illumina Inc. and the HNR sample was genotyped at the Department of Genomics at the Life & BrainCenter, University of Bonn. Genotyping of the follow-up sample was performed by primer extension reaction chemistry with MALDI-TOF mass spectrometry using the iPLEX® Assay (Sequenom, San Diego, USA) at the Life & BrainCenter, University of Bonn. Genotyping results were imported into a central computer system for statistical analysis.
Data analysis and quality control
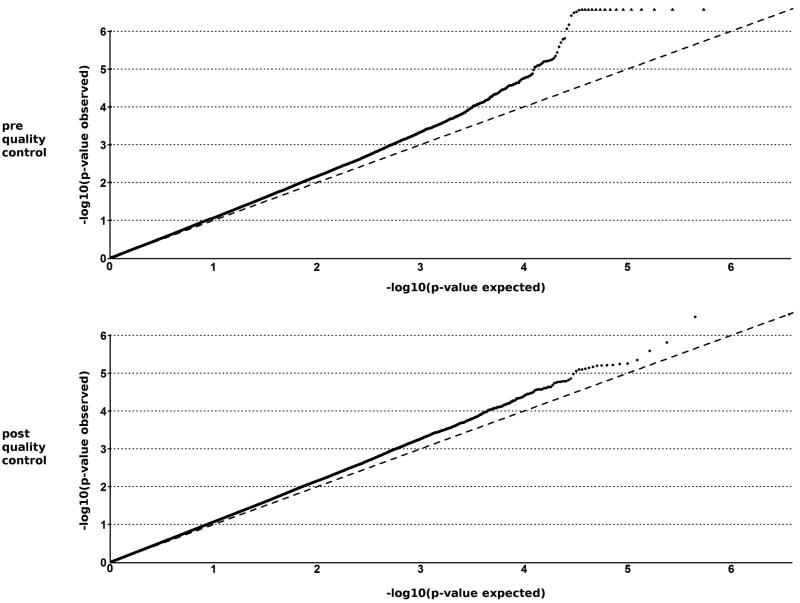
Data analysis and quality control (QC) was performed using the software packages R version 2.5.1 (http://www.R-project.org) and PLINK version 1.0.3.41 In the GWAS, genotype data were cleaned before analysis by removing SNPs or individuals that did not fulfil the QC criteria, which included: SNP call proportion ≥95%, subject completeness proportion ≥95%, SNP minor allele frequency ≥0.01 and SNP conformity with Hardy-Weinberg equilibrium expectations (p≥0.01 in controls). To correct for cryptic relatedness, all pairs of individuals displaying an identity-by-state (IBS) value larger than 1.6 were marked, and for each pair the individual with the lower typing rate was removed from the analysis. Cochran Armitage trend statistics were used to calculate significant association for autosomal SNPs. To visualize the outcome of the QC steps, Cochran Armitage p-values were depicted in a quantile-quantile plot (Figure 1). We observed good adherence of p-values to the line of expectance, which implies that potential spurious associations characterized by an inflation of highly significant p-values were successfully removed by our QC measures. The remaining slight deviations from the line of expectance are interpreted to include true genetic effects. Further correction for lambda42 improved the QQ-plot of Armitage p-values (eFigure1).
Animal Studies
Animals
Three groups of 2-3 months old alcohol preferring rats were used for long-term alcohol consumption and gene expression profiling: male P rats (n = 15; IndianaUniversity, Indianapolis), male HAD rats (n = 13; IndianaUniversity, Indianapolis) and male AA rats (n=14; National Public Health Institute, Helsinki). The rats were kindly provided by T.K. Li (Department of Psychiatry, Institute of Psychiatric Research, Indiana University School of Medicine, Indianapolis)and D. Sinclair (Department of Mental Health and Alcohol Research, National Public Health Institute, Helsinki). Each rat strain shows alcohol preference due to various neurochemical alterations, as indicated by the abbreviations P (preference), HAD (high alcohol drinking), AA (alcohol addicted).43,44 All experimental procedures were approved by the Committee on Animal Care and Use (Regierungspräsidium Karlsruhe), and carried out in accordance with the local Animal Welfare Act and the European Communities Council Directive of 24 November 1986 (86/609/EEC).
According to the protocol of Vengeliene et al.,45,46 8 P rats, 7 HAD rats, and 7 AA rats were given ad libitum access to tap water and to 5%, and 20% ethanol solution (v/v). All rats underwent a two-week deprivation cycle after 8 weeks of continuous alcohol availability. After the deprivation period, rats were given access to alcohol again and 3 more two-week deprivation periods were introduced in a random manner (the duration between deprivation periods varied between 4 and 16 weeks). The long-term voluntary alcohol drinking procedure, including all deprivation phases, lasted a total of 52 weeks. Total ethanol intake (g/kg of body weight/day) was calculated as the daily average across 7 measuring days. For comparison, 3 age- and weight-matched control groups, consisting of 7 P rats, 6 HAD rats, and 7 AA rats, experienced identical handling procedures for the entire duration of the experiment, but did not receive alcohol.
