Abstract
Free full text
Role and species–specific expression of colon T cell homing receptor GPR15 in colitis
Abstract
Lymphocyte recruitment maintains intestinal immune homeostasis but also contributes to inflammation. The orphan chemoattractant receptor GPR15 mediates regulatory T cell homing and immunosuppression in the mouse colon. We show that GPR15 is also expressed by mouse TH17 and TH1 effector cells, and is required for colitis in a model that depends on their trafficking to the colon. In humans GPR15 is expressed by effector cells including pathogenic TH2 cells in ulcerative colitis, but is not expressed by regulatory T (Treg) cells. The TH2 transcriptional activator GATA-3 and the Treg–associated transcriptional repressor FOXP3 robustly bind human, but not mouse, GPR15 enhancer sequences, correlating with expression. Our results highlight species differences in GPR15 regulation, and suggest it as a potential therapeutic target for colitis.
Recruitment of lymphocytes from the circulation is a tissue– and cell–specific process that is mediated by adhesion and chemoattractant receptors1,2. The interaction of lymphocyte adhesion receptors with and their ligands on vascular endothelium allows endothelial capture of blood borne lymphocytes and mediates lymphocyte rolling, but arrest and firm adhesion of rolling cells, as well as subsequent diapedesis, require the engagement of lymphocyte receptors of the chemoattractant GPCR family3. Signaling via these chemoattractant receptors triggers rapid integrin-dependent lymphocyte adhesion on the endothelium, and activates and drives programs of lymphocyte motility. Chemoattractant receptors thus play critical roles in the recruitment of lymphocyte subsets from the blood, and they direct the trafficking of lymphocyte subsets in both homeostatic and inflammatory states3,4. As examples, CCR7 helps target naive lymphocytes and subsets of memory and effector cells to lymph nodes and tertiary lymphoid tissues in chronic inflammation; CCR10, CCR8 and CCR4 participate selectively though not exclusively in skin homing by memory/effector T cells; and CCR9 serves as a specific T cell and plasmablast homing receptor for the small intestines5. Interestingly, CCR10 also targets IgA plasmablast homing to mucosal tissues including the bronchial tree and the colon, but is not expressed by gut homing T cells: CCR10 expression on T cells is largely mutually exclusive with that of the integrin intestinal homing receptor α4β7 (ref. 5), which is required for efficient lymphocyte homing to the intestines through its recognition of the mucosal vascular addressin MAdCAM1 (refs. 3,6). Although inflammatory chemokines and their receptors may participate7,8, whether colon effector and memory T cells have specific chemoattractant trafficking receptors analogous to skin and small intestinal T cell–selective chemokine receptors has remained unclear.
GPR15 is an orphan GPCR and an HIV co–receptor that is structurally related to known lymphocyte trafficking receptors9,10. A recent study has implicated this receptor in colon homing of Treg cells in the mouse11, but its role in effector T cell trafficking and function is not known. Here we examine the expression and function of GPR15 on effector T cells in mouse and man. Our results show that GPR15 is important for effector as well as regulatory T cell localization to the mouse colon and implicate GPR15-dependent effector cell recruitment in murine colitis. We also describe substantial differences in GPR15 expression in the human, with expression by TH2 cells but not Treg cells in the normal and inflamed human colon. We relate the expression of GPR15 by human TH2 cells and by mouse, but not human Treg cells, to inter–species differences in binding of transcriptional regulators to Gpr15 enhancer sequences.
Results
Activated colon CD4+ T cells express GPR15
Using Gpr15–GFP knock–in mice11, we confirmed that GFP+ CD4+ are most frequent among T cells in colon lamina propria (LP), as compared to small intestine (SI) and peripheral lymphoid tissues. They were infrequent among the intraepithelial lymphocytes of the colon or SI, and absent in naive CD4+ T cells of all tissues (Supplementary Fig. 1a;11). GFP+ CD4+ T cells in the colon display an activated phenotype, characterized by high expression of activation antigens CD44, CD69 and CD25, and low expression of CD45RB and CD62L (Supplementary Fig. 1b); and they express the integrin homing receptor α4β7 for intestinal vascular adhesion ligand MAdCAM1 but lack the SI trafficking receptor CCR9 (Supplementary Fig. 1b). Consistent with a prior report11, under homeostatic conditions most, although not all, colon LP GFP+ CD4+ T cells express markers of Treg differentiation, including Foxp3, CD25, GITR and Helios, and lack effector cytokine expression (Supplementary Fig. 1c).
Gpr15 mediates CD4+ TEM accumulation in the colon
To assess the importance of GPR15 to effector/memory T cells (TEM) in different tissues, we reconstituted irradiated mice with a mixture of allotype–marked Gpr15gfp/gfp (Gpr15–knockout; KO) and Gpr15gfp/+ (Gpr15–heterozygous; het) bone marrow cells, allowing Gpr15–KO and Gpr15-het lymphocytes to compete for niches within a common host environment. Gpr15–KO CD4+ repopulated the colon LP with significantly lower efficiency than their Gpr15–het counterparts (Supplementary Fig. 2a,b and Fig. 1a). In contrast, GPR15 deficiency had no effect on the reconstitution of CD4+ T cells in lymphoid tissues or SI LP; nor did it alter colon naive CD4+ T cell frequency (Fig. 1a). Importantly, TEM cell (CD45RBlo, CD44hi, Foxp3−) accumulation was reduced to the same extent as that of Treg cells (CD25high, Foxp3+; Fig. 1a). This accumulation is likely mediated by GPR15–dependent recruitment, because GPR15 effects colon homing of in vitro–generated TEM (Supplementary Fig. 3), Treg cells as well as transduced T cells11; however, effects on retention, survival or proliferation cannot be excluded. These results demonstrate an important role for GPR15 in long–term TEM as well as Treg reconstitution and homeostasis in the mouse colon.
