Abstract
Free full text

Targeting Latent TGFβ release in muscular dystrophy
Abstract
Latent TGFβ binding proteins (LTBPs) bind to inactive TGFβ in the extracellular matrix. In mice, muscular dystrophy symptoms are intensified by a genetic polymorphism that changes the hinge region of LTBP, leading to increased proteolytic susceptibility and TGFβ release. We have found that the hinge region of human LTBP4 was also readily proteolyzed, and that proteolysis could be blocked by an antibody to the hinge region. Transgenic mice were generated to carry a bacterial artificial chromosome encoding the human LTBP4 gene. These transgenic mice displayed larger myofibers, increased damage after muscle injury, and enhanced TGFβ signaling. In the mdx mouse model of Duchenne muscular dystrophy, the human LTBP4 transgene exacerbated muscular dystrophy symptoms and resulted in weaker muscles with an increased inflammatory infiltrate and greater LTBP4 cleavage in vivo. Blocking LTBP4 cleavage may be a therapeutic strategy to reduce TGFβ release and activity and decrease inflammation and muscle damage in muscular dystrophy.
INTRODUCTION
In Duchenne muscular dystrophy (DMD), the loss of dystrophin weakens the sarcolemma of muscles rendering them susceptible to contraction-induced damage. Mutations in the dystrophin-associated proteins, the sarcoglycans, lead to a similar weakened sarcolemma and myofiber injury. A genomewide scan for modifiers of muscular dystrophy in mice identified the Ltbp4 gene, encoding latent TGFβ binding protein 4, as an important determinant of sarcolemma fragility and fibrosis (1). In humans with DMD, polymorphisms in the LTBP4 gene were shown to correlate with extended ambulation in DMD patients and reduced TGFβ signaling (2)
LTBP4 encodes an extracellular matrix-associated protein and is highly expressed in skeletal muscle. LTBPs are anchored to extracellular matrix fibrils through the N-terminus, whereas the C-terminus binds to latent TGFβ to form a large latent complex (3–5). A proline-rich hinge domain separating the N- and C-terminal domains is the target of proteolysis, and this cleavage is associated with latent TGFβ release and activation (6–9). TGFβ activity regulates injury and repair in muscle, kidney, lung, heart and brain (10–14). TGFβ activity is triggered in both chronic and acute muscle injury (10, 15). TGFβ mRNA levels are elevated in human DMD (16–18), and systemic administration of neutralizing TGFβ antibody or the angiotensin II type 1 receptor blocker losartan helps to normalize muscle architecture, repair and function in the mdx mouse model of DMD, suggesting a direct role for excessive TGFβ signaling in muscle disease (19, 20).
The modifier polymorphism in murine Ltbp4 encodes a deletion or insertion of 12 amino acids within LTBP4’s hinge region (1). The Ltbp4 allele with the smaller hinge is more susceptible to proteolysis and is associated with increased release of latent TGFβ and TGFβ signaling (1). Because human LTBP4 has an even smaller hinge, human LTBP4 is predicted to release more latent TGFβ. Here, we tested this notion in vitro and in vivo. We found that the human LTBP4 hinge was more readily proteolyzed than the murine LTBP4 hinge. Further, an antibody that blocked proteolytic cleavage of LTBP4 demonstrated that the proline-rich hinge is the site of proteolysis. The human LTBP4 gene was inserted into mdx mice using a bacterial artificial chromosome (BAC). Mdx mice with the human LTBP4 BAC showed increased muscle membrane leakage and fibrosis. Furthermore, the presence of the human LTBP4 gene was associated with weaker muscles, greater infiltration of muscles by macrophages and increased TGFβ signaling. These data identify cleavage of LTBP4 as a potential target for treating muscular dystrophy and provide a biological strategy for regulating TGFβ release.
RESULTS
LTBP4 is organized in a striated pattern around muscle fibers
Ltbp4 was identified as a modifier of muscular dystrophy in mice from an unbiased genomewide screen where it strongly associated with increased membrane fragility and fibrosis (1). LTBP4 mRNA is expressed in human muscle (21). We examined LTBP4’s pattern of protein expression using anti-LTBP4 antibodies and confocal microscopy. Two distinct extracellular pools of LTBP4 were seen using four different anti-LTBP4 antibodies. One pool of LTBP4 was parallel to the long axis of myofibers, and a second pool was observed closely apposed to the sarcolemma in a striated pattern in muscle (Fig. 1A). The LTBP4 localized near the sarcolemma was in a striated pattern (Fig. 1A). This pattern reflected an organized matrix immediately surrounding muscle fibers and suggested that LTBP4 is positioned to stabilize the myofiber.