Gene expression profiling in alcohol-preferring rat strains
Preparation of brain samples and RNA isolation are described in the Supplemental Text. Target preparation was done for individual samples from caudate putamen and amygdala using 5 μg of total RNA. Hybridization to RG U34A arrays, staining, washing and scanning of the chips were performed according to the manufacturer′s technical manual (Affymetrix, Santa Clara, CA).
Data mining
Micro Array Suite 5.0 (Affymetrix) derived cell intensity files (CEL) were processed in R 2.1.1 language and environment (http://www.R-project.org) using Bioconductor 1.6 packages.47 Each array was inspected for regional hybridization bias and quality control parameters as recently described.48 Fifty-three arrays (27 from caudate putamen and 26 from amygdala) passed the quality filter and were included in the statistical analysis. Of the 8799 probe sets on the RG_U34A array, only those with intensity values > 100 in at least 25 % of the samples were retained (6344 probe sets). A 3-way ANOVA was used to identify differentially expressed genes across strain, brain region and treatment. The list of genes affected by long-term ethanol consumption included those with a p < 0.05 for treatment from the 3-way ANOVA. Posthoc analysis was performed via template matching across strains. Correlation coefficients (r) for consistent up or down regulation by ethanol in either caudate putamen or amygdala or both regions were calculated.
Integration of animal data into the human study
SNPs which have been found to be associated with the disorder in the GWAS with p<1×10-3 or which were found to lie in a SNP cluster (cluster definition: ≥2 SNPs with p ≤1×10-2 and at least one SNP with p<1×10-3 in a distance of <30kb) (see, e.g., eFigures 2-4) were taken forward to the follow-up study if they were located in a homolog of a rat gene showing differential brain expression with long-term high ethanol consumption compared to their respective controls without ethanol access. Only one SNP was selected for each gene. If more than one SNP in the gene fulfilled the above mentioned criteria, the SNP with the highest number of assigned transcripts (Sullivan et al., annotation file: https://slep.unc.edu/evidence/) was taken forward. In cases of similar transcript numbers, the SNP with the lowest p-value was chosen.
In the animal study, we have used a convergent approach with two strategic lines to integrate the rat data:
a qualitative approach looking for orthologous or paralogous genes in GWAS and expression profiling in rats (see list of genes in eTables 2 and 3).
a (semi-quantitative) ranking strategy as theoretically described by Bertsch et al.,28 where those orthologous or paralogous genes were included. In this strategy a weighing ratio of 1 (both datasets with equal weights) was employed, multiplying the p-values of respective rat and human genes.
With strategy a) a total number of 19 additional SNPs were identified and ranked according to strategy b). Since the total number of SNPs derived through the animal approach was only 19, we eventually decided to include all of them in the follow-up study instead of selecting a small subset based on the ranking order of genes. The three genes which were confirmed by the follow up study (Tab. 1) had been ranked # 7 (ADH), #12 (CDH), and # 19 (GATA) in approach b). We want to conclude from these data, that it is hard to introduce quantitative evaluations in those convergent approaches. Nevertheless, convergent approaches as pursued in this study can provide very valuable additional information and apparently become increasingly more popular.
We searched Gene Ontology (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi) for the annotation of gene products for those genes in which confirmed SNPs of the follow up study are located. Gene Ontology provides a standardised vocabulary describing gene products in order to ensure uniformity across databases.49 Weinvestigated whether these genes share distribution patterns in Gene Ontology (GO) categories by mapping them to GO annotations.
Results
Human Studies
We excluded 11 of the total 487 GWAS cases with alcohol dependence from further analyses. Of these, six DNA samples failed to genotype, and, in five subjects, the genome-wide IBS score was above 1.60, indicating possible cryptic relatedness. Of the 561,466 SNPs genotyped per individual, 99.6% passed quality control (QC). The final data set was comprised of 524,396 SNPs (511,701 autosomal and 12,695 sex-linked markers) in a total of 476 male cases and 1,358 male controls. The mean call rate of the final set of SNPs (cases and controls) was 99.76% (±0.45%).
