Abstract
Free full text

Mitochondrial dynamics and inheritance during cell division, development and disease
Preface
During cell division, it is critical to properly partition functional sets of organelles to each daughter cell. The partitioning of mitochondria shares some common features with other organelles, particularly in their interactions with cytoskeletal elements to facilitate delivery to the daughter cells. However, mitochondria have unique features – including their own genome and a maternal mode of germline transmission – that place additional demands on this process. We discuss the mechanisms regulating mitochondrial segregation during cell division, oogenesis, fertilization and tissue development. The mechanisms that ensure the integrity of these organelles and their DNA include fusion-fission dynamics, organelle transport, mitophagy, and genetic selection of functional genomes. Defects in these processes can lead to cell and tissue pathologies.
Introduction
Organelles are a distinguishing feature of eukaryotic cells. During somatic cell proliferation, they must segregate properly to daughter cells, and during germline inheritance, a highly functional population of organelles must be transmitted to the offspring. One such organelle is the mitochondrion1, which is best known for its critical function in energy production via oxidative phosphorylation (OXPHOS). The OXPHOS pathway generates many more adenosine triphosphate (ATP) molecules per glucose molecule than the glycolysis pathway. Mitochondria also have important roles in other types of metabolism, in regulating intracellular calcium concentration and signalling in neurons, in assembly of iron-sulfur clusters that are important for oxidation-reduction reactions2, in apoptosis3, and in innate immunity4.
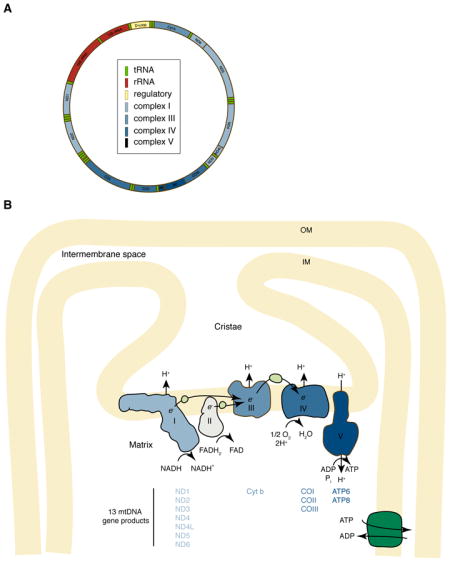
According to the endosymbiotic theory, mitochondria are descendants of ancient bacteria that entered into a symbiotic relationship with primitive host cells5. Mitochondria retain several characteristics of their putative bacterial ancestors: a double-membrane, a proteome similar to that of α-proteobacteria, and the ability to synthesize ATP via a proton gradient created across its inner membrane (Box 1). In addition to these prokaryotic characteristics, mitochondria also undergo membrane remodelling through cycles of fusion (two mitochondria joining to form a single mitochondrion) and division (or fission; a single mitochondrion dividing into two)6 (Box 2). The balance of fusion and fission controls mitochondrial structure, and depending on the cell type, the numerous separate mitochondria in the cell can shift to form a single, interconnected membranous structure.
Mitochondria have two features that constrain their segregation. First, they have a genome (mitochondrial DNA or “mtDNA”) that encodes critical bioenergetic functions (Box 1). Each mitochondrion contains one or more mtDNA molecules that are organized into mtDNA-protein complexes termed nucleoids. In proliferating cells, the partitioning of mitochondria to daughter cells must also result in proper distribution of mtDNA. Second, mitochondria in mammals are transmitted to subsequent generations exclusively through the maternal lineage. Because of this uniparental inheritance pattern, mtDNA lacks the benefits of recombination arising from sexual reproduction. However, mechanisms have evolved to reduce the chance of transmitting pathogenic mtDNA mutations to offspring. In some cases, these mechanisms fail, and maternally inherited disorders termed mitochondrial encephalomyopathies arise when a significant load of mtDNA mutations are passed to the offspring. These diseases are characterized by reduced mitochondrial function, with clinical signs being most pronounced in tissues with high energy consumption.
With mitochondria functioning in diverse cellular processes and their dysfunction implicated in several diseases, an understanding of mitochondrial dynamics, segregation and germline inheritance is of great interest. In this Review we first outline the molecular mechanisms that ensure proper partitioning of mitochondria during cell division: the fusion/fission cycle, mitochondrial interactions with cytoskeletal elements and the endoplasmic reticulum (ER), and mitophagy. In the second part of the Review we discuss the specific regulation of mtDNA inheritance and its uniparental transmission through the maternal lineage, including the evidence for genetic bottlenecks during oogenesis and embryonic development, and the mechanisms preventing transmission of paternal mitochondria to offspring. Throughout, our main focus is on mammalian mitochondria, but studies performed in yeast, worms and flies are discussed in cases where they have provided substantial insight to our understanding of those issues.