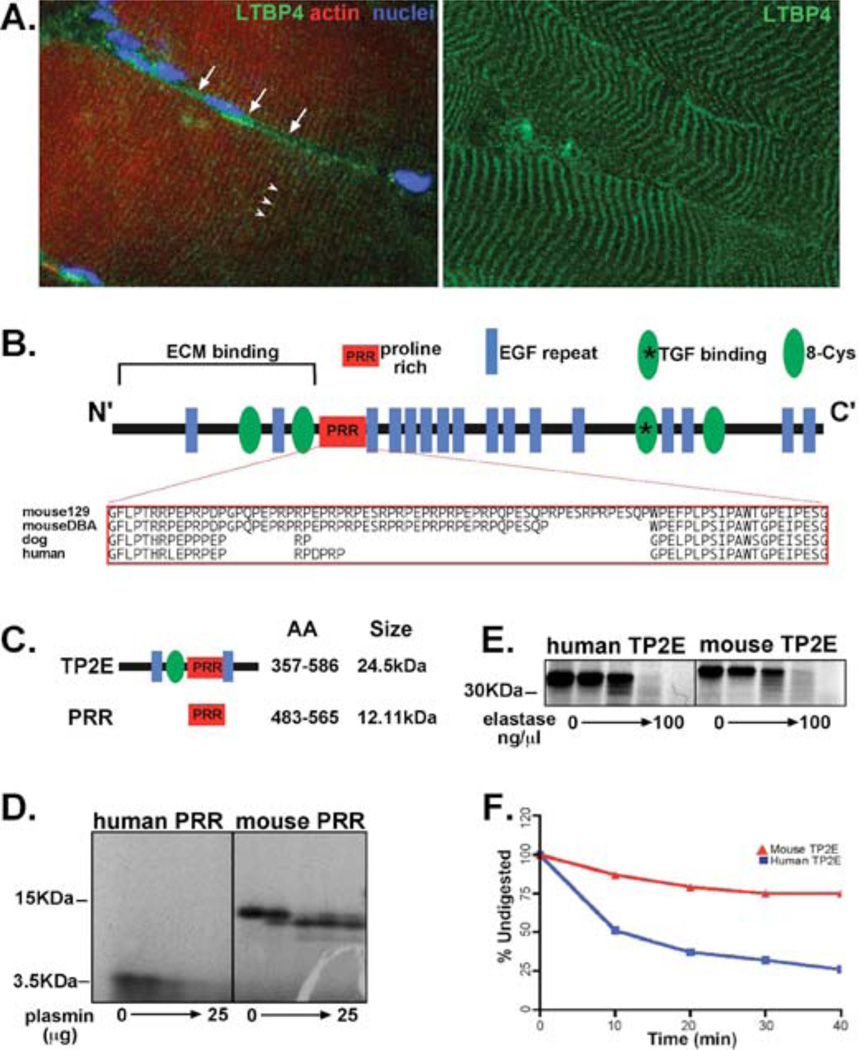
(A) Individual muscle fibers were isolated from murine muscle and stained with multiple antibodies to LTBP4. Longitudinal muscle fibers have two pools of LTBP4 protein, which run parallel (white arrows) and perpendicular (white arrow heads) to the long axis of the muscle. The perpendicular or costameric LTBP4 is closely apposed to the muscle membrane (white arrows). The right panel shows the costameric LTBP4 pattern in striated muscle. (B) Mice have an insertion/deletion polymorphism that alters the proline-rich region (PRR) of LTBP4. The majority of mouse strains carries the insertion allele shared by the 129T2/SvEmsJ strain (129), while a minority of mouse strains shares the deletion seen in the DBA/2J strain (D2). This polymorphism regulates the severity of muscular dystrophy in mice (1) and alters proteolytic cleavage of LTBP4, a process that is linked to TGFβ release and signaling. Human LTBP4 has a smaller PRR hinge compared to murine LTBP4. (C) Fragments containing either human or murine LTBP4 PRR hinge were expressed in vitro and digested with either plasmin (D) or elastase (E), two serine proteases know to cleave LTBPs. Human LTBP4 was more readily digested compared to murine PRR (F).
The hinge region of human LTBP4 is a target of serine proteases
The insertion/deletion polymorphism that modifies muscular dystrophy in mice alters the proline rich region (PRR) of LTBP4 by 12 amino acids. The shorter PRR hinge was associated with greater LTBP4 proteolytic susceptibility, increased TGFβ signaling, and worsening of muscular dystrophy (1). The exacerbated muscle membrane fragility and muscle fibrosis seen with a shorter PRR hinge was associated with decreased grip strength (1). Human LTBP4 has a smaller LTBP4 hinge region compared to the murine LTBP4 sequence (Fig. 1B). The canine LTBP4 hinge is also small, and it is notable that DMD mutations in these two species cause severe disease that is associated with accelerated loss of ambulation (22–24). To test whether human LTBP4 protein was more susceptible to proteolytic cleavage than murine LTBP4, the PRR hinge from LTBP4 was expressed in vitro and digested with serine proteases. With increasing concentrations of plasmin, the human LTBP4 PRR hinge was rapidly digested whereas the murine PRR hinge was comparatively resistant to plasmin treatment (Fig. 1C–F). A larger fragment of LTBP4 containing the PRR and flanked by an 8 cysteine repeat and two EGF motifs was similarly rapidly digested compared to the comparable murine LTBP4 sequence (Fig. 1E, F). Results were similar for plasmin and elastase (Fig 1D, E), two serine protease family members that have been shown to cleave LTBPs in vitro (25–27).
To evaluate cleavage of full length LTBP4, HEK293 cells were cultured under conditions that promoted extracellular matrix formation and LTBP4 expression (1, 2). Cell lysates were treated with plasmin for 30 minutes under conditions that produced partial proteolysis and reduction of full length LTBP4 (Fig. 2A). We generated an antibody to the PRR of LTBP4, called anti-PRR 831, and this antibody recognized both human and mouse LTBP4 PRR on immunoblotting and ELISA (Fig. S1). Anti-PRR 831 antibody blocked plasmin-mediated cleavage of intact LTBP4 demonstrating that PRR is the target of proteolysis in LTBP4 (Fig. 2B). A non-specific antibody, raised in the same species as the anti-PRR 831 antibody, did not protect against proteolytic cleavage, confirming that human LTBP4 processing occurs specifically at the PRR (Fig. S1). Human and murine LTBP4 are highly related sequences with over 90% identity; the regions of difference are at the extreme N-terminus and the PRR hinge. Every LTBP4 antibody that was tested recognized both human and murine LTBP4 (Santa Cruz, sc-30360 and sc-33144, custom made chicken polyclonal antibodies 829, 830, 831, 832, 870 and 871, custom made rabbit polyclonal antibodies 24226, 24227, 28199 and 28200). The anti-PRR antibody showed high affinity for human LTBP4 as measured by ELISA, but did show cross-reactivity with the murine protein (Fig. S1).
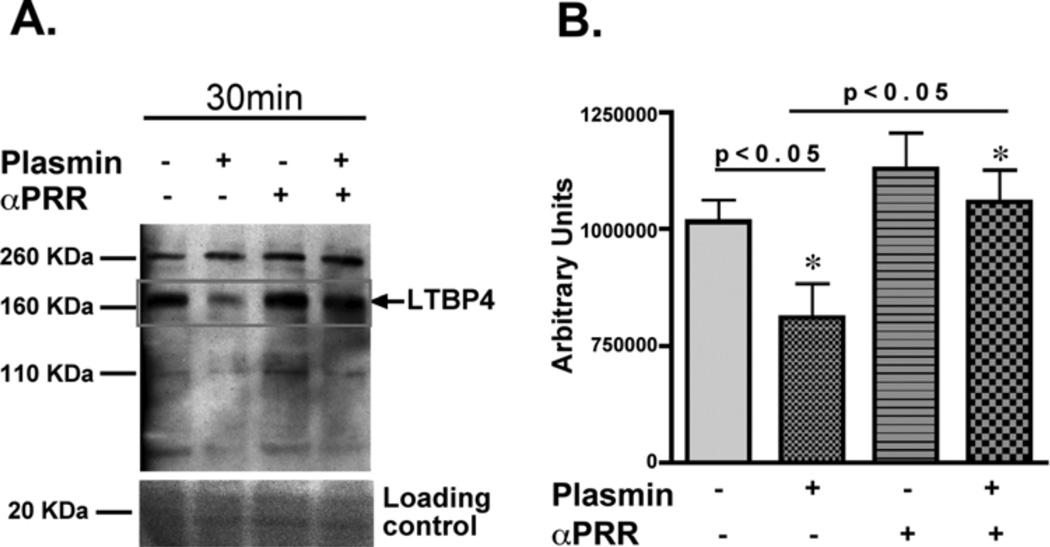
The anti-PRR 831 antibody was generated against the human LTBP4 PRR hinge region and was tested in an in vitro proteolysis assay. (A) Full-length LTBP4 was tested using cells grown to confluence under conditions that enrich for LTBP4 expression. Plasmin-mediated cleavage of human LTBP4 was reduced in the presence of the anti-PRR antibody. (B) Experiments were repeated in triplicate and demonstrated significantly different cleavage (one way ANOVA).