We used two complementary SNP selection strategies, a ‘lowest p-value’ strategy and a ‘rodent candidate gene’ strategy, to prioritize SNPs for follow-up. In total, 139 SNPs were carried forward for genotyping in the follow-up sample. These included all SNPs with a p-value <10-4 (Armitage trend test for autosomes and allelic test for x-chromosome), n=121, of which an assay design was possible for n=120 (see Supplementary Online Content eTable 1), and additional 19 SNPs with at least nominal significance, which would not have been accounted for by the ‘lowest p-value’ approach. These 19 SNPs are among 22 SNPs (three of which were already represented in the aforementioned ‘lowest p-value’ selection) located in human homologs of rat genes showing differential expression in the rat brain following chronic alcohol consumption (for more details see Supplementary Online Content) and therefore have a higher a priori probability of being involved in the aetiology of alcohol dependence.
In the follow-up study, 16 SNPs showed association with at least nominal significance (p<0.05, two-sided), 15 of which (9 intragenic, 6 intergenic) were associated with the same allele as in the GWA study. The number of significantly replicated SNPs is higher than expected by chance (p=2.45×10-6). Three of the intragenic SNPs are derived from the 22 SNPs selected on the basis of the animal model (Table 1). This number is also significantly higher than that expected by chance (p=1.69×10-2).
Table 1
| SNP | Chromosomal band | Genesa |
|---|---|---|
| rs1344694 | 2q35 | |
| rs7590720 | 2q35 | |
| rs705648 | 2q35 | Peroxisomal trans-2-enoyl-CoA reductase (PECR) |
| rs1614972b | 4q23 | Alcohol dehydrogenase 1C (class I), gamma polypeptide (ADH1C) |
| rs13362120 | 5q15 | Calpastatin (CAST) |
| rs13160562 | 5q15 | Endoplasmic reticulum aminopeptidase 1 (ERAP1); calpastatin (CAST) |
| rs1864982 | 5q32 | Protein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform (PPP2R2B) |
| rs6902771 | 6q25.1 | Estrogen receptor 1 (ESR1) |
| rs729302 | 7q32.1 | |
| rs13273672b | 8p23.1 | GATA binding protein 4 (GATA4) |
| rs1487814 | 11p14.3 | |
| rs7138291 | 12q22 | Coiled-coil domain containing 41 (CCDC41) |
| rs36563 | 14q24.2 | |
| rs11640875b | 16q23.3 | Cadherin 13, H-cadherin (heart) (CDH13) |
| rs12388359 | Xp22.2 |
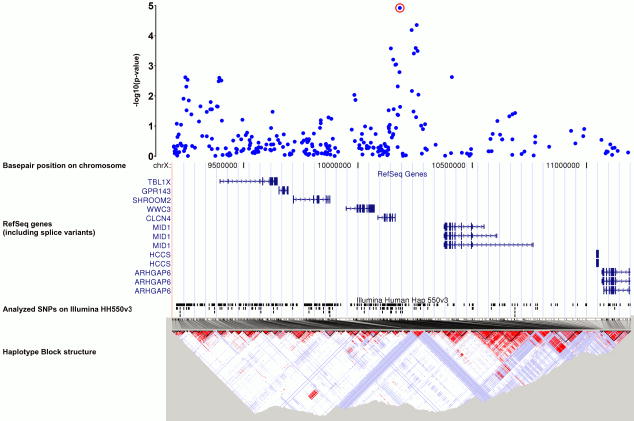
Combining the data across the two samples showed genome-wide significance (Bonferroni adjusted α-level of 0.05/524k)50,51 for two SNPs, i.e. rs7590720 (p=9.72×10-9) and rs1344694 (p=1.69×10-8). These SNPs map to the 3′-flanking region of the gene encoding peroxisomal trans-2-enoyl-coA reductase (PECR), located on chromosome 2q35. They are in linkage disequilibrium with each other and with another SNP, rs705648 (p=1.78×10-6),located within the PECR gene (rs1344694 – rs7590720: D′=1.0, r2=0.739; rs1344694 - rs705648: D'= 0.943, r2=0.568; rs7590720 – rs705648: D′=0.948, r2=0.776; HapMap).
A further eight SNPs are located within genes: rs13362120 (combined p-value 1.85×10-5) in the calpastatin (CAST) gene; rs13160562 (combined p-value 7.09×10-6) in the endoplasmatic reticulum aminopeptidase 1 (ERAP1)/calpastatin (CAST) genes; rs1864982 (combined p-value 3.46×10-6) in the protein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform (PPP2R2B)gene; rs6902771 (combined p-value 8.30×10-6) in the estrogen receptor 1 (ESR1) gene; rs7138291 (combined p-value 3.68×10-5) in the coiled-coil domain containing 41 (CCDC41) gene; rs1614972 (combined p-value 1.41×10-4) in the alcohol dehydrogenase 1c (ADH1C) gene; rs13273672 (combined p-value 4.75×10-4) located in the GATA binding protein 4 (GATA4) gene and rs11640875 (combined p-value 1.84×10-5) in the cadherin 13 (CDH13) gene. The first five SNPs were selected for the follow-up study through the ‘lowest p-value’ strategy and the latter three via the ‘rodent candidate gene’ approach. These findings and those of the intergenic SNPs are provided in Table 1, Table 2 and eTable 5.