Mitochondrial dynamics and segregation
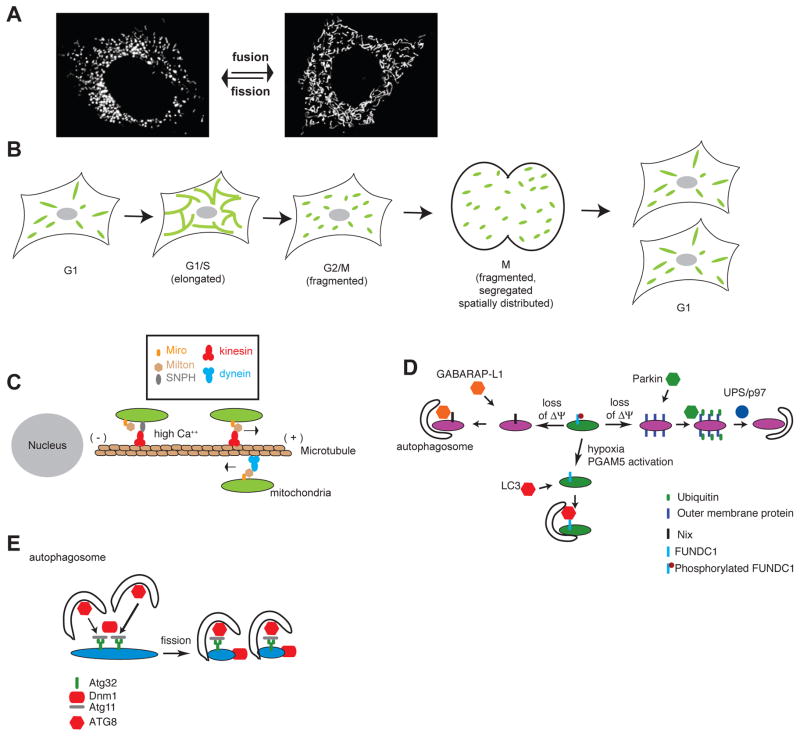
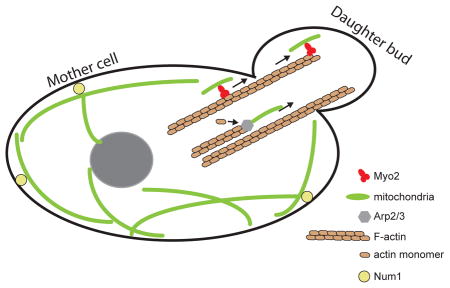
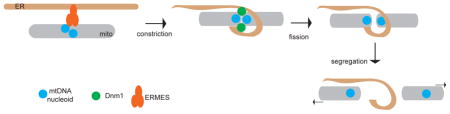
In discussing mitochondrial inheritance, “proper partitioning” of the mitochondria implies that mitochondria and mtDNA are distributed equally to daughter cells (in the case of symmetric cell division), and that the inherited mitochondria are highly functional. In mammalian cells that divide symmetrically, segregation could, in principle, be a largely passive process, as long as mechanisms exist to ensure a homogenous and evenly dispersed spatial distribution of organelles throughout the cell soma (Figure 1A). As discussed below, faithful segregation is facilitated by the induction of mitochondrial fission during the G2 and mitosis phases of the cell cycle (Fig. 1B) and by the interaction of mitochondria with the cytoskeleton and endoplasmic reticulum (ER) elements that are spread throughout the cell soma. Active mechanisms that deliver mitochondria to daughter cells are not known in mammalian cells; however, this may simply reflect a lack of suitable model systems to study this process. In particular, little is known about mitochondrial segregation in mammalian cells that divide asymmetrically. In budding yeast, which undergo asymmetric cell division, mitochondrial segregation is driven by mechanisms that actively deliver a subset of mitochondria in the daughter cell, while retaining another subset in the mother cell (Box 3).
Microtubule-based mitochondrial motility
Mitochondrial distribution within the cell soma depends on mitochondrial trafficking along the cytoskeleton. In mammals, microtubule filaments, created by the polymerization of α- and β-tubulin subunits, form a cytoskeletal platform for the distribution and transport of mitochondria The colocalization of mitochondria and microtubules has been well documented, and treatment of cells with microtubule depolymerizing agents results in misalignment and redistribution of mitochondria7, 8. Depending on the cellular context, actin and intermediate filaments can also regulate mitochondrial distribution. For example, the actin cytoskeleton plays a prominent role in organizing the presynaptic terminal and dendritic spines of neurons, and actin filaments help to recruit mitochondria to these regions9.
Motor proteins transport mitochondria along microtubules: Kinesins transport mitochondria in the plus-end (anterograde) direction, and dyneins transport mitochondria in the minus-end (retrograde) direction. Numerous kinesin family members have been implicated in mitochondrial anterograde transport. For instance, deletion of KIF5B, a Kinesin-1 family member, in mammalian placental cells leads to a dramatic redistribution of mitochondria - rather than being evenly distributed throughout the cell, they accumulate into a single perinuclear cluster10. This observation suggests that kinesin-based motility in the microtubule plus-end direction is required for proper mitochondrial distribution throughout the cell body. Other studies have provided evidence for the involvement of kinesins in mitochondrial anterograde motility in mammalian neurons11–13. In contrast to yeast (Box 3), deficiencies in mitochondrial motor proteins have not yet been causally linked with mitochondrial segregation defects in mammalian cells.
Additional components of the kinesin-based transport system have been identified in genetic screens of Drosophila melanogaster neuronal function14. The C-terminal cargo-binding domain of kinesin-1 interacts with an adaptor protein called Milton, which in turn binds to Miro, an integral membrane protein localized to the mitochondrial outer membrane. In fly neurons, anterograde mitochondrial transport is severely impaired in the absence of either Milton or Miro15, 16. Interestingly, Miro has calcium-binding EF hand domains that are involved in stalling mitochondria in response to local elevation of calcium concentration at active synapses within axons17. In mouse axons, an additional important factor that regulates mitochondrial localization is local stalling of mitochondria - at any given time, most mitochondria in axons are immobile. This pausing results from anchoring of axonal mitochondria to the microtubule cytoskeleton by syntaphilin (SNPH)18. When calcium levels are high, Miro releases Kinesin-1, which is then bound and inhibited by SNPH19. This close interplay between SNPH and the Milton-Miro transport complex regulates the saltatory movement of axonal mitochondria. Moreover, data from proliferating mammalian cells suggest that the mitochondrial kinesin complex plays an important role in regulating the mitochondrial distribution throughout the cell soma. In mammalian fibroblasts, co-overexpression of Milton and KIF5B, a member of the Kinesin-1 family, results in clustering of mitochondria at the cell periphery20, and KIF5B deletion results in mitochondrial accumulation around the nucleus. Thus, the Miro–Milton–KIF5B complex can regulate mitochondrial distribution through the promotion of anterograde motility (Figure 1C). Recent data suggests that Miro and Milton can also regulate retrograde motility through their association with dynein motor proteins21 (Figure 1C).
Because cells lacking Miro or Milton function show dramatic changes in mitochondrial distribution, it is possible that these molecules play a role in mitochondrial partitioning during cell division. However, this point remains to be experimentally addressed, because most studies have utilized non-dividing neurons. In contrast to mammals, mitochondrial motility and distribution in budding yeast is dependent on the actin cytoskeleton, and mutants in these processes show clear segregation defects (Box 3).