hLTBP4 BAC transgenic mice showed increased SMAD signaling and myofiber hypertrophy
A BAC encoding the complete human LTBP4 gene was used to generate transgenic mice (hLTBP4 BAC transgenic mice). The hLTBP4 BAC spanned base pairs 41089002 to 41240302 of the human chromosome 19 genomic region (UCSF working version GRCh37/hg19, February 2009), and the human LTBP4 gene was from 19891bp to 57891bp of this BAC (Fig. 3A). The BAC contained approximately 30 kb of sequences 5’ of the LTBP4 initiation site such that it included regulatory regions. Multiple founder lines were generated and verified for transmission and integration of the full-length human LTBP4 gene, and one line was selected for breeding mice and further analysis. RT-PCR using material from hLTBP4 BAC Tg+ mice with human LTBP4 specific primers confirmed mRNA expression of the transgene in heart and muscle (Fig. 3B). Immunoblotting of uninjured muscle and heart from transgene and littermate controls showed very slight upregulation of LTBP4 protein (1.2x wildtype). None of the 12 antibodies tested distinguished between mouse and human LTBP4, so immunoblotting detected both murine and human LTBP4 (Fig. 3C). This hLTBP4 BAC resulted in enhanced TGFβ signaling with an increase in pSMAD (Fig. 3D).
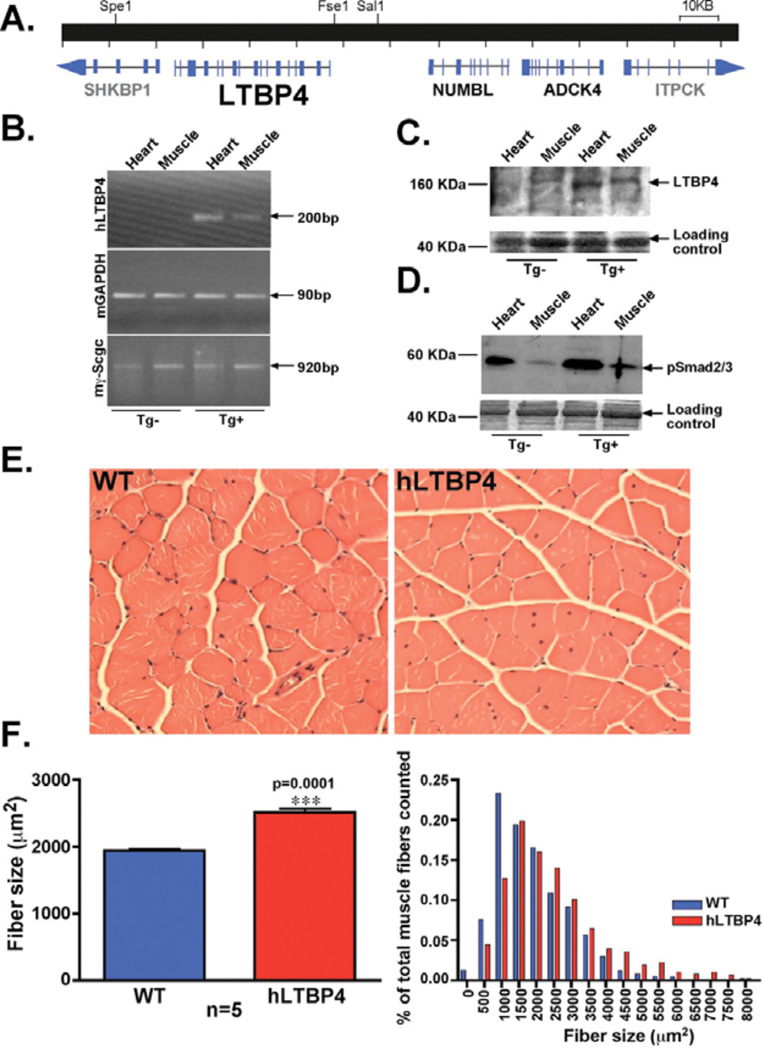
The human LTBP4 gene was expressed in a BAC transgene that was inserted into mice and propagated through breeding. Multiple lines were generated and one was characterized in detail. (A) Genomic map of the BAC containing the full-length human LTBP4 gene. NUMBL and ADCK4 genes were also present in the BAC and were expressed at very low levels, whereas SHKBP1 and ITPCK were truncated genes and were not expressed. (B) RT-PCR using primers specific to human LTBP4 demonstrated expression in cardiac and skeletal muscle. (C) Immunoblot from whole muscle lysates demonstrated minimal upregulation of total LTBP4 (1.2x normal muscle). Notably, antibodies did not distinguish between human and murine LTBP4. (D) hLTBP4 BAC transgene expression resulted in elevated pSMAD in hLTBP4 mouse muscle compared to muscle from wildtype littermate controls. (E) Staining of mouse quadriceps muscle with hematoxylin and eosin revealed larger myofibers in hLTBP4 mice compared to wildtype (WT) littermate controls. (F) Cross-sectional area of muscle myofibers was larger in hLTBP4 BAC mice compared to littermate controls. The distribution of fiber size was shifted rightward with larger myofibers across the entire spectrum. P=0.0001, (t-test).