Table 2
SNPs confirmed in the follow-up study – Comparison of results of GWAS, replication and pooled analysis.
| GWA Scana | Replication Sampleb | Pooled Samplec | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| SNP | Allele Ad | Allele Bd | Frq A Ca | Frq A Co | P-valuee | Odds Ratio (95% CI) | Frq A Ca | Frq A Co | P-valuee | Odds Ratio (95% CI) | Frq A Ca | Frq A Co | P-valuee | Odds Ratio (95% CI) |
| rs1344694 | G | T | 0.603 | 0.685 | 5.57E-6 | 0.70 (0.60–0.81) | 0.613 | 0.659 | 3.36E-3 | 0.82 (0.72–0.93) | 0.610 | 0.674 | 1.69E-8 | 0.76 (0.69–0.83) |
| rs7590720 | A | G | 0.645 | 0.724 | 5.70E-6 | 0.69 (0.59–0.81) | 0.661 | 0.713 | 6.68E-4 | 0.79 (0.68–0.90) | 0.656 | 0.719 | 9.72E-9 | 0.74 (0.67–0.82) |
| rs705648 | C | T | 0.313 | 0.244 | 2.68E-5 | 1.41 (1.20–1.67) | 0.292 | 0.256 | 1.24E-2 | 1.19 (1.04–1.37) | 0.299 | 0.249 | 1.78E-6 | 1.29 (1.16–1.43) |
| rs1614972 | C | T | 0.754 | 0.690 | 2.84E-4 | 1.37 (1.16–1.61) | 0.721 | 0.690 | 3.62E-2 | 1.16 (1.01–1.33) | 0.732 | 0.690 | 1.41E-4 | 1.23 (1.10–1.36) |
| rs13362120 | C | T | 0.258 | 0.329 | 5.72E-5 | 0.71 (0.60–0.84) | 0.296 | 0.333 | 1.25E-2 | 0.84 (0.73–0.96) | 0.283 | 0.331 | 1.85E-5 | 0.80 (0.72–0.88) |
| rs13160562 | A | G | 0.252 | 0.326 | 2.31E-5 | 0.70 (0.59–0.83) | 0.283 | 0.316 | 2.47E-2 | 0.85 (0.75–0.98) | 0.273 | 0.322 | 7.09E-6 | 0.79 (0.71–0.88) |
| rs1864982 | A | C | 0.181 | 0.129 | 9.71E-5 | 1.49 (1.22–1.82) | 0.163 | 0.130 | 4.47E-3 | 1.30 (1.09–1.56) | 0.169 | 0.130 | 3.46E-6 | 1.36 (1.20–1.55) |
| rs6902771 | C | T | 0.591 | 0.517 | 9.31E-5 | 1.35 (1.16–1.56) | 0.552 | 0.505 | 3.15E-3 | 1.20 (1.06–1.37) | 0.565 | 0.512 | 8.30E-6 | 1.24 (1.13–1.36) |
| rs729302 | A | C | 0.599 | 0.671 | 6.19E-5 | 0.73 (0.63–0.85) | 0.641 | 0.677 | 1.84E-2 | 0.85 (0.75–0.97) | 0.627 | 0.673 | 4.02E-5 | 0.82 (0.74–0.90) |
| rs13273672 | C | T | 0.364 | 0.312 | 2.18E-3 | 1.27 (1.09–1.47) | 0.342 | 0.311 | 3.61E-2 | 1.15 (1.01–1.32) | 0.350 | 0.312 | 4.75E-4 | 1.19 (1.08–1.31) |
| rs1487814 | A | G | 0.589 | 0.511 | 3.58E-5 | 1.37 (1.18–1.59) | 0.544 | 0.508 | 2.48E-2 | 1.15 (1.02–1.30) | 0.559 | 0.510 | 3.49E-5 | 1.22 (1.11–1.34) |
| rs7138291 | C | T | 0.142 | 0.093 | 2.46E-5 | 1.61 (1.28–2.00) | 0.115 | 0.095 | 4.10E-2 | 1.23 (1.01–1.54) | 0.124 | 0.094 | 3.68E-5 | 1.36 (1.18–1.58) |
| rs36563 | A | C | 0.210 | 0.147 | 6.21E-6 | 1.54 (1.28–1.85) | 0.184 | 0.159 | 3.83E-2 | 1.19 (1.01–1.41) | 0.192 | 0.152 | 4.57E-6 | 1.33 (1.18–1.50) |
| rs11640875 | A | G | 0.389 | 0.326 | 5.25E-4 | 1.32 (1.12–1.54) | 0.372 | 0.332 | 9.85E-3 | 1.19 (1.04–1.35) | 0.377 | 0.329 | 1.84E-5 | 1.24 (1.12–1.36) |
| rs12388359 | G | T | 0.811 | 0.900 | 1.21E-5 | 0.47 (0.34–0.67) | 0.840 | 0.879 | 1.25E-2 | 0.72 (0.56–0.93) | 0.831 | 0.888 | 3.57E-6 | 0.62 (0.50–0.76) |
Specific GO biological process, cellular component and molecular function terms remained largely unique for each of the genes and were not found to be enriched among the genes analysed. No specific coherent underlying process could be deduced from GO term associations. As a consequence, we considered the literature-derived molecular function of each of the gene products of intragenic SNPs separately.