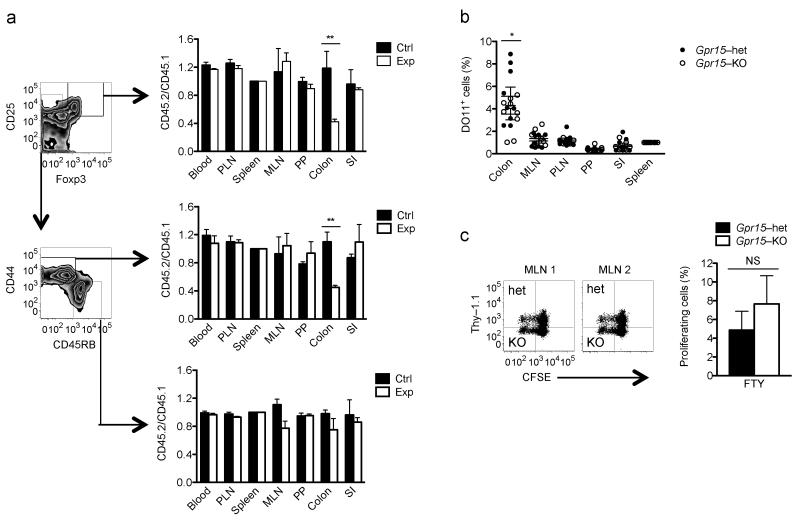
GPR15 mediates accumulation of TEM and Treg cells in the colon. (a) Gpr15–KO (CD45.2) or het (CD45.2) bone marrow cells were combined with Gpr15-het (CD45.1) cells and transferred into irradiated hosts (CD45.1 × CD45.2 F1) to generate experimental (Exp) or control (Ctrl) chimeras, respectively, as in Supplementary Figure 2a. Ratios of Gpr15–KO to Gpr15–het derived cells (CD45.2/CD45.1) among Treg cells (CD4+, CD25hi, Foxp3+; top), Treg–excluded TEM cells (CD4+, Foxp3−, CD44hi, CD45RBlo; middle), and naive CD4+ T cells (CD4+, Foxp3−, CD44lo, CD45RBhi; bottom) in Exp mixed bone marrow chimeras (white bars); ratios of Gpr15- het to het cells (CD45.2/CD45.1) in control chimeras (black bars). Peripheral lymph nodes (PLN), mesenteric lymph node (MLN), Peyer’s patches (PP), small intestine (SI). n = 3, mean + SEM, **p <0.01 (2–way ANOVA, Bonferonni posttest). Representative of 3 experiments. (b) Splenocytes from DO11.10 Gpr15–het or DO11.10 Gpr15–KO were i.v. injected into Balb/c (Thy–1.1) mice. After 24 h, recipients were treated intra-rectally with ovalbumin (OVA) and cholera toxin (CT). Accumulation of OVA-specific CD4+ T cells in tissues was assessed 5 days later, as in Supplementary Figure 2c. The percent antigen–specific (DO11+) of Gpr15-het or KO CD4+ TEM cells (CD44hi, CD45RBlo) 5 days after antigen exposure was normalized to the ratio in the spleen. Data from 9 mice from 3 independent experiments are shown. Mean ± SEM, *p <0.05 (2–way ANOVA, Bonferroni posttest). (c) Similar proliferation rate of antigen–reactive Gpr15–het and Gpr15–KO cells. CFSE–labeled splenocytes from Gpr15–het (Thy–1.1, Thy–1.2) and KO (Thy–1.2) DO11 mice were co–injected i.v. in equal proportion into congenic non–DO11 hosts (Thy–1.1) on day 0. Recipients were treated with OVA and CT per rectum on day 1, FTY720 or PBS i.p on day 3, and tissues were collected on day 5, as in Supplementary Figure 2e. (d) Recipient MLN gated on donor DO11+ CD4+ T cells. n = 3; mean + SEM. Not significant (NS).
We next evaluated GPR15 involvement in colon antigen–specific CD4+ T cell generation. Allotype–marked, ovalbumin (OVA)–specific (DO11.10) Gpr15–KO or Gpr15–het splenic CD4+ T cells were transferred into recipient mice that were immunized intra–rectally 24 h later with ovalbumin (OVA) and cholera toxin (Supplementary Fig. 2c), a procedure that selectively induces T effector cell responses12. Activated T cells arising in the colon or draining mesenteric lymph node (MLN) then home via the blood to the colon13. OVA–specific Gpr15–het T cells were more efficient than their Gpr15–KO counterparts at populating the recipient mice colons (Fig. 1b and Supplementary Fig. 2d). This difference was not due to differences in the overall OVA response between the het and KO T cells, as indicated by similar antigen–specific proliferation in the MLN of FTY720–treated mice14, in which lymphocyte exit from lymph nodes (MLN) is controlled (Supplementary Fig. 2e and Fig. 1c). Thus GPR15 supports the accumulation of antigen–reactive CD4+ effector T cells in the colon.
Gpr15 required for CD45RBhi T cell transfer colitis
Having shown that GPR15 contributes to colon effector T cell recruitment and accumulation, we next assessed its importance in colon inflammation. We used a well–established model of colitis, the CD45RBhi CD4+ T cell transfer model15, in which colitis is dependent on effector T cell expression of intestinal trafficking receptors16,17. In the absence of Treg cells, transferred T cells develop into pathogenic effector T cells that home to the colon and induce disease15. Rag2−/− mice that received naive Gpr15–KO T cells were protected from developing colitis (Fig. 2), consistent with a critical function for GPR15 on pathogenic effector T cell colon trafficking. Inhibition of colitis by Treg cells in this model does not require colon homing18, and accordingly, GPR15–deficient Treg cells and WT Treg cells prevented or delayed colitis in a comparable manner (Fig. 2). In the acute DSS–induced colitis model, where innate immunity plays a predominant role and colitis is not dependent on T cell colon homing19,20, as well as the acute TNBS model, Gpr15–KO and WT mice were similarly susceptible to intestinal inflammation. (Supplementary Fig. 4). The inefficiency of Gpr15–deficient cells to induce colitis in the CD45RBhi transfer model thus reveals a role of GPR15 in the colon trafficking of pathogenic proinflammatory T cells.