Hematoxylin and eosin staining of quadriceps muscle from 8 week-old hLTBP4 Tg+ mice and wildtype littermate controls showed a larger cross-sectional area of hLTBP4 muscle fibers: average diameter 2141 µm2 for hLTBP4 Tg+ mice compared to 1648 µm2 for wildtype littermate controls (p<0.0001; Fig. 3F). The distribution of myofiber cross-sectional area showed an increase in the largest myofibers and a decrease in the smallest myofibers in hLTBP4 Tg+ mice compared to wildtype littermate controls (Fig. 3F). The total number of muscle fibers did not differ between hLTBP4 mice and wild-type littermate controls, consistent with muscle fiber hypertrophy rather than hyperplasia: average myofiber number per quadriceps at maximum diameter was 4898 for transgene-positive mice and 4493 for wildtype littermate controls (p=0.59). We measured membrane permeability, a reflection of sarcolemmal disruption, by quantifying Evans blue dye uptake and hydroxyproline content, which is a reflection of collagen content. As expected, in the basal state, dye uptake and hydroxyproline content were comparable between hLTBP4 Tg+ mice and wildtype littermate controls (Fig. S2).
LTBP4 protein expression is increased in injured muscle
To induce injury, we injected cardiotoxin into the tibialis anterior muscle and examined the muscle 7 days later. Cardiotoxin injection into the tibialis anterior muscle of hLTBP4 BAC Tg mice was associated with an increased mononuclear cell infiltrate, increased fibrosis and fatty deposits (Fig. 4A) compared to non-transgenic controls. Notably, injury was associated with a marked increase in LTBP4 protein in both nontransgenic mice and hLTBP4 BAC tg mice consistent with the role of LTBP4 and TGFβ in the injury response (Fig. 4B). Cardiotoxin injection of hLTBP4 BAC Tg muscle induced a dystrophic process with fatty deposition and an increase in pSMAD staining that could be detected in centrally positioned nuclei of the muscle (Fig. 4C). Nuclei positioned in the center of fibers were disproportionately pSMAD-positive, and this was increased in HLTBP4 BAC Tg+ muscle compared to wildtype (Tg-) (Fig. 4D).
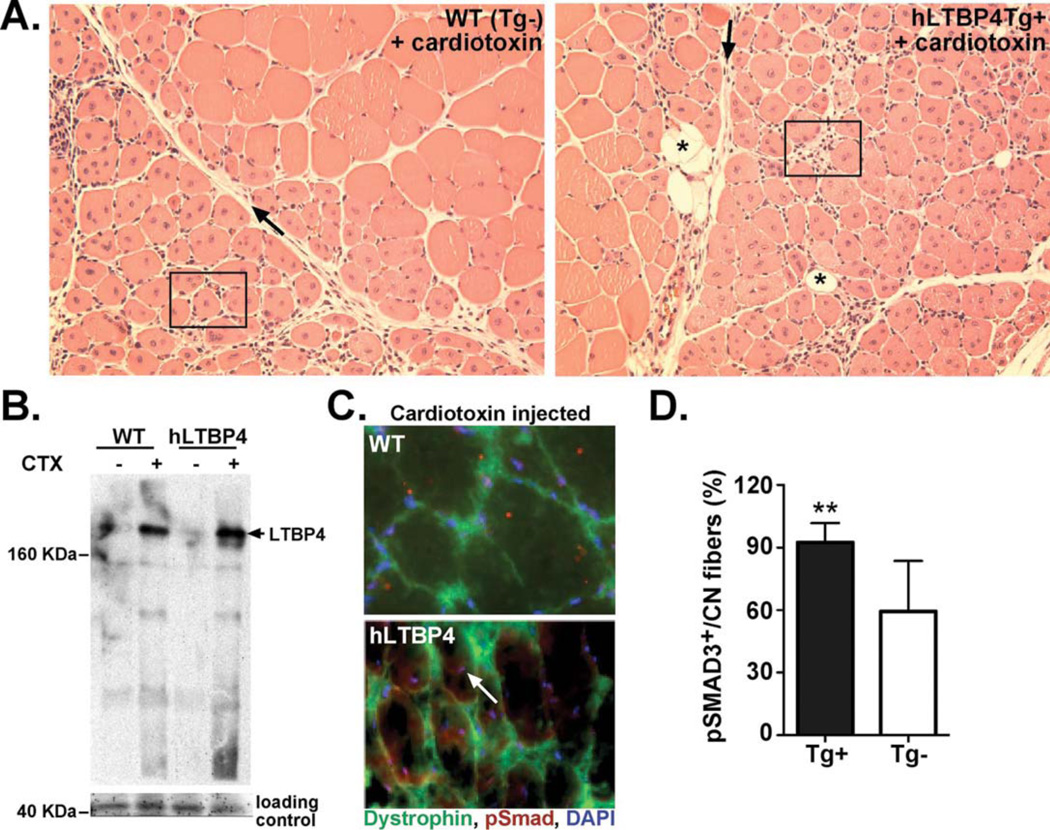
(A) Shown is the tibialis anterior muscle after cardiotoxin (CTX)-induced injury to wildtype (left) and littermate hLTBP4 Tg+ mice (right). Muscle is stained with hematoxylin and eosin. hLTBP4 muscle showed enhanced mononuclear cell infiltration (boxed), fibrosis (black arrow) and fatty deposition (black asterisk). (B) Immunoblotting showed elevated LTBP4 in muscle after cardiotoxin injury in both wildtype and hLTBP4 transgenic muscle consistent with a role for LTBP4 in mediating muscle injury. (C) Immunofluorescence microscopy showed elevated nuclear pSMAD (white arrow) in hLTBP4 muscle compared to wildtype (WT) muscle, consistent with enhanced proteolysis of LTBP4 and enhanced TGFβ activity. (D) Centrally-located nuclei were pSMAD3-positive and this was increased in hLTBP4 Tg+ muscle after cardiotoxin injection compared to wildtype (Tg-) muscle. **p = 0.04, n=4 in each group, (t-test).
hLTBP4 expression enhanced skeletal muscle membrane damage and fibrosis in mdx mice
To test whether human LTBP4 exacerbated muscular dystrophy in the mdx mouse, the hLTBP4 transgene was introduced into the mdx mouse background. For these experiments, all comparisons were made between hLTBP4 Tg+ littermates and mdx littermates so as to avoid potential effects from the two genetic backgrounds C57Bl6/J and C57Bl10/J; male mice were compared unless otherwise stated. Histopathological features of muscular dystrophy were more evident in hLTBP4/mdx mice compared to mdx mice (Fig. 5). hLTBP4/mdx mouse muscle had smaller myofibers and an increase in internal myonuclei compared to littermate mdx mouse muscle (Fig. S3). hLTBP4/mdx mice displayed worse histopathology with larger areas of muscle fibrosis and cellular infiltrate (Fig. 5A). Increased uptake of Evans blue dye could be seen in mdx/hLTBP4 mouse muscle (Fig. 5A, lower panels; Fig. S4A) and was quantitated (Fig. 5B, p=0.01). We also found that fibrosis and hydroxyproline were increased in hLTBP4/mdx mouse muscle compared to the muscle of mdx littermate controls consistent with an enhanced dystrophic phenotype induced by human LTBP4 sequence (Fig. 5B; Fig. S4B). hLTBP4/mdx muscle was also weaker compared to mdx muscle showing reduced single, acute twitch force and as well as a lower summated tetanic specific force (Fig. 5C).