Fusion and fission ensure mitochondrial homogenization
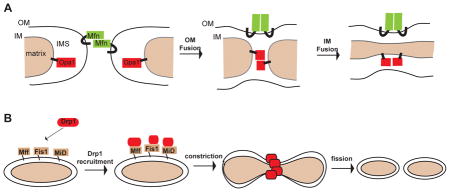
During mitochondrial fusion, two mitochondria merge both their outer and inner membranes, resulting in a single, larger mitochondrion. Mitochondrial fission is the opposite process, whereby a mitochondrion divides into two organelles. Fusion of mitochondria is mediated by three dynamin-related GTPases: the Mitofusins 1 and 2 (Mfn1 and Mfn2) on the mitochondrial outer membrane and Optic atrophy 1 (Opa1) on the inner membrane, whereas fission is mediated by the Dynamin-related protein 1 (Drp1)6, which is also a large GTPase (Box 2). Complete deficiencies in any of these proteins is embryonic lethal in mice, and mutations in some of these proteins are associated with neuromuscular disease in humans6, 22. Studies in cell culture and tissues suggest that fusion and fission are critical for maintaining the health of the mitochondrial population23–26. For example, when mitochondrial fusion is blocked, Purkinje neurons in the cerebellum degenerate due to defects in electron transport and mtDNA maintenance27.
How could fusion and fission maintain the proper function of mitochondria? Cells lacking the mitofusins or Opa1 show a striking heterogeneity between mitochondria in protein content, mtDNA nucleoid content and membrane potential23, 27–29. As a result, mutant cells show defects in respiratory chain function28 and accumulation of mtDNA deletions23. Thus, cycles of fusion and fission promote mixing of mitochondrial content and homogenization of mitochondrial protein and DNA in the cell. Whereas functional mitochondria continually mix their contents, quality control mechanisms exist to prevent grossly dysfunctional mitochondria from fusing with healthy mitochondria30. For example, mitochondria that lose their inner membrane potential undergo proteolytic inactivation of Opa131, resulting in loss of inner membrane fusion that prevents their re-incorporation into the cellular mitochondrial population network30. In addition, many forms of mitochondrial dysfunction result in activation of mitochondrial fission or inactivation of fusion31, 32. These dynamic changes promote the conversion of dysfunctional mitochondria into smaller physical units that can be targeted for degradation via autophagy (see below). Conversely, mitochondria that elongate due to inhibition of the fission activity of Drp1 are protected from autophagy, presumably due to their enlarged size33, 34.
Mitochondrial fusion and fission dynamics are regulated during the cell cycle35. Enhanced fusion is associated with the G1-to-S phase transition, when the mitochondria tend to appear as large, interconnected networks (Figure 1B). Mitochondrial fission is activated during mitosis35, 36 through the phosphorylation of Drp1, resulting in individual mitochondrial units. Because they are associated with cytoskeletal elements and with the ER (Box 4), these individual mitochondria are distributed evenly throughout the cell soma, which ensures that they are passively partitioned in equal numbers to daughter cells during cytokinesis. In cells lacking Drp1, the mitochondria exist constitutively as elongated networks due to lack of mitochondrial fission. Nevertheless, mitochondria are segregated to daughter cells, presumably because the cytokinesis machinery is sufficient to forcibly cleave mitochondria25, 26. However, the partitioning of mitochondria to daughter cells in Drp1 mutant cells appears less uniform, perhaps because the mitochondria remain in large networks during mitosis25, although this observation remains to be quantified systematically. It is unclear whether this segregation defect has any physiological consequence in the daughter cells.
Thus, the combined effects of fusion and fission promote a homogenous and healthy population by mixing mitochondrial protein and DNA contents throughout the mitochondrial population of the cell, whereas enhanced fission at the onset of mitosis facilitates equal passive segregation mitochondria to daughter cells.
Mitochondrial clearance by mitophagy
Mitophagy is the sequestration of mitochondria by autophagosomes followed by their degradation in lysosomes. Thought to be the primary mechanism for degradation of mitochondria, mitophagy controls the composition of the mitochondrial population in the cell by culling subpopulations of mitochondria that are dysfunctional. Although mitochondria can be non-selectively removed as part of a bulk autophagy response, mitophagy can be selective for damaged mitochondria under certain cellular contexts. For instance, mitochondria that are experimentally depolarized attract the autophagy machinery and undergo mitophagy40, 41. In yeast, nitrogen-starvation leads to bulk cellular autophagy that includes mitophagy. However, if cells starved for nitrogen are kept on a carbon source requiring mitochondrial metabolism, mitophagy (but not cellular autophagy in general) is reduced, indicating that mitophagy can be differentially regulated from bulk autophagy42. By selectively targeting dysfunctional mitochondria, mitophagy could serve to maintain the quality of the mitochondrial population. However, how efficiently mitophagy is in achieving this remains unclear, as there are many pathological states, such as mitochondrial encephalomyopathies, in which highly dysfunctional mitochondria or pathogenic mtDNA mutations persist in cells and tissues43.
The Pink1-Parkin Mitophagy pathway
The mitophagy pathway most studied involves Pink1 and Parkin; mutations in their encoding genes are involved in familial cases of Parkinson’s disease44. As first shown in fly mutants, Parkin acts downstream of Pink1 to maintain mitochondrial function45–47. In mammalian cell culture models, these two proteins promote mitophagy of compromised mitochondria. In response to depolarization of the mitochondrial inner membrane, Parkin, which is an E3 ubiquitin ligase, translocates from the cytosol onto the mitochondrial outer membrane in a Pink1-dependent manner48, 49, where it causes widespread polyubiquitination of mitochondrial outer membrane proteins50, 51. This polyubiquitination promotes degradation of mitochondrial outer membrane proteins by the 26S proteasome50, 52. Protein degradation is also dependent on the AAA ATPase p9753, which may have a role in extracting proteins from the mitochondrial outer membrane in order to make them more accessible to the proteasome54. Degradation of mitochondrial outer membrane proteins is necessary for the mitophagic response50 (Figure 1D). Overexpression of Parkin in cytoplasmic hybrid (cybrid) cells which are heteroplasmic for a mutation in the mitochondrial COX1 gene reduces the level of the pathogenic mutation, suggesting that the Pink1-Parkin pathway can introduce bias in the maintenance of mtDNA genotypes55. The functions of Pink1 and Parkin in mitophagy have mostly been studied by overexpression in mammalian cell lines. However, their involvement in mitophagy is supported by proteomic analyses indicating that flies mutated in these genes have slower turnover of mitochondrial proteins56.