Animal Studies
The GeneChip® Rat Genome U34 Set provides gene expression data for more than 24,000 known genes and EST clusters for comprehensive coverage of the rat genome. The two brain regions were evaluated separately. 3-way ANOVA revealed 542 genes differentially expressed at a p<0.05 in either caudate putamen or amygdala or both brain regions of alcohol preferring P, HAD, and AA rats after one year of ethanol consumption. A detailed description of the ‘rodent candidate gene’ strategy for selection of markers for the follow-up study is given in paragraph ‘Integration of animal data into the human study’.
Comment
Our GWA and replication study of alcohol dependence has detected and replicated evidence for association with 15 markers. Two of these markers, rs7590720 and rs1344694, remained significant after genome-wide correction for multiple testing in the combined sample of 1460 patients and 2332 control persons.
These two markers are located ~5kb apart in chromosomal region 2q35, which has been implicated in alcohol dependence through previous linkage studies. Linkage to this region was found in a genome-wide search in 2282 individuals from 262 families with a high density of alcohol dependence by the Collaborative Study on the Genetics of Alcoholism (COGA).52 Near the microsatellite D2S1371, the highest LOD score for comorbid alcoholism and depression phenotype was 4.12 (this LOD score however, was only obtained in one dataset, i.e. in the replication dataset, while in the initial and combined datasets the LOD scores were 0.00 and 2.16, respectively).52 Marker D2S1371 is located ~1.4Mb away from rs7590720 and rs1344694. Another COGA project53 found linkage with a maximum LOD score of 2.4 between a low level of response (LR) to alcohol and the microsatellite marker D2S434 which is only ~1.7Mb away from our markers rs7590720 and rs1344694. The LR to alcohol is an endophenotype related to heavy drinking and alcohol problems.54 It is genetically influenced in both animals55-57 and humans58,59 with an estimated heritability in humans of between 40-50%60. Two further linkage studies of the COGA project61,62 reported linkage with LOD scores of 3.28 and 3.0, respectively, between the same marker D2S434 and the P3(00) component of the event related potential (ERP). Heritability of the P3 amplitude ranges between 50-80% (for review see 62) and P3 deficits have been repeatedly reported to be a strong vulnerability factor for alcohol dependence, especially in males with a high familial loading of alcohol dependence.63,64 In a linkage scan conducted by Hill et al.65 in 330 members from 65 families with a high density of alcohol dependence, linkage to 2q35 with marker D2S2382 was identified. This marker is located only ~0.2Mb from our two main SNPs. A LOD score of 3.68 was observed in a four-parameter model which included measurement of age, gender and alcohol dependence along with measurement of constraint - a personality construct66 which appears to overlap with risk taking behaviour.
These linkage findings, which all satisfied the Lander and Kruglyak67 criteria for suggestive linkage, underline the potential importance of this chromosomal region. However, since no meta-analysis of linkage studies in alcohol addiction exists, the possibility that these findings are merely due to random noise cannot be ruled out.
The gene which is closest to the peak association signal and which also harbours another associated SNP, rs705648 (p=1.78×10-6), is the peroxisomal trans-2-enoyl-coA reductase (PECR). PECR appears to be a key enzyme of fatty acid metabolism as it catalyzes the reduction of medium chain (unsaturated) enoyl-CoAs to saturated acyl-CoAs which may then undergo oxidation in mitochondria for energy production. This pathway, originating from triglycerides, is particularly important in conditions of starvation when energy supply is switched from utilization of glucose to fatty acids. In this condition, PECR needs to be up-regulated. Triglycerides are typically increased in the blood of alcohol dependent individuals.68,69 PECR reduces C-C double bonds in not only even-chain, but also in odd-chain fatty acids. On degradation to acetyl-CoA, odd-chain fatty acids end up with propionyl-CoA, which requires carboxylation to an even-chain fatty acid. This step is mediated through methyl-malonyl-CoA, an intermediate formed by the essential cofactors methyl-tetrahydrofolate as the methyl-donor, and vitamin B12. Folate deficiency, often observed in alcohol-dependent patients,70 may therefore interfere with this pathway and prevent the availability of a sufficient supply of acetyl-CoA. Expression of PECR is highest in the liver, followed by the kidney, muscle tissue, the lungs and the heart, but barely – if at all – in the brain.71If this gene is involved in alcohol dependence, it must act in the periphery rather than in the CNS. It could therefore be expected that we detected PECR only in our GWAS, which identifies susceptibility genes independent of organ-specific expression, but not in the animal approach, which is brain-specific.