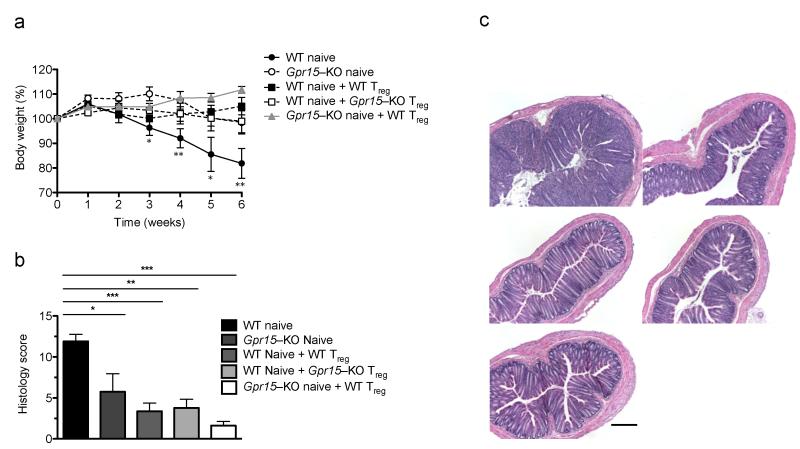
GPR15–deficient naive T cells fail to induce colitis in the CD45RBhi CD4+ T cell transfer model. (a) Weight loss of Rag2−/− mice that received either WT naive CD4+ T cells, Gpr15–KO naive CD4+ T cells, WT naive plus WT Treg cells, WT naive plus KO Treg cells, or KO naive plus WT Treg cells. n = 5, mean + SEM, *p <0.05, **p <0.01 between WT naive (disease control) and Gpr15 KO naive (2-way ANOVA, Bonferroni posttest). Representative of 3 experiments. (b) Clinical score for histological inflammation in colon. *p <0.05, **p < 0.01, ***p <0.001 (1–way ANOVA, Tukey’s posttest). (c) H&E staining of distal colon section: WT naive (top left), Gpr15–KO naive (top right), WT naive + WT Treg (middle left), WT naive + Gpr15–KO Treg (middle right) and Gpr15–KO naive and WT Treg (bottom). Scale bar denotes 10 μm.
Colitis in the naive T cell transfer model depends on the generation of pathogenic IFN–γ- and IL17–producing TH1 and TH17 effector CD4+ cells21-23. To determine whether a deficiency in generation or localization of such effector cells might underlie or contribute to the reduced ability of KO T cells to induce colitis, we transferred allotype–marked naive Gpr15–KO and het CD4+ T cells together into Rag2−/− mice and assessed effector T cell presence in recipient tissues 2 weeks later, prior to clinical colitis onset (Supplementary Fig. 5a). Compared to GFP+ Gpr15–KO T cells, GFP+ Gpr15–het (GPR15–expressing) T cells preferentially accumulated in the colon of Rag2−/− recipients, consistent with a prominent role for the receptor in effector T cell localization (Fig. 3a). On the other hand, GPR15 deficiency had no significant effect on the frequency of TH1 and TH17 effector T cells among the donor populations (Fig. 3b and Supplementary Fig. 5b-d), or on the fraction of proliferating T cells as indicated by the extent of BrdU incorporation (Fig. 3c), or on that of apoptotic CD4+ T cells as indicated by Annexin V staining (data not shown). In related Rag2−/− transfer studies (Supplementary Fig. 5e), we found that GFP–expressing subsets of colon–localized Gpr15–KO and het derived cells also displayed similar cytokine, Foxp3 and Ki67 expression (Supplementary Fig. 5f). These data suggest that the failure of GPR15–deficient T cells to induce colitis is not due to alterations in T cell activation or production of proinflammatory cytokines.

GPR15 deficiency inhibits effector T cell accumulation in the colon, but not TH1 or TH17 differentiation. Naive CD45RBhi CD4+ T cells sorted from Gpr15–KO (CD45.2) or het (CD45.1, CD45.2) mice were co–injected into Rag2−/− recipients in equal proportion, as in Supplementary Figure 5a. (a) Ratios Gpr15–het to Gpr15–KO GFP+ or GFP− CD4+ T cells in recipient tissues after 2 weeks, normalized to those in the spleen. n = 5, mean + SEM, *p <0.001. (2–way ANOVA, Bonferroni posttest). (b) Similar percentage of Gpr15–het- and KO-derived IFN-γ and IL–17+ CD4+ T cells in recipient colon after 2 weeks (left). Ratio of Gpr15– het- to KO-derived IFN–γ+ or IL–17+ CD4+ T cells, normalized to that of the spleen (right). Representative of 2 experiments. (c) BrdU incorporation 2 weeks after cell transfer as in (a); baseline (grey shade). n = 3; mean + SEM.
GPR15 expression on human CD4+ T cells
To determine the relevance of these mouse studies to humans, we assessed GPR15 expression by colon CD4+ T cells in ulcerative colitis (UC; a “TH2–like” disease in which inflammation is limited to the colon24), in Crohn’s disease (typically a TH1 and TH17 disease25), and in colons from non–UC patients that included healthy or normal margins of cecal or colorectal adenocarcinomas. Surprisingly, GPR15 was highly enriched among UC colon TH2 cells (IL-5+ or IL–13+; Fig. 4a). Few IFN–γ+ or IL–17+ effector T cells expressed the receptor (Fig. 4a) and, in contrast to the mouse, there was little or no GPR15 expression on Treg cells (CD127lo, Foxp3+, CD4+) from either UC or non–UC colons (Fig. 4, Supplementary Fig. 6). Treg cells lacked GPR15 expression whether resting or activated (Supplementary Fig. 6b). Consistent with this differential expression in vivo, GPR15 is preferentially induced on in vitro–generated human TH2 effector cells (IL–4+), but not on Treg cells under conventional polarizing conditions (Fig. 5a; Foxp3+ cells are CD127lo; not shown). In contrast, GPR15 expression was observed in mouse cells under Treg and TH17 cells (Fig. 5b; and see11), but not TH2–polarizing conditions (Fig. 5b). These results highlight GPR15 expression by pathogenic T cells in colitis, and reveal dramatic differences in the patterns of cell type GPR15 expression in humans versus mice.