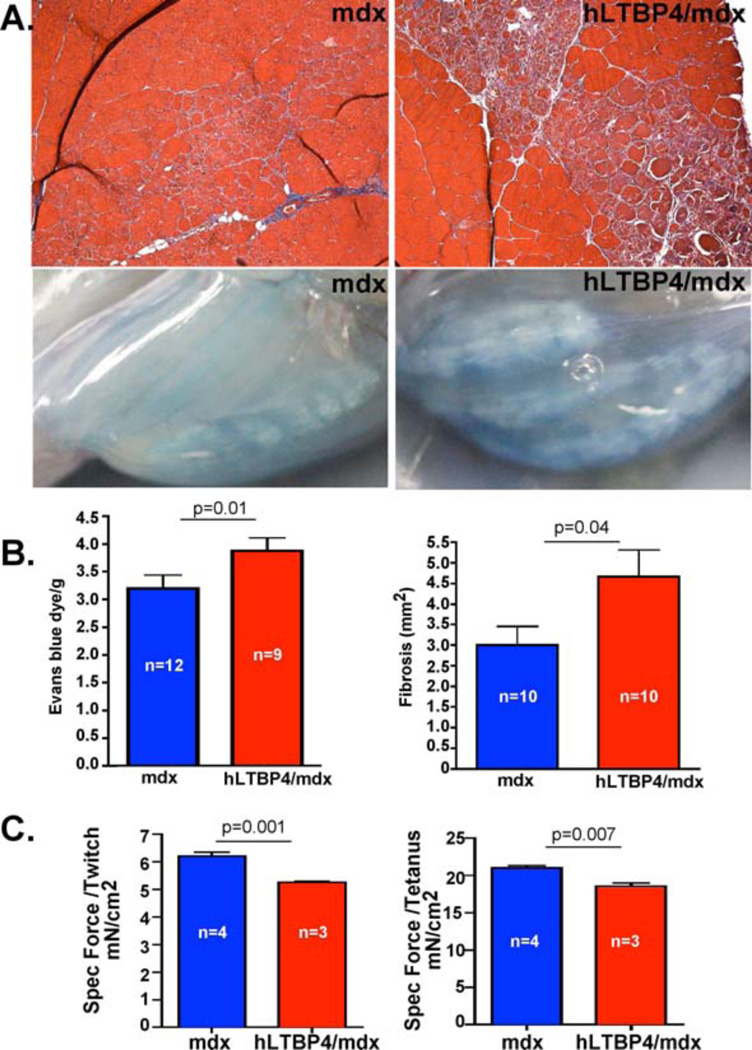
(A, top) Masson Trichrome staining showed a marked replacement of myofibers by fibrotic tissue (blue areas) in the quadriceps muscle of hLTBP4/mdx mice (right) compared to the quadriceps muscle of mdx mice (left). (A, bottom) hLTBP4 transgene expression exacerbated membrane damage in hLTBP4/mdx mice compared to mdx littermate controls as shown by increased uptake of Evans blue dye, a marker of membrane fragility and permeability. (B) Quantitation of increased Evans blue dye uptake and increased areas of fibrosis in hLTBP4/mdx mouse muscle compared to mdx littermate muscle. (C) HLTBP4/mdx muscles were weaker compared to mdx muscle as shown by reduced acute twitch and the summated tetanic specific forces (p=0.001 and p=0.007, respectively, t-test).
hLTBP4/mdx mouse muscle showed increased macrophage infiltration that was dispersed throughout the muscle compared to the muscle of mdx mice, as indicated by F4/80 staining (Fig. 6A, Fig. S5). Macrophage infiltration is known to be an important trigger for muscle degeneration (28, 29), and may be one source of the proteases that cleave LTBP4. An antibody to the amino-terminus of LTBP4 revealed increased cleavage of LTBP4 in vivo, seen as the amino-terminal ~80 KDa fragment (Fig. 6B). The smaller, more readily cleaved hinge region in human, as compared to mouse, LTBP4 was associated with greater TGFβ release and increased TGFβ signaling (Fig. 6C). We conducted a pilot study with anti-LTBP4 antibody to determine whether stabilizing the LTBP4 hinge could protect against cardiotoxin-injury in hLTBP4/mdx mice in vivo. Animals injected with anti-PRR 831 antibody displayed less muscle damage with reduced inflammatory infiltrate and larger myofibers (Fig. 6D). Taken together, these results demonstrate that human LTBP4 transgene expression aggravates muscle disease in the mdx mouse by increasing membrane damage and worsening skeletal muscle fibrosis and that blockade of LTBP4 cleavage is a useful approach to prevent such damage.