Other mitophagy pathways
During erythrocyte maturation, intracellular organelles such as mitochondria are completely degraded. It has been suggested that the removal of damaged mitochondria is important in preventing downstream pathways that lead to erythrocyte cell death57. The mitochondria are eliminated by sequestration into autophagosomes, but the process may involve both canonical and non-canonical autophagy pathways. At least some mitochondrial degradation persists in the absence of core components of the autophagic machinery, such as the ATG558 or ATG759 proteins. The BH3 (Bcl2 homology domain 3)-containing protein Nix, located on the mitochondrial outer membrane, has been shown to be important for elimination of mitochondria from erythrocytes (Figure 1D). In Nix knockout mice, about a third to a half of the circulating erythrocytes show aberrant persistence of mitochondria, even though other cellular components, such as ribosomes, are degraded60, 61. During maturation, erythroid mitochondria normally show loss of membrane potential, which likely acts as a signal to promote mitophagy. Mitochondrial membrane depolarization fails to occur in Nix-deficient erythroid cells, and artificial dissipation of the membrane potential can restore mitophagy61. Nix contains an N-terminal LIR (LC3-interacting region) motif that binds to LC3 (Microtubule-associated proteins 1A/1B light chain 3)-like proteins, especially GABARAP-L162, which are ubiquitin-related proteins that are located on autophagosomes and are important for their maturation. The LIR motif is essential for the ability of Nix to mediate mitophagy62. These observations suggest that Nix is a selective mitophagy receptor that physically connects the autophagy machinery to the mitochondrial surface in erythroid cells.
In cultured mammalian cells, another mitophagy receptor is the outer membrane protein FUNDC1, which regulates autophagic degradation of mitochondria in response to hypoxia63 (Figure 1D). Overexpression of FUNDC1 reduces total mitochondrial mass in cells, and FUNDC1 depletion conserves mitochondrial mass during hypoxia63. FUNDC1 has a LIR motif that is important for recruitment of LC3 and for mitochondrial degradation. Under hypoxic conditions, the mitochondrial phosphatase PGAM5 (phosphoglycerate mutase family member 5) dephosphorylates the LIR motif, which increases its physical association with LC3 and consequently promotes mitophagy64.
In yeast, genetic screens have identified the outer membrane protein ATG32 as a mitophagy receptor65, 66 (Figure 1E). ATG32 contains an LIR motif that binds ATG8, the yeast LC3 ortholog that is located on the surface of autophagosomes. In addition, it binds ATG11, an adaptor for several selective forms of autophagy. ATG11 has been shown to interact with Dnm167, the yeast ortholog of Drp1 that is involved in mitochondrial fission. Based on these observations, it has been suggested that once ATG11 has been recruited by ATG32 to mitochondria, Dnm1 is recruited to these marked mitochondria to facilitate fission67, and LC3 is recruited to target them to autophagosomes. The process of fission may be important for generating smaller mitochondrial fragments that can be segregated into autophagosomes. Mitochondrial fission has also been shown to facilitate Parkin-mediated mitophagy53. Nevertheless, a study in yeast indicates that Dnm1 and other components of the mitochondrial fission machinery are dispensable for mitophagy68.
Taken together, these results suggest that mitochondrial degradation depends on specific mitophagy receptors present on the mitochondrial outer membrane that function to target mitochondria to the autophagic machinery. It is also possible that in some cases ubiquitination serves to mark mitochondria for autophagy. In addition, mitophagy can also involve outer membrane protein degradation by the ubiquitin-proteasome system and the AAA ATPase p97, and reduction of mitochondrial size by mitochondrial fission50, 53. By culling damaged mitochondria, particularly mitochondria with defective mtDNA, mitophagy has the potential to regulate the functionality of the mitochondrial populations inherited by daughter cells; however more work is necessary to understand the physiological impact of this process.
mtDNA germline dynamics and inheritance
A typical human cell contains hundreds of copies of mtDNA and until a few years ago it was assumed that individuals exhibit homoplasmy for a single mtDNA genotype. This assumption has certain consequences, for example when analysing mtDNA genotypes in forensic cases and for tracing human lineages during evolution. Significant heteroplasmy was known to occur in patients with mitochondrial encephalomyopathies (see below), but these cases were thought to be the exception rather than the rule. With the advent of deep sequencing, however, it has become clear that low-level heteroplasmy in humans is normally present in most tissues69, 70. The source of this heteroplasmy is two-fold. A portion is maternally inherited, as evident by the observation that some of the low frequency alleles found in an individual can be found also in the mother. Another portion, not present in the mother, is presumed to arise from de novo mutations that occur during tissue development69, 70.
Because the mtDNA encodes for respiratory chain proteins and for the tRNAs and rRNAs that are necessary for their translation (Box 1), mutations in the mitochondrial genome result in OXPHOS defects that cause a large collection of diseases termed encephalomyopathies. These diseases have a surprisingly broad range of clinical phenotypes, but tissues that are highly metabolically active, such as skeletal muscle, heart muscle, nerve, and brain, tend to be most commonly affected43. Encephalomyopathies can be familial or sporadic. When familial they have a maternal inheritance pattern, because mtDNA is passed from the mother to her offspring. The most common familial encephalomyopathy is Leber’s hereditary optic neuropathy (LHON), which is caused by Complex I deficiency due to mutations predominantly in the ND1, ND4, or ND6 genes of the mitochondrial genome (Box 1). LHON typically causes bilateral deterioration of central vision due to loss of retinal ganglion cells and atrophy of the optic nerve71, 72. In sporadic encephalomyopathies, respiratory chain defects originate from a de novo mutation. For example, in chronic progressive external ophthalmoplegia (CPEO), mtDNA genomes containing a partial deletion are found in muscle tissue only, implying that the mutation arose in precursor cells of this lineage43. CPEO causes weakness in the eye-associated muscles, resulting in inability to raise the eyelids (ptosis) and difficulty in moving the eyeballs (ophthalmoplegia).