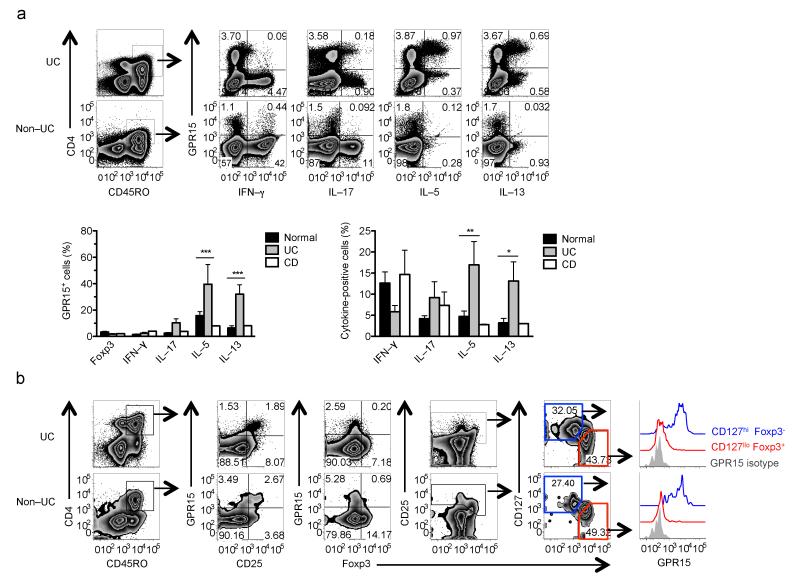
Human proinflammatory CD4+ T cells express GPR15 in the colon. (a) Expression of GPR15 on cytokine–producing CD45RO+CD4+ T cells from ulcerative colitis (UC) or non–UC colons (top). In UC colons, GPR15 is expressed on IL–5+ and IL–13+ CD45RO+CD4+ T cells (bottom). n = 5 UC specimens, 11 Non–UC or normal samples, and 2 Crohn’s disease (CD) colons, which showed similar cytokine expression among GPR15+ cells as normal samples. Mean + SEM; *p <0.05, **p < 0.01, ***p <0.001 (2–way ANOVA, Bonferroni posttest). Statistics not determined for CD samples. (b) Lower expression of GPR15 on Treg cells (CD127lo, Foxp3+; red histogram) as compared to non–T memory CD4+ Treg, cells (CD127hi, Foxp3−; blue).
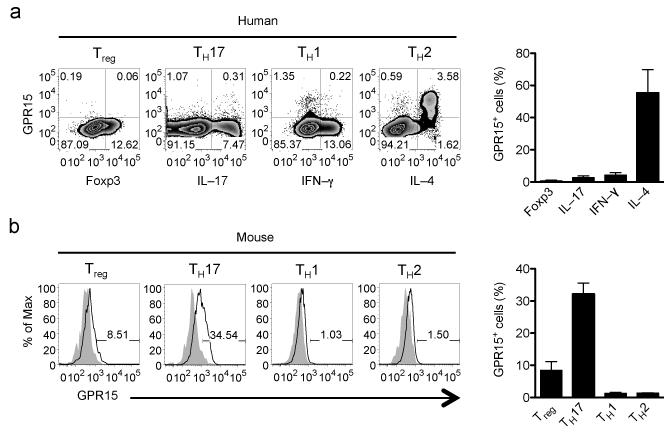
Distinct patterns of GPR15 expression by in vitro–polarized human and mouse T cells. Naive CD4+ T cells enriched from human PBMC (a) or mouse Gpr15–het spleen (b) were polarized as described in the online methods. GPR15 is expressed on human but not mouse in vitro–polarized TH2 cells. Human cells are gated on CD45RO+ CD4+ T cells, and mouse cells are gated on CD44hi CD4+ T cells. Combined results of 3 mouse and 4 human experiments; mean + SEM.
Binding of GATA–3 to GPR15 enhancer region
GATA–3 is the master regulator of TH2 cell differentiation and cytokine expression26 and it interacts with Foxp3 to regulate TH2 versus Treg gene expression27-29 cell. In an attempt to identify mechanisms underlying T cell subset– and species–specific GPR15 regulation, we analyzed GATA–3 binding at the human and mouse loci using previously generated ChIP–Seq data30,31 (Fig. 6a,b). We found that GATA–3 bound to a highly conserved site downstream of GPR15 in humans, but not in mice (site marked “Downstream” in Fig. 6a and b). In contrast, GATA–3 bound to a site upstream of GPR15 close to CLDND1 in humans and Cldn25 in mice (marked “Upstream” in Fig. 6a,b). The significance of these interactions was confirmed using model–based analysis of ChIP–Seq (MACS), which identifies significant numbers of sequence reads indicative of transcription factor binding sites32. MACS identifies both the upstream and downstream GATA–3 binding sites in humans with p values of 2.6 × 10−101 and 1.1 × 10−63, respectively (83 and 111 sequence reads, respectively), but only the upstream Cldn25–I associated upstream site was significant in mouse (p = 6 × 10−37 with 80 reads compared to only 9 reads for the downstream site, determined as not significant by MACS). A GATA–3 binding site within the IL4 CNS2 was identified as significant in both human and mouse (1.7 × 10−85 with 83 reads and 5.1 × 10−33 with 97 reads, respectively). This species–specific binding of GATA–3 to the downstream site was confirmed by ChIP–qPCR, with binding to the upstream site and to the IL4 CNS2 element remaining constant between human and mouse (Fig. 6c). Consistent with this species–specific binding, consensus GATA–3 sites could be identified at this site in human but not in the mouse (Supplementary Fig. 7). Notably, a tandem GATA–3-STAT–6 motif is conserved in multiple species, but contains an A–to–G nucleotide difference in the mouse (Supplementary Fig. 7), a mutation known to attenuate GATA–3 binding33 (and TTD and JP, unpublished).