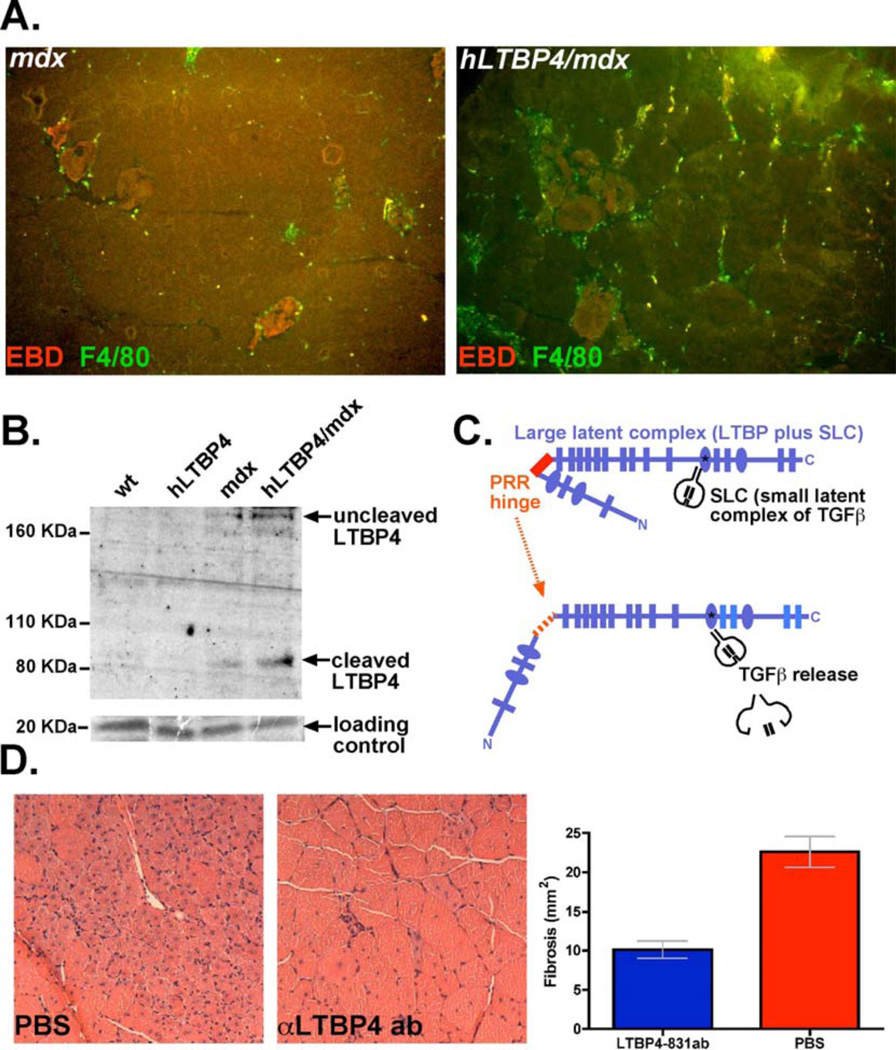
(A) An increased mononuclear cell infiltrate was seen in muscle on histopathology and was further examined with F4/80 staining. F4/80 staining for macrophages showed an increase in macrophage infiltration into hLTBP4/mdx muscle compared to mdx muscle (white dashed boxes) (B) Immunoblot for LTBP4 demonstrating LTBP4 cleavage products in hLTBP4/mdx muscle compared to muscle of age-matched mdx, hLTBP4, and wildtype mice. (C) Model of LTBP4 action illustrating cleavage and subsequent release of TGFβ, which may activate inflammatory macrophages, and worsen myofiber membrane damage and fibrosis. (D) hLTBP4/mdx muscle showed less injury and less fibrosis after cardiotoxin injection by hematoxylin and eosin staining if the mice received anti-PRR LTBP4 antibody.
DISCUSSION
LTBP4 resides in the extracellular matrix where it forms a complex with inactive TGFβ. The release of latent TGFβ from this large latent complex has been linked to cleavage of LTBP (6, 8, 9, 26, 30). An insertion/deletion in Ltbp4’s hinge region was previously found to be a modifier for muscular dystrophy. The smaller hinge region was linked to increased release of TGFβ, increased TGFβ signaling and exacerbation of muscular dystrophy, see as increased pathological changes and reduced strength (1). Human LTBP4 has a contracted hinge region compared to the murine sequence, and the smaller hinge domain is more readily targeted by serine proteases. Antibodies specific to the PRR hinge region blocked proteolytic cleavage. LTBP4 is enriched between myofibers in the extracellular matrix running parallel to the long axis of muscle. LTBP4 was also seen close to the sarcolemmal surface. This pattern mirrored the underlying striation pattern in muscle. Notably, dystrophin and the dystrophin-associated proteins are concentrated in a striated pattern that essentially links the extracellular matrix to the membrane in a rivet-like pattern along the sarcolemma. The observation that extracellular matrix components reflect membrane structure implies that signaling may be regionally restricted within the myofiber.
TGFβ activation is tightly regulated by the small latent complex, which includes the TGFβ prodomain, and by the large latent complex, which includes LTBPs. The prodomain of TGFβ binds to integrin RGD domains (31). Furthermore, the interaction between LTBP1 and TGFβ is sensitive to mechanical forces because of the manner in which the prodomains of TGFβ interact with LTBPs (32). We hypothesized that the hinge domain of LTBP4 may be critical to stabilizing TGFβ through a conformational change and proteolytic cleavage process. The precise TGFβ family members bound by LTBP4 in muscle injury and disease are not known. Abnormal BMP4 signaling is seen in the absence of LTBP4 suggesting that the interaction may not be limited to TGFβ1, 2, and 3 (33). LTBPs, including LTBP4, bind to other extracellular matrix proteins including fibrillin and fibronectin (34–38). It is possible that the LTBP4 hinge region changes conformation with contraction or injury, thereby altering the affinity of LTBP4 for other extracellular matrix proteins, which in turn affect LTPB4’s binding latent TGFβ.
hLTBP4/mdx mice showed enhanced muscle membrane damage, weaker muscles, increased skeletal muscle fibrosis, and greater macrophage infiltration of muscle. Enhanced inflammation of muscle is a hallmark of DMD pathogenesis (28, 29). An increase in F4/80 macrophages was seen in hLTBP4/mdx muscle. An altered extracellular matrix may inhibit muscle regeneration because muscle precursor cells respond to TGFβ signaling by showing redirected differentiation along the fibroadipogenic lineages (39–42). Notably, fatty deposits were seen after injury in hLTBP4 transgenic muscle, a feature not typically seen in normal mouse muscle after injury. The use of a BAC to drive human LTBP4 transgene expression may not restrict expression of LTBP4 to myofibers; hLTBP4 may be expressed in inflammatory cells, the vasculature and other cell types.
The invasion of inflammatory cells into dystrophic muscle is likely to further disrupt cell integrity through release of cytokines. As a modifier, Ltbp4 acts to stabilize the sarcolemma, and may work through both direct and indirect effects. LTBP4 has TGFβ-independent effects on the extracellular matrix, and proteolytic cleavage of LTBP4 could generate fragments that could directly contribute to inflammation (43, 44). The extracellular matrix and the sarcolemma are tightly coupled to stabilize and protect muscle from contraction-induced damage. Therefore, sarcolemmal integrity and proper extracellular matrix organization are critical for healthy and fully functional muscles. Our data suggest that the sarcolemma can be stabilized at the extracellular face by targeting extracellular matrix-associated proteins such as LTBP4 resulting in reduced TGFβ release from the large latent complex leading to reduced muscle inflammation and damage in muscular dystrophy. Taken together, these data support that blocking LTBP4 cleavage, as a means of regulating latent TGFβ release, is a therapeutic approach for muscular dystrophy.