Understanding mtDNA segregation and inheritance has biomedical implications for understanding how mutations in mtDNA can cause the wide variability and diversity of tissue defects found in mitochondrial encephalomyopathies. Such diseases also often have a progressive and variable clinical outcome. Patients inherit a load of pathogenic mtDNA from their mothers, but typically not at high enough levels to cause an immediate OXPHOS deficiency in cells. As an individual ages, genetic drift during cell proliferation results in variability in the levels of pathogenic mtDNA between cells. Only cells or tissues that acquire very high levels of pathogenic mtDNA will exhibit an OXPHOS deficiency. The threshold for this bioenergetic breakdown is quite high: most mtDNA mutations need to accumulate to greater than 60–90% of total mtDNA in the cell before OXPHOS activity is compromised73–75. In both mouse76 and human cells77, the complementation of mtDNA mutations was proposed to depend on mixing of genomes by mitochondrial fusion. In a mouse genetically engineered to have high mtDNA mutation rates, reduction in mitochondrial fusion indeed exacerbated the effects of mtDNA mutations23.
mtDNA undergoes a genetic bottleneck during oogenesis
Analysis of mtDNA transmission in animal pedigrees has provided evidence that heteroplasmic mtDNA genotypes can rapidly segregate in the offspring. This phenomenon has been particularly well-documented in Holstein cows78, where offspring from a heteroplasmic mother can have mtDNA haplotype ratios that differ significantly from each other or from the mother. In extreme cases, a mother with low levels of heteroplasmy can give rise to offspring that are homoplasmic for the rare haplotype78. These observations support the occurrence during oogenesis of a severe mtDNA genetic “bottleneck”, such that only a small fraction of mtDNA molecules are ultimately represented in the mature egg (Figure 2). Experimental and theoretical analyses suggest that this bottleneck in mice consists of a couple hundred segregating molecules of mtDNA79. A similar situation occurs in humans and has implications on our understanding of familial mitochondrial encephalomyopathies, as clinical studies indicate that a heteroplasmic mother can have offspring whose pathogenic mtDNA content varies substantially from her own80, 81.
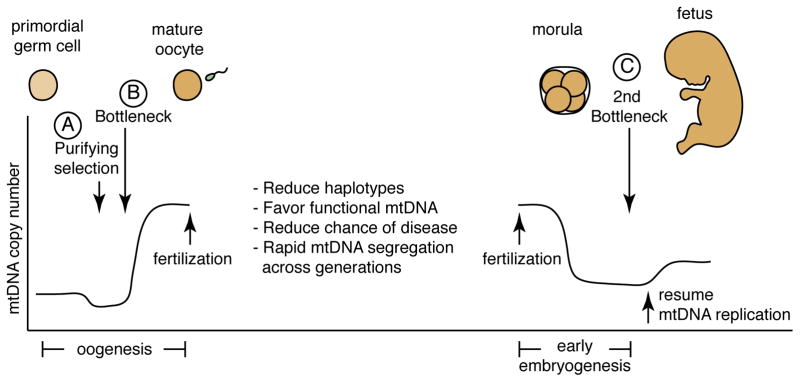
Two major mechanisms affect mtDNA genotypes during oocyte development — purifying selection (A) and the mtDNA genetic bottleneck (B). This is followed by a second bottleneck in early embryogenesis (C). By eliminating oocytes containing deleterious mtDNA mutations, purifying selection reduces the possibility of severe mtDNA disease and favors the preservation of functional mtDNA. Little is known about the exact timing of this purifying selection, but one study suggests that it occurs during oogenesis prior to the genetic bottleneck84. The mtDNA genetic bottleneck may occur due to reduction of mtDNA copy number or due to amplification of a subset of mtDNA molecules. Regardless of the mechanism, the result of the bottleneck is to decrease the number of haplotypes within an egg. This phenomenon also increases genotypic variance between mature eggs, a feature that facilitates the rapid segregation of mtDNA variants between generations. During very early embryonic development, mtDNA replication is not active, and the mtDNA content per cell is diluted due to an increase in cell number. It has been proposed that this reduction in mtDNA content per cell followed by the resumption of mtDNA replication results in a second bottleneck (C) during early embryonic development91. The graph of mtDNA copy number is meant to illustrate trends during development; direct quantification is available for only selected stages.
Although the occurrence of an mtDNA bottleneck during oogenesis is widely accepted, its molecular mechanism is unclear as recent studies have provided conflicting data about it. During oocyte maturation, a dramatic increase occurs in mitochondrial content and mtDNA copy number, culminating in mature (“metaphase II”) oocytes containing >100,000 copies of mtDNA, presumably in preparation for the metabolic demands of fertilization and implantation. The mtDNA bottleneck is though to occur prior to this period of rapid mtDNA accumulation. Several mechanisms could potentially lead to rapid segregation of mtDNA haplotypes. The bottleneck may be numerical, that is, the level of mtDNA might be transiently constricted earlier in oogenesis, thereby limiting the diversity of mtDNA molecules that will be present in the mature oocyte. In this case, rapid segregation of haplotypes between oocytes is achieved by the reduction of the number of mtDNA molecules present in each oocyte and by random genetic drift. Quantitative measurements of mtDNA numbers during oogenesis provide evidence for82 and against83, 84 the severe developmental contraction of mtDNA content predicted by this model. Alternatively, selective replication and amplification of only a subpopulation of mtDNA molecules84 may take place. The timing of mtDNA replication occurs independently of the cell cycle85. As a result, it is possible for a few mtDNA genomes in the developing oocyte to be preferentially replicated at the expense of others, leading to an apparent bottleneck. In this case, it is unclear whether this selective replication is stochastic or biased towards particular mtDNA haplotypes.
Mitochondrial quality control in the female germline
Genetic bottlenecks and genetic drift would result in oocytes with randomly skewed mtDNA genomes compared to the mtDNA population of primordial germ cells. However, two lines of research involving mice with pathogenic mtDNA mutations indicate the presence of a purifying selection process that monitors the quality and functionality of mtDNA genomes (Figure 2). In the first set of studies, female mice that are heteroplasmic for a severe mtDNA mutation, such as a large deletion86 or a frameshift in the ND6 gene87, show selective reduction of the pathogenic mitochondrial genomes during oogenesis. Offspring of such females have a lower mutation burden, and this tendency to reduce the mutational load is enhanced with maternal age. In contrast, offspring born to mice heteroplasmic for non-pathogenic polymorphisms show mutation burdens that are similar to the maternal burden. This suggests that oocytes containing severe mtDNA mutations are selectively eliminated from the germline in an age-dependent manner. Further evidence for the existence of an mtDNA quality control filter comes from the second set of studies, which analysed mice containing high levels of mtDNA mutations generated by a knock-in of an error-prone mtDNA polymerase. Female mice harboring high levels of mtDNA mutations were mated to a wildtype male, and the spectrum of mtDNA mutations in the offspring was compared to that in the mother. The offspring showed a strong suppression of mutations that change the coding sequence of protein-coding genes88, a clear signature of genetic selection. Collectively, these studies point towards a process of “purifying selection” that helps maintain the functionality of mtDNA in the female germline and therefore in the offspring. The presence of this filter likely explains why, in humans, most of the mutations causing familial encephalomyopathies are relatively mild, and most encephalomyopathies caused by large deletions are sporadic rather than familial43. mtDNA genomes with large deletions do arise de novo in somatic tissues with age89, but these mutations are not passed through the germline.