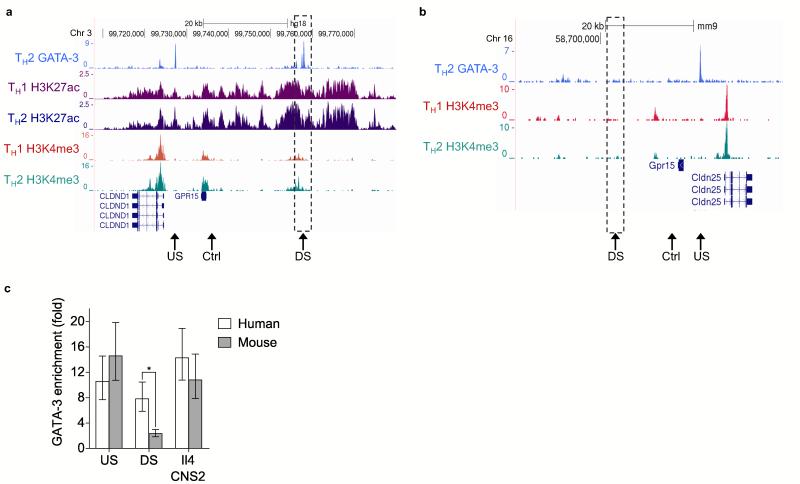
Differential binding of GATA–3 to human and mouse Gpr15 enhancer sequences. (a) ChIP–Seq data showing binding of GATA–3 at the human GPR15 locus in TH2 cells. The number of sequencing reads from ChIP–enriched DNA are plotted per million background–subtracted total reads and aligned with the Hg18 build of the human genome. Association with H3K4me3 and H3K27ac in human TH1 and TH2 cells are shown below. The position of the upstream enhancer is marked with a dashed box. The positions of qPCR primers used in (c) are marked by arrows. (b) As (a), except for Gata–3 and H3K4me3 at the mouse Gpr15 locus (Mm9 build). (c) Enrichment of DNA corresponding to the human and mouse upstream and downstream GATA–3 binding sites marked in (a) and (b), relative to input DNA and a control non–bound region (also marked in (a) and (b)), measured by ChIP–qPCR. Mean and SD from triplicate wells from one ChIP per species. *p < 0.05, unpaired Student’s t–test.
Active chromatin marks characterize this region in the GPR15–expressing ENCODE cell line GM12878 (Supplementary Fig. 7), whereas repressive chromatin modifications are observed in cell lines that do not express GPR15. We therefore hypothesized that the downstream site is an enhancer, active in human TH2 cells. To test this, we performed ChIP–Seq for the active enhancer mark H3K27ac and the transcriptional initiation mark H3K4me3 in in vitro polarized human TH1 and TH2 cells (Fig. 6a). We found that the site was indeed associated with H3K27ac and more so in TH2 than TH1 cells, mirroring the expression of GPR15. The upstream site and the GPR15 gene were also associated with greater amounts of H3K4me3 in TH2 cells. In contrast, no H3K4me3 is present at the downstream site or at the GPR15 gene in mouse (Fig. 6b), consistent with the lack of GATA–3 binding. We conclude that GATA–3 exhibits differential binding to an enhancer at the GPR15 locus between human and mouse and that this contributes to altered patterns of enhancer activation and GPR15 expression in TH2 cells between the species.
Foxp3 is responsible for the repression of T effector genes in Treg cells34. We therefore considered that differential binding of this factor could be responsible for the differential expression of GPR15 between mouse and human Treg cells. ChIP–Seq for FOXP3 in human and mouse Treg cells identified significant binding to the GPR15 enhancer in human but not mouse (Supplementary Fig. 8). In human Treg cells, the GPR15 enhancer was one of the most significant binding sites, with p values between 1.7×10−11 and 1.7×10−80 and in the top 1% of sites in 4 different experiments. By contrast, in replicate experiments in mouse, the p values for Foxp3 binding to the Gpr15 enhancer were 1.4×10−4 and 2.6×10−4, below the default MACS significance threshold of 10−5 and only within the top 13% or 23% of all sites with p <10−3. Therefore, the low expression of GPR15 in human Treg cells may reflect efficient binding and repression by FOXP3.
Discussion
GPR15 is an HIV co–receptor and an orphan GPCR with structural homology to known chemokine receptors. Recently, GPR15 was shown to be expressed by colon Treg cells, and to contribute to their homing to and function in the colon. Our studies reveal that GPR15 is also an effector/memory CD4+ T cell receptor with a role in CD4+ TEM cell homing and colitis pathogenesis, whose expression is differentially regulated in CD4+ T cell subsets between humans and mice. We show that GPR15 expression is enriched on human TH2 colon cells, but in striking contrast to the mouse, most human colon Treg cells (whether in IBD or non–IBD human colons) fail to express GPR15. We identified mutations that disrupt GATA–3 binding and GPR15–enhancer activation in the mouse enhancer region, and described differences in GATA–3 and FOXP3 binding to the enhancer in mice versus humans that may underlie the species–specific regulation of GPR15 on effector versus regulatory T cell subsets. Moreover, ChIP–Seq data indicate that FOXP3, a repressor of TH2–associated genes in Treg cells, binds with high efficiency to the human but not mouse enhancer in Treg cells, correlating with inhibition of GPR15 expression in human regulatory T cells.
Our results are consistent with, but extend, those of another study11. They used transfected naive lymphocytes to show convincingly that GPR15 functions as a colon homing receptor, and demonstrated its role in Treg cell localization and function in the colon as well. Our results confirm the receptor’s role in Treg cell localization in the mouse, but also demonstrate a similarly important role in TEM cell homing and accumulation in the large intestine. Although GPR15 participation as a T cell trafficking receptor for the colon is now established, additional roles for the receptor in cell retention or survival are not excluded, nor is its involvement in homing to other epithelial surfaces35. Expression of GPR15 on Tregcells was previously shown to be required for transferred Treg cells to prevent mortality and severe colitis associated with Citrobacter rodentium11, a widely accepted model of infectious colitis with properties similar to the colitis caused by human pathogen enteropathogenic E. coli. We, however, employed the widely used CD45RBhi CD4+ T cell transfer colitis model that does not require Treg cell homing to the colon for disease control. Treg cell suppression of colitis in the CD45RBhi CD4+ T model does not appear to require gut homing: β7–deficient Treg cells are fully capable of suppressing disease18, whereas Treg cell suppression of colitis in this model requires Treg cell migration to lymph nodes; CCR7–deficient Treg cells were less efficient at preventing colitis as compared to CCR7–sufficient Treg cells36. In contrast, the pathogenic T cells in the CD45RBhi CD4+ T cell transfer model do require intestinal trafficking receptors for colitogenesis: colitis in this model is inhibited by antibody blockade of the gut homing receptor α4β7 or by genetic deficiency of integrin β17,37; and we showed that it is also inhibited by GPR15 deficiency on transferred T cells. β7 integrin deficiency does not alter the course of DSS–induced colitis19,20, correlating with the lack of effect of GPR15 deficiency, and likely due to the non–critical role of T cells in the acute chemically–induced models. Together, the results highlight the importance of GPR15 expression for T cell localization and function in the colon. GPR15–dependent T cell homing can mediate immune pathogenesis in T effector–cell driven colitis when TEM cell activity is required locally in the colon, but it can contribute to immune regulation in models in which Treg cell responses are important locally in the gut wall. In this context, we note that the CD45RBhi CD4+ T cell transfer model has been used to identify and validate antibody inhibitors of intestinal lymphocyte trafficking that have led to effective therapeutics for human colitis17,38,39.