A further 4 SNPs are intergenic and further eight SNPs are located in genes. No ontological relatedness was detected between these genes. That is not surprising, given the assumed genetic heterogeneity of alcohol addiction. In the Supplemental Text, we discuss data from linkage findings, biochemistry and animal studies that have provided evidence that these loci may be involved in alcohol dependence. However, none of these markers achieved genome-wide significance in our study. This does not necessarily disqualify them. Alcohol dependence is a complex and heterogeneous disorder that is influenced by multiple genetic, biological, psychological, and sociocultural factors. Formal genetic and linkage studies do not indicate the existence of genes with major effects in the European population. Thus, genome-wide significant findings in moderately-sized samples will be rare, but the number of replicable associations may actually be much larger. One replicated association that did not withstand correction for multiple testing was obtained with the SNP rs1614972 in the ADH1C gene. The ADH gene cluster is located on chromosomal region 4q, which is among the most consistently replicated loci contributing to alcohol phenotypes.72 The ADH1C gene belongs to the biologically relevant alcohol-metabolising alcohol dehydrogenase (ADH) genes. The p-value of this SNP, which was at position 282 of our top-down ranking, achieved a p=2.84×10-4 in the GWAS. It would not have been carried forward into the follow-up study if it had not been supported by evidence from our animal study. In order to achieve a power of 80% to detect significant association withstanding correction for multiple testing, we would have needed to include 5573 patients and 8900 controls (regarding actual OR and MAF of this SNP). The same is true for our replicated association to marker rs11640875 which had rank no. 510 in our GWAS findings. It is located in the cadherin 13 (CDH13) gene, a gene which has been described in previous studies as a susceptibility locus for both alcohol dependence25 and methamphetamine dependence.73 Increasing sample sizes is often not feasible and bears the risk of increasing clinical as well as genetic heterogeneity. Experience from GWAS into diabetes, for example, has shown that differences in assessment can jeopardize even the most robust findings.74 Obtaining very large sample sizes may necessitate inclusion of samples from different cultural and ethnic backgrounds which can also increase heterogeneity. The ADH1C gene belongs to the ADHclass I genes (ADH1A, ADH1B and ADH1C) which have been extensively investigated for the risk of alcohol dependence, initially in Asian75,76,77and subsequently in African78,79and European80-84populations. A metaanalysis85 reported a significantly higher risk of alcohol dependence among carriers of the ADH1C*2 allele (OR=1.91) in East Asian populations, but its role in the European population is less clear. Our associated marker rs1614972 is in complete LD (D'=1.0; r2=0.311, HapMap) with one of the two polymorphisms of the ADH1C*2 protein isoform that has recently been investigated in 575 patients and 530 controls from the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) sample. The two marker haplotype comprised of markers rs1614972 and rs1693482 yielded evidence for association with alcohol dependence. Single marker analysis with rs1614972, however, produced no evidence at all. While the allele frequencies in Irish and German patients are the same, they differ between the Irish and German control samples; the allele frequencies between the German GWAS and the follow-up study being the same. This finding underlines the importance of sample homogeneity.
In order to increase the genetic homogeneity of the sample, and thus increase the power of the study, we attempted to select patients as rigorously as possible. Patients of German descent were recruited within a joint research project and were stratified for male gender and an early age at onset, a combination of traits which is associated with increased heritability.32,33 Prior studies have shown that population stratification in Germansare small.40,86,87 Thus, at the time when SNPs for the follow-up study were chosen, we had not applied a formal test to control for population stratification. A post-hoc principal components analysis (PCA)88(EigenSoftv1.01; http://www.genepath.med.harvard.edu/~reich/Software.htm) and genomic control analysis42 (showing a lambda of 1.099) had no major impact on the significance level of our 15 SNPs (see Supplementary Online Content, eTable 4), thus confirming these prior obervations.