Little is known about the molecular mechanism of this quality control filter. Even severe mtDNA mutations, such as deletions, do not cause OXPHOS defects until they accumulate beyond high thresholds73–75. In the studies described above using heteroplasmic mice, purifying selection is observed at mutation loads far lower than these thresholds86, 87, at least when the thresholds are assessed in commonly used cell models. Therefore, if the filter operates at the cellular level, it is surprisingly sensitive and likely involves a mechanism assessing a feature other than total cellular OXPHOS capacity. Because some mtDNA mutations increase the levels of reactive oxygen species (ROS), ROS sensing is one possible mechanism for this quality control filter. Another possibility is that the filter is operating on the level of individual organelles, so that dysfunctional organelles containing pathogenic genomes might be segregated and eliminated, for example by mitophagy.
A second genetic bottleneck during early embryogenesis
Between the zygote and the blastocyst stages90 there is no mtDNA replication, which results in an mtDNA copy number reduction during early embryogenesis due to dilution as cells rapidly divide. Later in development, mtDNA replication provides an increase in copy number, depending on tissue type. Experiments in rhesus monkeys suggest the existence of an additional genetic bottleneck during this early embryonic period prior to the resumption of mtDNA replication91 (Figure 2). When heteroplasmic oocytes are fertilized, the resulting 8-cell embryo shows a broad range of heteroplasmy in its individual blastomeres. However, later in development, fetuses arising from heteroplasmic oocytes show a tendency toward homoplasmy91. These observations suggest that somatic cells undergo an mtDNA bottleneck sometime after the formation of the blastocyst. In contrast, female germline cells in the foetus retain a broad range of heteroplasmy, suggesting that mtDNA inheritance in somatic and germline lineages are differentially regulated during embryogenesis. Another experiment with heteroplasmic mice has revealed that some somatic tissues can rapidly change the composition of their mtDNA92: In mice heteroplasmic for wildtype Balb/c and NZB mtDNA haplotypes, blood and spleen invariably show progressive accumulation of the Balb/c haplotype, whereas the kidney and liver show accumulation of the NZB haplotype. The basis for this is unknown, but the two haplotypes do not result in differences in respiratory activity of the mitochondria93. These observations suggest that the genetic bottlenecks in somatic cells may include a considerable degree of complexity and tissue specificity.
Depletion of paternal mtDNA
A dramatic example of bias in mitochondrial segregation is the exclusively maternal inheritance of mitochondria in mammals. This has been recently further validated in humans by deep sequencing techniques, which enable the detection of even minor maternal genotypes, but failed to detect any signatures of paternal mtDNA in the tissues of offspring69.
Although uniparental inheritance of mtDNA is widespread in eukaryotes, the reason for its evolutionary conservation remains unclear. Most naturally occurring polymorphisms in mtDNA have negligible effects on mitochondrial function, and it is not obvious why it would be detrimental to have an equal mixture of two different, wildtype mtDNA genotypes (though as mentioned above, humans normally do have low levels of heteroplasmy). A recent mouse study has provided striking evidence that substantial germline-inherited heteroplasmy can result in metabolic and behavioural aberrations97. Cytoplasts from the NZB mouse strain were fused with female embryonic stem cells from the 129S6 mouse strain to generate heteroplasmic stem cells, which were used to produce lines of heteroplasmic mice. When passed through the female germ line, a marked tendency towards the reduction of the NZB mtDNA genotype was observed, indicating that a bias in transmission between generations can occur even with two wildtype mtDNA genotypes. The NZB and 129S6 mtDNA genotypes are presumably equivalent in function, because mice homoplasmic for either show indistinguishable levels of activity, fertility, or behaviour. However when NZB-129S6 heteroplasmic animals were compared to their homoplasmic NZB or 129S6 littermates, they exhibited a number of metabolic and behavioural changes, including reduced physical activity and lower food intake. In addition, they showed aberrations in tests that measure anxiety and spatial learning. This mouse model therefore provides evidence that heteroplasmy can have metabolic and behavioural effects and indicates that even apparently equivalent wild type mitochondrial genomes can be distinguished during germline inheritance97. Uniparental inheritance of mtDNA may therefore be advantageous by promoting homoplasmy and avoiding such phenotypes in the offspring. The molecular mechanisms underlying these heteroplasmic defects are unknown, but may be related to subtle differences in bioenergetic properties or ROS generation between the two mtDNA haplotypes.
An early hypothesis suggested that uniparental inheritance of mitochondria may be a passive consequence of the asymmetry in male and female gamete size; for example, in mammals, the male gamete is much smaller and contains many fewer mitochondria compared to the oocyte. However, some organisms, such as the unicellular green alga Chlamydomonas reinhardtii, have equal-sized gametes but nevertheless show uniparental inheritance of mtDNA98. As discussed below, recent studies indicate that active mechanisms that function pre- and post-fertilization have evolved to ensure uniparental inheritance of mtDNA. There are indications that these mechanisms can occasionally go awry or be bypassed. For example, there is a single clinical case reported of a patient with exercise intolerance who inherited mtDNA haplotypes from both his mother and father, with the paternal haplotype accumulating to high levels in skeletal muscles99. The patient’s myopathy resulted from an apparent de novo mutation that arose in the paternal mtDNA.