These considerations may have significant implications for translation of mouse studies to the human. Our data show that patterns of GPR15 expression are quite different in man. In sharp contrast to the mouse, few if any human colon Treg cells isolated from patients with colon cancer (normal adjacent tissue), ulcerative colitis, or Crohn’s disease expressed the receptor. The highest frequency of GPR15 expression was found on TH2 effector T cells, an important pathogenic TEM cell population in the colon of ulcerative colitis patients24,40,41. Notably, in vitro–generated human, but not mouse TH2 cells, expressed GPR15. Although it is not feasible to carry out functional studies of the orphan receptor in humans, GPR15 is highly conserved in mammalian species including man. Thus our results suggest that GPR15 may help target pathogenic TH2 cells to the colon in man, but is probably less important in man than in mouse for the homing and function of regulatory T cells in the gut wall.
We identified an active GPR15 enhancer region with marked differences in sequence and cell type specific transcription factor binding and chromatin marks in human and mouse. The human enhancer comprises canonical GATA–3 binding motifs that are conserved in multiple species but missing in the mouse. GATA–3 plays key roles in TH2 cell differentiation and cytokine expression26. GATA–3 ChIP–Seq data30,31 and targeted ChIP-qPCR revealed that GATA–3 binds to the 3′ (downstream) GPR15 enhancer in human but not mouse TH2 cells, whereas binding to a site upstream at CLDND1/Cldn25 and to the established IL4 CNS2 locus is conserved between the two species. The active enhancer mark H3K27ac and the transcriptional initiation mark H3K4me3 in human TH1 and TH2 cells confirmed the increased activity of the enhancer and increased expression of GPR15 in human TH2 cells. Unlike in the human, H3K4me3 is not observed at Gpr15 or its enhancer in mouse TH2 cells. We conclude from these experiments that GATA–3 only binds, and the enhancer is only activated, in human TH2 cells and not in mouse TH2 cells.
We also found that Gpr15 expression in Treg cells is species–specific, with expression suppressed in human cells. Potentially explaining this, ChIP–Seq for Foxp3, a transcriptional regulator necessary for repression of T effector cell genes in Treg cells42,43 demonstrates significant binding to the GPR15 enhancer only in human and not mouse Treg cells. Foxp3 is thought to bind DNA with co–factors, including GATA–329, thus the reduced binding of Foxp3 to the Gpr15 enhancer in mouse Treg cells may be due to differences in GATA motifs and loss of GATA–3 binding. Additional factors are also likely to be involved in the differential expression of GPR15 between human and mouse T cell subsets, including those responsible for the increased expression in mouse TH17 cells.
Although differences in transcription factor binding between species have been reported44, to our knowledge, the differential binding of GATA–3 at GPR15 between human and mouse is the first example in which defined differences in transcription factor binding have been associated with species differences in targeting mechanisms of functionally distinct T cell populations, differences expected to result in altered patterns of local (TH–cell derived) cytokine production and thus mechanisms of pathogenesis. Manipulation of the enhancer sequences in T cells in vitro and in vivo will be required to elucidate the specific consequences of the species differences in GATA–3 binding shown here.
In conclusion, our studies show that GPR15 can control the colon localization of T effector cells, and we demonstrate its key role in a model of colitis that requires effector cell homing to and function in the gut wall. Our studies also identify significant differences in Treg versus T effector cell expression of the receptor between mice and humans, differences that suggest a predominant proinflammatory role for GPR15 in man. These species differences may fundamentally restrict the relevance of mouse models for the study of Treg trafficking in colitis. Indeed, given its preferential expression by pathogenic effector cells in man, inhibition of GPR15 may present a therapeutic approach to controlling inflammatory T cell recruitment in ulcerative colitis.
Online Methods
Animals
BALB/c, C57BL/6 and B6.SJL mouse strains were purchased from Jackson Laboratory and bred in house. Gpr15–GFP mice, generated as described11, were backcrossed 10 times onto the C57BL/6 and BALB/c backgrounds prior to use in experiments, or for mating with B6.SJL and DO11.10 breeders to obtain CD45.1+ and DO11.10 Gpr15 Het (Gpr15gfp/+) and KO (Gpr15gfp/gfp) mice. Rag2 constitutive knockout (Rag2−/−) mice were obtained from Taconic and housed for at least 1 week prior to being used as recipients in colitis studies. Animals were maintained in accordance to National Institutes of Health guidelines and experiments were approved by Stanford University Institutional Animal Care and Use Committee.
Cell Isolation
Spleen, Peyer’s patches, mesenteric and peripheral (including inguinal, brachial and axillary) lymph nodes were mechanically dispersed through a stainless steel 200 μm wire mesh to yield single–cell suspensions in Hank’s buffered salt solution (HBSS; Mediatech, Inc.) containing 2% BCS. Splenocytes were incubated in Red Blood Cell Lysing buffer (Sigma–Aldrich) and the cells washed by centrifugation for 5–10 min at 300 g, 4 °C. Intraepithelial and lamina propria lymphocytes were isolated from the small intestine and colon as described previously45. After excision of the Peyer’s patches, the intestines were cut into ~0.5 cm pieces and cleansed in HEPES–buffered HBSS containing 2% BCS. To harvest IEL, the tissues were incubated twice with 1 mM dithiothreitol at 37 °C for 20 min and the supernatant collected. To isolate LP lymphocytes, the tissue pieces were incubated in 2 mM EDTA for 15 min (× 2 rounds) followed by 3 rounds of collagenase–digestion at 37 °C (30–45 min each), and the supernatant was collected, pooled and washed. Centrifugation in a 30/70% percoll step–gradient produced enriched IEL or LP lymphocytes at the interface, and the cells were then washed, counted and stained for flow cytometry analysis.