Alcohol dependence as defined by DSM-IV is complex, and association studies with more biologically refined (endo) phenotypes will help to disentangle this heterogeneity. Our approach of selecting more genetically homogeneous groups by stratifying for male sex and and an earlier age at onset is a first step in this direction. We must point out, however, that in order to achieve our overall sample of more than 1500 patients, it was necessary to relax our early age at onset criteria. While in the GWAS patients the upper inclusion limit was 28 years with a median of 20 years, the upper inclusion limit in the replication sample had to be extended to 45 years with a median of 30 years. Although, this cannot be considered as a specifically early age at onset, the sample as a whole and especially the GWAS sample has clearly been stratified for early age at onset.
In conclusion, we have identified genome-wide significant association with two SNPs located in a chromosomal region for which linkage and animal studies provide compelling evidence for the presence of susceptibility variants predisposing to alcohol addiction. Further independent studies are required to confirm these findings. The gene, which is located close to these SNPs - PECR- is a plausible candidate and merits further investigation. We have shown that the GWA approach is a powerful tool for producing valuable findings in even a moderately-sized sample, when it has been carefully selected for genetically homogeneous cases. In addition, our study underlines the value of a convergent approach between animal studies and systematic genetic screens in humans in order to deal with the multiple testing problem in GWAS.

Genome-wide overview of GWAS findings. SNPs at which the test statistic exceeds 30, are represented by triangles at the top of the plot.
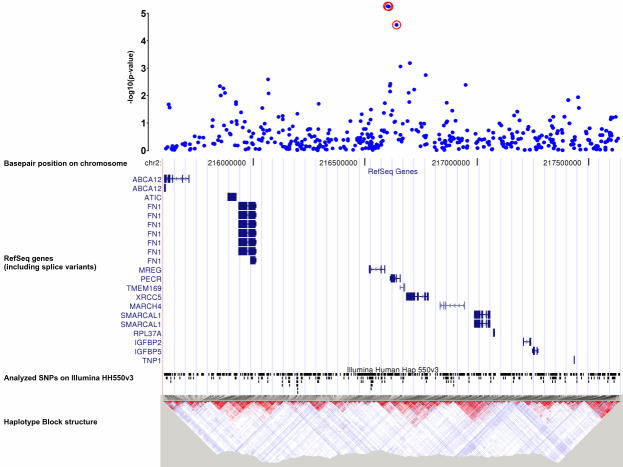
Illustration of p-values in a +/− 1Mb interval around the most significant findings at 2q35: rs1344694, rs7590720 and rs705648. Gene annotations from UCSC RefSeq genes, LD-maps from HapMap.
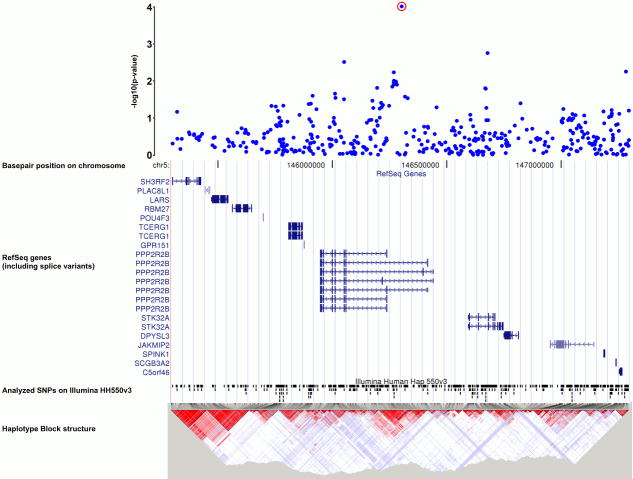
Illustration of p-values in a +/− 1Mb interval around the most significant finding rs1864982 at 5q32.Gene annotations from UCSC RefSeq genes, LD-maps from HapMap.
Supplementary Material
Supplement
eTable1. Markers with p<1E-4 from GWAS selected for follow-up
eTable2. Cross section of human genes with nominal significance in GWAS and rat homologs differentially expressed after chronic ethanol consumption
eTable3. SNPs derived from comparison of GWAS data with animal studies
eTable4. Comparison of analyses without correction for stratification vs. two different methods to control for population stratification
eTable 5. p-values of tests for deviation from Hardy-Weinberg-Equilibrium for case and control samples in both stages
Acknowledgments
Funding Support: This work was supported by a grant from the National Genome Research Network (NGFN) (to MR, MMN and HEW) of the German Federal Ministry of Education and Research (BMBF), and by grants FKZ 01GS0117/NGFN and FKZ EB 01011300; and 01EB0410 from the Bundesministeriumfür Bildung und Forschung (RS and KM). MMN and SC received support from the Alfried Krupp von Bohlen und Halbach-Stiftung. The Heinz Nixdorf Recall Study was supported by a grant of the Heinz Nixdorf Foundation.