Pre-fertilization depletion of paternal mtDNA
In Drosophila melanogaster, mtDNA are eliminated from spermatozoa as they develop from the “onion stage” spermatid to fully elongated spermatids that are subsequently individualized (Figure 3A). Spermatid maturation takes place within a syncytium; sperm individualization is a process necessary for generating individual gametes, whereby each spermatid is packaged into a separate plasma membrane, separating it from the syncytium. During the final stage of spermatid elongation, the mtDNA is progressively removed from the nuclear end towards the tail100. By the time spermatids are fully elongated, few mtDNA nucleoids persist. As a result, mature sperm in Drosophila melanogaster normally have little if any mtDNA by the time of fertilization.
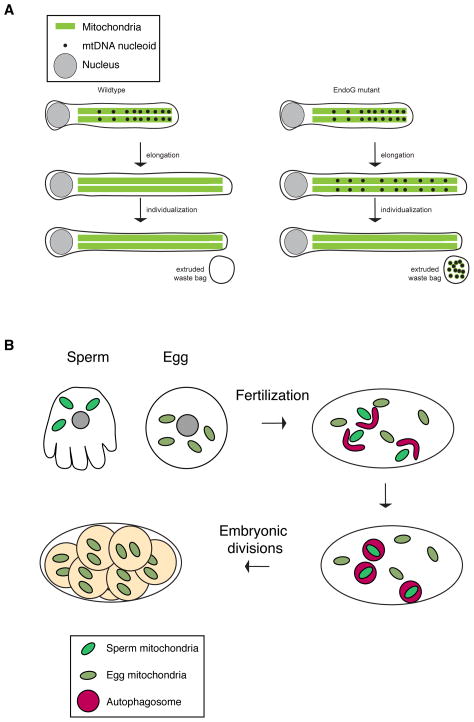
(A) Pre-fertilization mechanisms in flies. In Drosophila melanogaster, two mechanisms remove mtDNA from sperm. Left: mtDNA is normally degraded during sperm elongation by the mitochondrial endonuclease EndoG. Right: In EndoG mutant flies, the mtDNA persists beyond the elongation stage but is ultimately removed by a second mechanism (extrusion) during the individualization stage, in which mtDNA and cellular debris are sequestered into a waste compartment that is extruded from the sperm body. (B) Post-fertilization mechanism in the nematode Caenorhabditis elegans. Shortly after fertilization, paternal mitochondria (bright green) co-localize with autophagy markers (not shown) and are eliminated by mitophagy by the 16–64 cell stages. Worms deficient for autophagy show persistence of paternal mitochondria long beyond this stage.
This degradation process is dependent on EndoG, one of five nucleases with mitochondrial targeting signals100. In EndoG mutants, mtDNA nucleoids aberrantly persist in the tail regions of fully elongated and individualized spermatids. Nevertheless, eggs fertilized by EndoG mutant males do not contain detectable paternal mtDNA. Analysis of EndoG mutants reveals a redundant process in later spermatid development that further eliminates mtDNA from the sperm. When sperm undergo individualization in EndoG mutant flies, the residual mtDNA in the spermatid tail is transported, along with other cellular components, into a waste compartment that is culled from the spermatid (Figure 3A). It will be interesting to determine whether similar pre-fertilization mechanisms occur in mammals. Indeed in mice, there is a 10-fold decrease in mtDNA levels as sperm mature101, but the molecular basis of this is unknown.
Post-fertilization depletion of paternal mtDNA
In bovine and primate zygotes, the paternal mitochondria can be selectively stained using ubiquitin antibodies, and ubiquitin has been proposed to be a mark that targets the paternal organelles for proteasomal destruction102. According to this hypothesis mitochondrial ubiquitination is performed in the sperm and no active mechanism in the egg would be needed to mark the paternal mitochondria for post-fertilization destruction102.
Genetic studies in Caenorhabditis elegans clearly indicate a role for mitophagy in removal of paternal mitochondria after fertilization103–105. Shortly after fertilization, autophagosomes are recruited to paternal mitochondria and consequently they disappear by the 16-cell or the 64-cell stages103, 104 (Figure 3B). Mutants in the autophagy pathway show persistence of paternal mitochondria well beyond these stages103, 104. Similar results were obtained in embryos treated with inhibitors of lysosomal function105. In contrast to mammals, the sperm mitochondria in nematode embryos are not polyubiquitinated103, 104. Nevertheless, nematodes with mutations in proteasome ubiquitin receptors are impaired in the degradation of their paternal mitochondria105.
Fertilized mouse oocytes show co-localization of the autophagic markers LC3, GABARAP, and p62 with the paternal mitochondria103, suggesting that autophagy is a conserved mechanism for paternal mtDNA depletion in mammals as well. However, this view has been challenged by the recent observation that the association of LC3 and p62 with paternal mitochondria is transient and occurs well before the loss of mitochondria106. As a result, it has been suggested that elimination of mtDNA in mammalian sperm is a passive process, largely accomplished prior to fertilization by reduction of mtDNA in sperm106. In addition, mitochondrial partitioning in the dividing blastomeres is uneven, and this may remove the remaining paternal mtDNA from most tissues of the embryo106. More work is needed in to resolve the mechanism of mammalian uniparental mtDNA inheritance.
Concluding remarks
In mammalian cells, the partitioning of mitochondria to daughter cells during cell division appears to involve passive mechanisms. Mitochondria undergo fragmentation during mitosis and are well distributed throughout the cell soma due to their interactions with the ER and cytoskeleton. As a result, cytokinesis can result in the partitioning of roughly equal amounts of mitochondria to daughter cells. The role of active transport processes, such as the Milton-Miro-Kinesin complex, in mitochondrial partitioning during cell division has not been examined. Active partitioning mechanisms have been delineated in budding yeast; it seems likely that proliferating mammalian cells would utilize analogous mechanisms, but thus far there are no data to support this. Because cells do not survive without mitochondria, the availability of temperature-sensitive mutants in S. cerevisiae has been instrumental in identifying mitochondrial transport mechanisms to the bud. To make further progress in our understanding of mammalian mitochondrial partitioning, new assays and model systems will need to be developed to analyse mitochondrial distribution, heterogeneity and segregation in mammalian cells, and to study asymmetric cell division.
During maternal transmission of mitochondria, mtDNA genetic bottleneck and purifying selection during oogenesis have major influences on the physiology of offspring and potentially on the overall evolution of species. The molecular mechanisms underlying these complex phenomena are poorly understood, and it is unknown whether the mechanisms regulating mitochondrial partitioning in cell division are relevant to understanding mitochondrial transmission between generations. A future challenge will be to synthesize these parallel lines of research. Mechanisms of mitochondrial quality control are another area for future exploration. Several pathways of mitophagy have been identified so far, and it will be important to understand the extent to which these pathways contribute to maintenance of mitochondrial function in health and disease.