MATERIALS AND METHODS
Study design
To assess the role of LTBP4’s hinge region, in vitro studies were conducted to compare human versus mouse LTBP4 hinge regions, as the murine sequence has a large insertion of additional amino acids. To assess the contribution of LTBP4’s hinge region in vivo, mice were generated to carry the human LTBP4 gene in order to test whether the human LTBP4 sequence induces greater TGFβ release and signaling. To document enhanced muscular dystrophy pathology in mice, hydroxyproline was quantified to reflect fibrosis and Evans blue dye content was measured to document muscle membrane leakage. These methods were applied to multiple muscle groups. To assess the affect human LTBP4 on muscle strength, ex vivo muscle mechanical studies were conducted to quantify the difference in acute contraction forces of mdx mice with and without the human LTBP4 gene.
In vitro expression and proteolysis
Constructs representing the proline-rich region, amino acids 483–565 (PRR) or amino acids (357–586 (TP2E) (numbering based on NM_001042544) were generated by PCR from either human cDNA or from mouse cDNA and ligated into pcDNA3.1His vector. In vitro transcription/translation was carried out using the reticulocyte lysate system (Promega, cat. #L4610) in the presence of 35S cysteine, and peptide fragments were cleaved as described (1). Ten µl of the 50 µl reaction was exposed to plasmin 1–10mM (Sigma Aldrich, cat. # P1867- 150UG) or elastase 1nM (Sigma Aldrich, cat. # E7885) for 30 minutes at 37°C. The digestion products were separated by 12% polyacrylamide electrophoresis, dried, and quantified by autoradiography using Quantity One (Bio-Rad, version 4.6.2).
Antibody generation
The peptide sequences EPRPEPRPDPRPGPELPLPC and EPRPEPRPDPRPGPELPC were conjugated to KLH and used as antigens in rabbits or chickens (Pocono Animal Farms). Indirect ELISA assays were performed as follows: high binding 96 well plates (Immunolon 4HBX flat bottom polystyrene plate, ThermoLab, cat. #3855) were coated with 1.05µg/ml of either human or mouse peptides (PRR) or a non-specific peptide overnight at room temperature. Unbound antigen was aspirated and wells were washed with buffer (R&D, cat. # 895126), blocked with 0.5% BSA for 3 hours at room temperature, and then washed again. Fifty µl of purified antibody at varying concentrations in blocking buffer was added to each lane or blocking buffer without antibody solution was used. The primary antibody was aspirated and wells were washed with washing buffer. The wells were incubated with 50µl of secondary antibody conjugated to HRP (at 1:25K dilution) for 1hr at room temperature. Following washing 3X with 250µl of washing buffer, 100µl substrate reagent (A+B) (R&D, cat. # Dy999) was added to each well. The wells were incubated at room temperature until the assay developed and the reaction was stopped with 50µl of 2N sulfuric acid (Fisher Scientific, cat. #SA818-1). The plate was read at 450 nm using a BMG LABTECH, FluoStar Optima plate reader. Raw absorbances were saved and enzymatic curves and Kd values were calculated using Prism (GraphPad). Percent identity between LTBP4 protein sequences was calculated using http://web.expasy.org/cgibin/sim/sim.pl?prot.
Cell culture and human LTBP4 digestion
HEK293T/17 cells were grown to confluence, and then an additional 5 days to allow for matrix deposition, rinsed in cold PBS, and lysed in whole tissue lysis buffer (50mM HEPES, pH 7.5, 150mM NaCl, 2mM EDTA, 10mM NaF, 10mM Na-pyrophosphate, 10% glycerol, with protease inhibitors (Roche, cat. # 11873580001). Whole lysates were freeze-thawed three times on dry-ice and cellular debris was removed by spinning the lysates for 5 minutes at 14000 RPM at 4°C. Total supernatant protein content was determined and 100µg was digested with 2.64nM plasmin for 30 minutes at 37C.
Immunoblotting
Lysates were separated using polyacrylamide gel electrophoresis. The lower portion of gels was removed and stained with Gel-Code (Thermo Scientific, cat. #24590) as loading controls, while the upper portion of the same gel was transferred to PVDF membranes. After blocking, PVDF membranes were incubated with primary antibodies as follows: anti-LTBP4 (Santa Cruz, SC-33144 at 1:500 dilution), pSMAD2/3 (Calbiochem cat. # 566415) or pSMAD3 (abcam cat. # ab514451). Secondary antibodies were from Jackson Immunochemicals (1:2500). Signals were detected using ECL plus (GE Healthcare, RPN2133). Densitometry was quantified using Quantity One (Bio-Rad, version 4.6.2).
Transgenic mice and breeding
The bacterial artificial chromosome (BAC) AC010412.9 (Open Biosystems, Cal-Tech H-BAC, CDT-2166J9-BHS1214) was injected into fertilized oocytes by the University of Chicago Transgenic Core Facility. Founder mice were identified by PCR of tail DNAs. Mice were bred to C57Bl6/J or mdx/C57Bl10/J for analysis. All procedures were carried out under the guidance and approval of the University of Chicago Institutional Animal Care and Use Committee.
Cardiotoxin injection
10µl of 10mM cardiotoxin in PBS (Chalbiochem, 217504) was injected into the right tibialis anterior muscle at day zero. The left tibialis anterior muscle was mock injected with PBS alone (Gibco, 14190-144). At day 7 post injection, the mice were sacrificed and muscles were quickly harvested for analysis. For histology, 7µm thick paraffin embedded sections of quadriceps muscles were stained with H&E and Masson Trichrome (MT), and images were collected using a Zeiss Axiocam epifluorescence microscope and recorded and processed using AxioVision 2.0.
Myofiber size analysis
Histological images were used for myofiber size analysis using ImageJ, version 1.43u, and statistical analysis was done using Prism (GraphPad). Fibrotic areas of muscle were scored blinded to genotypes or treatment using sections from quadriceps muscle.