Footnotes
The content of the paper was/will be presented as an oral presentation at the following meetings: Deutscher Suchtkongress, Mannheim, Germany, 11.-14. 06 2008; Drei-Länder-Symposium für Biologische Psychiatrie, Göttingen, Germany, 09.–11.10.2008; XVIth World Congress on Psychiatric Genetics, Osaka, Japan, 11.-15.10.2008; 1st Annual Meeting NGFN-Plus and NGFN-Transfer in the Program of Medical Genome Research, Helmholtz ZentrumMünchen/Neuherberg, Germany, 12.-13.12.2008.
Financial Disclosure: None reported.
Additional Contributions: We thank Marina Füg, Christine Hohmeyer and Rosa Ferrando for their expert technical assistance. We also thank Thomas G. Schulze, MD, CIMH/NIMH Mood and Anxiety Disorders Program, for his helpful discussions and Christine Schmael, MD, CIMH, for critically reading the manuscript.
Access to Data: The three listed first and three listed senior authors are responsible for distinct, but integral, parts of the study: the clinical study with homogenous assessment of the patients, the animal study and the genetic study. They had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
References
Full text links
Read article at publisher's site: https://doi.org/10.1001/archgenpsychiatry.2009.83
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4229246?pdf=render
Free to read at archpsyc.ama-assn.org
http://archpsyc.ama-assn.org/cgi/content/abstract/66/7/773
Subscription required at archpsyc.ama-assn.org
http://archpsyc.ama-assn.org/cgi/reprint/66/7/773.pdf
Subscription required at archpsyc.ama-assn.org
http://archpsyc.ama-assn.org/cgi/content/full/66/7/773
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Human genetics and epigenetics of alcohol use disorder.
J Clin Invest, 134(16):e172885, 15 Aug 2024
Cited by: 0 articles | PMID: 39145449 | PMCID: PMC11324314
Review Free full text in Europe PMC
Generalized genetic liability to substance use disorders.
J Clin Invest, 134(11):e172881, 03 Jun 2024
Cited by: 0 articles | PMID: 38828723 | PMCID: PMC11142744
Review Free full text in Europe PMC
Neuronal-specific methylome and hydroxymethylome analysis reveal significant loci associated with alcohol use disorder.
Front Genet, 15:1345410, 03 Apr 2024
Cited by: 0 articles | PMID: 38633406 | PMCID: PMC11021708
Maternal pre-pregnancy BMI, offspring epigenome-wide DNA methylation, and childhood obesity: findings from the Boston Birth Cohort.
BMC Med, 21(1):317, 23 Aug 2023
Cited by: 6 articles | PMID: 37612641 | PMCID: PMC10463574
GABAA receptor subtypes and benzodiazepine use, misuse, and abuse.
Front Psychiatry, 13:1060949, 12 Jan 2023
Cited by: 12 articles | PMID: 36713896 | PMCID: PMC9879605
Review Free full text in Europe PMC
Go to all (256) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (Showing 16 of 16)
- (6 citations) dbSNP - rs705648
- (6 citations) dbSNP - rs1344694
- (5 citations) dbSNP - rs1614972
- (5 citations) dbSNP - rs7590720
- (3 citations) dbSNP - rs7138291
- (3 citations) dbSNP - rs13362120
- (3 citations) dbSNP - rs1864982
- (3 citations) dbSNP - rs13160562
- (3 citations) dbSNP - rs6902771
- (3 citations) dbSNP - rs13273672
- (3 citations) dbSNP - rs11640875
- (2 citations) dbSNP - rs729302
- (2 citations) dbSNP - rs36563
- (2 citations) dbSNP - rs1487814
- (2 citations) dbSNP - rs12388359
- (1 citation) dbSNP - rs1693482
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genome-wide association study of alcohol dependence implicates a region on chromosome 11.
Alcohol Clin Exp Res, 34(5):840-852, 01 Mar 2010
Cited by: 205 articles | PMID: 20201924 | PMCID: PMC2884073
Association of ADH4 genetic variants with alcohol dependence risk and related phenotypes: results from a larger multicenter association study.
Addict Biol, 16(2):323-333, 01 Apr 2011
Cited by: 11 articles | PMID: 20626721
Replication of genome wide association studies of alcohol dependence: support for association with variation in ADH1C.
PLoS One, 8(3):e58798, 13 Mar 2013
Cited by: 26 articles | PMID: 23516558 | PMCID: PMC3596339
Alcohol Dependence Genetics: Lessons Learned From Genome-Wide Association Studies (GWAS) and Post-GWAS Analyses.
Alcohol Clin Exp Res, 39(8):1312-1327, 25 Jun 2015
Cited by: 66 articles | PMID: 26110981 | PMCID: PMC4515198
Review Free full text in Europe PMC