Acknowledgments
Work in laboratory of D.C.C. is supported by NIH grants GM062967 and GM110039, the Muscular Dystrophy Association, and the Howard Hughes Medical Institute. P.M. is supported by a Baxter Postdoctoral Fellowship.
Glossary
| mtDNA nucleoid | A punctate structure within mitochondria containing one or more copies of the mtDNA genome in complex with proteins |
| Oxidative phosphorylation | A biochemical pathway within mitochondria that generates ATP through the oxidation of nutrients |
| Myosin | A family of ATP-dependent molecular motors that transports cargo along actin filaments |
| Kinesin | A family of ATP-dependent molecular motors that transports cargo along microtubule filaments, usually towards the plus-end |
| Dynein | A family of ATP-dependent molecular motors that transports cargo along microtubule filaments towards the minus-end |
| Endosymbiotic theory | A theory postulating that genome-containing organelles, such as mitochondria and chloroplasts, are derived from prokaryotes that have undergone endosymbiosis with the ancestral eukaryotic cells |
| Heteroplasmy | A state in which more than one haplotype of mtDNA exists in a cell or organism |
| Homoplasmy | A state in which a single haplotype of mtDNA exists in a cell or organism |
| dynamin-related GTPases | a family of large GTP hydrolyzing enzymes related to “classical” dynamin, which uses GTP hydrolysis to mechanically remodel membranes |
| microtubule directionality (plus/minus) | Describes the polarity of microtubule filaments, which are polymerized from asymmetric dimers of α-tublin and β-tubulin |
| formin proteins | a group of proteins that regulate the actin cytoskeleton and cell signaling |
| cytoplasmic hybrid (cybrid) cell | a hybrid cell created by the fusion of a cell with an enucleated second cell, thereby combining the nucleus and mitochondria from two distinct cells |
| genetic bottleneck | a drastic reduction in the genetic diversity of a population |
| genetic drift | a change in the allele or haplotype frequencies within a population due to stochastic forces |
| purifying selection | a mode of natural selection in which detrimental genetic variants are selected against |
| macroautophagy | The most commonly studied autophagic process, whereby cellular components are incorporated into autophagosomes and ultimately degraded or recycled through the lysosome. Macroautophagy is commonly referred to as simply “autophagy” |
| autophagosomes | a double-membrane structure that engulfs cellular components for delivery to the lysosome |
| lysosomes | organelles that degrade cellular components using acid hydrolases |
| motor proteins | molecular motors that use ATP hydrolysis to power movement of cargo along substrate surfaces |
Biographies
Prashant Mishra is a postdoctoral fellow in the laboratory of David Chan at the California Institute of Technology (Pasadena, CA, USA). After finishing his bachelor’s degree in Biochemical Sciences at Harvard University, he completed his M.D./Ph.D degrees at the University of Texas Southwestern Medical Center. His Ph.D. studies were carried out in the laboratory of Rama Ranganathan. His current research focuses on regulation of mitochondrial fusion.
David C. Chan is Professor of Biology in the Division of Biology and Biological Engineering at the California Institute of Technology (Pasadena, CA, USA). After M.D./Ph.D. studies at Harvard Medical School, he completed postdoctoral work at the Whitehead Institute at the Massachusetts Institute of Technology. His laboratory works on the biochemistry and physiological functions of mitochondrial dynamics.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nrm3877
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4250044?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nrm3877
Article citations
Mitochondrial Dynamics Drive Muscle Stem Cell Progression from Quiescence to Myogenic Differentiation.
Cells, 13(21):1773, 26 Oct 2024
Cited by: 0 articles | PMID: 39513880 | PMCID: PMC11545319
Review Free full text in Europe PMC
Mitochondrion-based organellar therapies for central nervous system diseases.
Cell Commun Signal, 22(1):487, 10 Oct 2024
Cited by: 0 articles | PMID: 39390521 | PMCID: PMC11468137
Review Free full text in Europe PMC
Cytoplasmic DNA and AIM2 inflammasome in RA: where they come from and where they go?
Front Immunol, 15:1343325, 10 Oct 2024
Cited by: 0 articles | PMID: 39450183 | PMCID: PMC11499118
Review Free full text in Europe PMC
Microcystin-LR in drinking water: An emerging role of mitochondrial-induced epigenetic modifications and possible mitigation strategies.
Toxicol Rep, 13:101745, 28 Sep 2024
Cited by: 0 articles | PMID: 39411183 | PMCID: PMC11474209
Review Free full text in Europe PMC
Myosin A and F-Actin play a critical role in mitochondrial dynamics and inheritance in Toxoplasma gondii.
PLoS Pathog, 20(10):e1012127, 07 Oct 2024
Cited by: 0 articles | PMID: 39374269 | PMCID: PMC11486366
Go to all (537) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Autophagosomal Sperm Organelle Clearance and mtDNA Inheritance in C. elegans.
Adv Anat Embryol Cell Biol, 231:1-23, 01 Jan 2019
Cited by: 3 articles | PMID: 30467692
Review
Biased placement of Mitochondria fission facilitates asymmetric inheritance of protein aggregates during yeast cell division.
PLoS Comput Biol, 19(11):e1011588, 27 Nov 2023
Cited by: 0 articles | PMID: 38011208 | PMCID: PMC10703421
Incompatibility between mitochondrial and nuclear genomes during oogenesis results in ovarian failure and embryonic lethality.
Development, 144(13):2490-2503, 02 Jun 2017
Cited by: 25 articles | PMID: 28576772 | PMCID: PMC5536873
Fusion, fission, and transport control asymmetric inheritance of mitochondria and protein aggregates.
J Cell Biol, 216(8):2481-2498, 14 Jun 2017
Cited by: 32 articles | PMID: 28615194 | PMCID: PMC5551707
Funding
Funders who supported this work.
Howard Hughes Medical Institute
NIGMS NIH HHS (4)
Grant ID: R01 GM062967
Grant ID: GM110039
Grant ID: GM062967
Grant ID: R01 GM110039