Antibodies and Fluorescence–activated cytometry
The following antibodies were used for flow cytometry in the mouse studies: CD62L–APC (MEL-14), CD103–PE (M290) (BD Pharmingen); CD25–PE–Cy7 (PC61), Foxp3–PE, Foxp3–FITC, Foxp3–APC (FJK–16s), α4β7–APC (DATK32), CCR9–PE (CW–1.2), and mouse IgG2a K isotype control–PE (eBioscience); CD4–Alexa Fluor 700 (GK1.5), CD45RB–APC–Cy7 (C363-16A), CD44–Pacific Blue (IM7) (Biolegend). For staining human blood and tissues, these antibodies were used: rabbit anti–human GPR15 (Abcam), rabbit IgG isotype control (Sigma–Aldrich) and goat anti–rabbit–AF488 secondary antibody (Invitrogen) and goat anti–mouse AF488 (Invitrogen); IL–13–PE (JES10-5A2), IFN-γ-PE (B27), IL-17A–PE–Cy7 (BL168), CD45RO–APC–Cy7 (UCHL1) and CD25–AF700 (BC96) were purchased from Biolegend; Foxp3–APC (PCH101) from eBioscience; CD4–Pacific Blue (RPA–T4) and IL–5–PE (TRFK5) from BD Biosciences. The mouse anti–human GPR15 MAb (Clone 367902; R&D Systems) and corresponding mouse IgG2B isotype control (R&D Systems) were used only in side-by-side assessment of the GPR15 PAb, as described in below. Where indicated, cells were activated with phorbol myristate acetate (PMA; 50 ng/mL), ionomycin (1 μg/mL) in the presence of Brefeldin A (to prevent cytokine release into media) for 4 h at 37 °C prior to surface staining46. Staining of mouse and human Foxp3 and intracellular cytokine antibodies was performed using Foxp3 fixation and permeabilization solutions as recommended (eBioscience). Data was acquired on an LSRII or Fortessa cytometer (BD Biosciences) and analyzed with FlowJo software (FlowJo, LLC).
CD45RBhi CD4+ T cell transfer model of colitis
CD4+ T cells were enriched from spleen of WT or Gpr15 KO mice by negative selection using the CD4+ T cell Isolation Kit II (Miltenyi Biotec). The bead–enriched CD4+ T cells were stained and sorted by flow cytometry using the ARIA II or ARIA III cell sorter (BD Biosciences) to separate naive cells (CD4+, CD45RBhi, CD25lo) and Treg cells (CD4+, CD45RBlo, CD25hi). Cells were washed with sterile HBSS, and 0.5 × 106 naive CD4+ T cells alone or with 0.1 × 106 Treg cells were transferred i.v. into each Rag2−/− mouse. The mice were weighed weekly and assessed for weight loss, stool consistency, bloody stools and general well being. The mice were euthanized at week 6 (or sooner if moribund or weight loss exceeded 20%). Following euthanasia, spleen, MLN and colon were harvested and colon length and stool consistency assessed. For histologic assessment, the distal–most 1 cm section of colons was fixed in 10% buffered formalin for tissue sectioning and hematoxylin & eosin staining47. The histology was assessed and scored in a blinded fashion as previously described48.
GPR15 expression in human colon samples
Colon resection samples were obtained from patients undergoing ulcerative colitis– or adenocarcinoma–indicated colectomies with approval by the Stanford University Institutional Review Board and obtaining informed patient consents. After removal of the serosa, mesentery fat and muscularis externa, lymphocytes were isolated from the lamina propria as described49, and the cells activated and stained for flow cytometric analysis.
Chromatin IP and ChIP–Seq data analysis
Human and mouse GATA–3 ChIP–Seq has previously been published30,31. Human and mouse H4K4me3 ChIP–Seq data have also been published50,51. Mouse GATA–3 and Foxp3 and human Foxp3 ChIP–Seq data were downloaded as fastq files from GEO (accessions listed in Supplementary Table 2), aligned to mm9 or hg18 with bowtie2 (default settings), bigwig files generated for visualization in the UCSC genome browser as described30 and binding sites identified with MACS (using input DNA as background, if available). ChIP for H3K27ac in in vitro polarized human TH1 and TH2 cells was performed as described30 using the antibody ab4729 (Abcam). ChIP for GATA–3 was performed with the D16 antibody (Santa Cruz Biotechnologies, Inc.) in mouse TH2 cells polarized as described52. ChIP for Foxp3 in mouse iTreg cells is described in the Supplementary information. DNA enriched by GATA–3 ChIP was measured by quantitative PCR relative to input DNA and normalized to a control region at the GPR15 locus that lacked GATA–3 binding. The primers used are listed in Supplementary Table 1.
Mixed bone marrow chimera
Bone marrow was extracted from femur and tibia of Gpr15–KO (Gpr15gfp/gfp; CD45.2) and Gpr15–het (Gpr15gfp/+; CD45.1) mice and red blood cells lysed. Cells were combined in equal ratios and injected into lethally irradiated F1 offspring of C57BL/6 × B6.SJL breeding (CD45.1, CD45.2) to generate experimental KO/Het mixed chimeras. In parallel, cells from Gpr15–het (CD45.2) and congenic Gpr15–het (CD45.1) mice were similarly transferred to generate Het/Het control chimeras. After 8–12 weeks, tissues from F1 recipients were harvested and prepared for analysis by flow cytometry. For each mouse, the ratio of CD45.2+ CD4+ to CD45.1+ CD4+ T cells recovered from each organ was calculated. Since bone marrow ’take’ from different donors can vary between recipients, the CD45.2/CD45.1 ratio of T cells in different organs was normalized to that of CD19+ B cells, and the relative ratio of cells in each tissue was additionally normalized to that of the spleen as described53.
Stable GPR15 transfectant generation
Murine pre–B cell lymphoma L1.2 cells were transfected (Nucleofector™, Lonza Inc.) with a mammalian expression vector pCMV6–AC–GFP) containing the human GPR15 cDNA with a C–terminal tGFP tag (Origene). G418–selected transfectants with highest levels of GFP expression were sorted for testing of antibody to human GPR15. The mouse Gpr15 ORF was subcloned into the pCMV6–AC–GFP vector 3′ of the Kozak consensus sequence to similarly generate stable L1.2 transfectants.