Immunofluorescence microscopy
7 µm thick tissue sections were fixed in 100% cold methanol for 2 minutes, rinsed in PBS (Gibco, 14190-144) and blocked in 0.1% PBS+Triton for 2 hours at 4°C. The tissue was then stained with the anti-dystrophin and anti–P-Smad2 antibodies (NCL-Dys1; Novocastra Leica, Biosystems, DYS1-CE-S, pSMAD2/3, Cell Signaling, 3101S, pSMAD3 Abcam ab51451) overnight at 4°C. Secondary antibodies were from Molecular Probes. Nuclei were visualized with Vectashield with DAPI (Vector Laboratories). Images were acquired on a Zeiss Axiocam and recorded and processed using AxioVision 2.0.
Evans blue dye uptake and hydroxyproline assays
Prior to harvesting, mice were injected intraperitoneally with Evans blue dye (EBD) at 5 µl/g body weight (Sigma-Aldrich, E2129), dissolved in PBS at 10 mg/ml. Dye uptake assays were conducted immediately, while tissues for hydroxyproline measurements were frozen at −80°C. Dye uptake assays were performed as described (45). Hydroxyproline (HOP) measurements were also performed as described (45). Fibrotic areas were quantified from Masson Trichrome staining blinded to genotype.
Ex vivo muscle mechanics
The extensor digitorum longus muscles were removed and studied as described (46).
In vivo antibody injections
5 mg/kg in PBS or PBS was injected into 8 week mice IP at day 0, day 3, and day 5. On day 1, the tibialis anterior muscle was injected in 20 µl of 5 mM cardiotoxin as described above. Animals were sacrificed on day 7 of the protocol and muscle was analyzed by Trichrome and H&E staining blinded to treatment strategy.
Statistical Methods
Data were analyzed with GraphPad Prism. Because it required multiple comparisons, the statistical analysis of plasmin digestion products used a one-way analysis of variance (ANOVA) with Tukey post-test analysis. Analysis of fibers size, central nuclei and pSMAD quantification were compared using Student’s t-test. Evans blue dye uptake and hydroxyproline content assessed in multiple muscle groups, and specific force measurements were compared using Student’s T test. Standard error of the mean (SEM) was reported.
Acknowledgements
The authors thank Se-Jin Lee (Johns Hopkins) for helpful comments
Funding: This work was supported by NIH NS047726 (EMM), AR052646 (EMM, ERB), HL61322 (EMM), T32 HD007009, F31 NS071848 (EC) and DOD USARMC MD120051/W81XWH-13-1-0234 (EMM).
Footnotes
Supplementary Materials
Fig S1. LTBP4 cleavage is specifically blocked by the anti-PRR antibody.
Fig S2. The BAC transgene does not alter membrane leak and fibrosis.
Fig S3. Human LTBP4 (hLTBP4) expression intensifies mdx muscle pathology.
Fig S4. Multiple muscles are worsened by the hLTBP4 transgene in the mdx mouse.
Fig S5. Increased macrophage infiltration in hLTBP4/mdx muscle.
Author contributions: EC performed proteolysis experiments, generated and analyzed transgenic mice and wrote the manuscript. SB analyzed data and examined SMAD signaling. BG performed Immunolocalization of LTBP4. TM and AD analyzed transgenic mice. JU performed histopathology analysis. MH generated transgenic mice. LS and EB performed and analyzed ex vivo muscle physiology. JD analyzed data and edited manuscript. EM oversaw the study, analyzed the data and wrote the manuscript.
Competing interests: EM and EC are inventors on the following patent related to this work: US Patent application 13/957,100 entitled “Mitigating tissue damage and fibrosis via latent transforming growth factor beta binding protein (LTBP4)“.
Data and material availability: Materials are available upon request and pending available supplies.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1126/scitranslmed.3010018
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4337885?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1126/scitranslmed.3010018
Article citations
The complex landscape of DMD mutations: moving towards personalized medicine.
Front Genet, 15:1360224, 26 Mar 2024
Cited by: 0 articles | PMID: 38596212 | PMCID: PMC11002111
Review Free full text in Europe PMC
The extracellular matrix differentially directs myoblast motility and differentiation in distinct forms of muscular dystrophy: Dystrophic matrices alter myoblast motility.
Matrix Biol, 129:44-58, 04 Apr 2024
Cited by: 1 article | PMID: 38582404
The super-healing MRL strain promotes muscle growth in muscular dystrophy through a regenerative extracellular matrix.
JCI Insight, 9(3):e173246, 04 Jan 2024
Cited by: 2 articles | PMID: 38175727 | PMCID: PMC11143963
Comprehensive analysis of m6A regulators characterized by the immune microenvironment in Duchenne muscular dystrophy.
J Transl Med, 21(1):459, 11 Jul 2023
Cited by: 0 articles | PMID: 37434186 | PMCID: PMC10334523
Microdystrophin Gene Addition Significantly Improves Muscle Functionality and Diaphragm Muscle Histopathology in a Fibrotic Mouse Model of Duchenne Muscular Dystrophy.
Int J Mol Sci, 24(9):8174, 03 May 2023
Cited by: 4 articles | PMID: 37175881 | PMCID: PMC10179398
Go to all (33) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Anti-latent TGFβ binding protein 4 antibody improves muscle function and reduces muscle fibrosis in muscular dystrophy.
Sci Transl Med, 13(610):eabf0376, 08 Sep 2021
Cited by: 17 articles | PMID: 34516828 | PMCID: PMC9559620
Overexpression of Latent TGFβ Binding Protein 4 in Muscle Ameliorates Muscular Dystrophy through Myostatin and TGFβ.
PLoS Genet, 12(5):e1006019, 05 May 2016
Cited by: 30 articles | PMID: 27148972 | PMCID: PMC4858180
Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice.
J Clin Invest, 119(12):3703-3712, 02 Nov 2009
Cited by: 137 articles | PMID: 19884661 | PMCID: PMC2786802
Non-Glycanated Biglycan and LTBP4: Leveraging the extracellular matrix for Duchenne Muscular Dystrophy therapeutics.
Matrix Biol, 68-69:616-627, 23 Feb 2018
Cited by: 22 articles | PMID: 29481844 | PMCID: PMC6015525
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1 TR000430
NHLBI NIH HHS (2)
Grant ID: HL61322
Grant ID: R01 HL061322
NIAMS NIH HHS (4)
Grant ID: P50 AR052646
Grant ID: U54 AR052646
Grant ID: AR052646
Grant ID: T32 AR053461
NICHD NIH HHS (1)
Grant ID: T32 HD007009
NINDS NIH HHS (4)
Grant ID: P01 NS072027
Grant ID: R01 NS047726
Grant ID: NS047726
Grant ID: F31 NS071848