Analysis of rabbit polyclonal antibody to human GPR15
To determine the specificity of the polyclonal GPR15 antibody (Abcam) used throughout the study, L1.2 transfectants (1 × 106 cells) of human (or mouse) GPR15 were incubated with 0.5 or 1 μg of the GPR15 polyclonal antibody for 30 min, followed by 30 min with PE–donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.; Supplementary Fig. 9a). The specific staining of the polyclonal antibody was also tested side–by–side with a monoclonal antibody to human GPR15 (Clone 367902; R&D Systems) on freshly isolated human colon cells (Supplementary Fig. 9b,c), and the two antibodies produced similar staining.
DSS–induced colitis
Two months old gender–matched Gpr15–KO and wild–type BALB/c mice were given drinking water containing 4.5% DSS (w/v; dextran sulfate sodium salt, 36,000-50,000 Da; MP Biomedicals) beginning day 0 (ref.54). The mice were weighed daily and assessed for the presence of bloody stool and general well being. They were euthanized on day 9.
TNBS–induced colitis
Two months old gender–matched Gpr15–KO and wild–type BALB/c mice were rectally instilled with 2 mg of TNBS (2,4,6–Trinitrobenzenesulfonic acid solution; Sigma–Aldrich) in 100 μL of 40% ethanol on day 0 (ref.54). The mice were assessed daily for weight loss, bloody stool and general well being, and they were euthanized on day 4.
Co–transfer of Gpr15 KO and WT naive CD4+ T cells into Rag2−/− mice
Similar to the cell transfer model of colitis, splenocytes from Gpr15gfp/gfp (Gpr15–KO; CD45.2) or Gpr15gfp/+ (Gpr15–het; CD45.1, CD45.2) mice were sorted by flow cytometry to enrich naive CD4+ T cells of >90% purity. The cells were combined at a 1:1 ratio, and a total of 1 × 106 cells were i.v. injected into each Rag2−/− recipient. After 2 weeks, lymphocytes from MLN, spleen and colon were isolated, and cells activated for 4 h prior to staining with surface markers and intracellular cytokines. Where indicated, GFP+ CD4+ T cells were sorted from pooled MLN or colon, activated and stained to assess GPR15 expression among effector/memory CD4+ T cells. For data analysis, normalization to spleen is used to control for variability between mice in injected cell numbers. The spleen lacks high endothelial venules, and unlike the other tissues examined, it is empirically less affected by homing or trafficking effects, making it a preferable (and commonly employed) internal reference tissue.
In vitro–polarized T helper cells
Naive CD62–L+ CD4+ T cells were enriched (Miltenyi Biotec) from human buffy coats and seeded into 96–well plates (0.2 × 106 cells/well, pre–coated with 1 μg/mL anti–CD3 antibody; clone HIT3a). To generate TH1 cells, IFN–γ (10 ng/mL) and recombinant IL–12 (5 ng/mL) were added. To polarize TH2 cells, IL–4 (10 ng/mL) and anti–IL–12 antibody (10 μg/mL) were used; for TH17 cells, IL–1β, IL–23, IL–6, TGF–β (10 ng/mL each) and 10 μg/mL of anti–IFN–γ and anti–IL–4 antibodies; for Tregcells, 5 ng/mL TGF–β. Cells were cultured for 5–6 days in the presence of IL–2 (10 ng/mL) and soluble anti–CD28 (1 μg/mL; clone CD28.2). Cells were washed, activated for 4 hours and stained with surface and intracellular markers as described above. Mouse cells from spleen of Gpr15gfp/+ mice were subjected to similar polarizing conditions as described previously55-57. Cytokines and antibodies were purchased from Peprotech Inc. or eBioscience.
Competitive short–term homing of effector CD4+ T cells
Bead–enriched CD4+ T cells (MACS kit; Miltenyi Biotec) from spleen of allotype–marked Gpr15–het and KO mice were cultured under TH17–polarizing conditions using plate bound anti–CD3 mAb (1 μg/mL; clone 145–2C11), soluble anti–CD28 mAb (1 μg/mL; clone 37.51), rmIL–2 (20 ng/mL), rmIL–6 (20 ng/mL), rhTGF–β1 (5 ng/mL) and retinoic acid (100 pM) as previously described11. After 3 days, cells were washed and i.v. injected into normal hosts. After 8–10 h, host lymphocytes were assessed for presence of donor cells.
Antigen–specific T cell accumulation
Splenocytes (25 × 106) from DO11.10, Gpr15–hetor DO11.10, Gpr15–KO were i.v. injected into Balb/c (Thy–1.1) mice. After 24 h, anesthetized recipients were treated intra–rectally with ovalbumin from chicken egg white (500 μg; Sigma–Aldrich) and cholera toxin (10 μg; Sigma–Aldrich) in a 150 μL volume. At day 5–post treatment, recipient tissues were collected and lymphocytes prepared for analysis by flow cytometry.
Realtime–PCR of human colon cells
Quantitative PCR was performed on sorted human colon lymphocytes from 1 UC and 2 non–UC samples to verify GPR15 and FOXP3 transcript levels in GPR15+ (Abcam) cells. GPR15+ subset (CD4+, CD45RO+, GPR15hi) contained <10% CD127lo, CD25hi cells CD4+ T cells. Total RNA was prepared using RNeasy Mini kit (Qiagen), and cDNA using GoScript reverse transcription system (Promega). Quantitative PCR was performed with SYBR Green PCR Master Mix and ABI–StepOnePlus Real–Time PCR system (Applied Biosystems) using commercially available primer sets (GPR15, VHPS–3771; FOXP3, VHPS–3398; IL2RA, VHPS–4553; GAPDH, VHPS–3541; RealTimePrimers.com).
Realtime–PCR of ChIP DNA
The enrichment of DNA by GATA–3 ChIP was quantified relative to input DNA using primers listed in Supplementary Table 1.
Blockade of lymphocyte egress with FTY720
To determine whether GPR15 is induced in the MLN, CFSE–stained (1 μM; Life Technologies) lymphocytes from Gpr15–het DO11 (Thy–1.1, Thy–1.2) and KO DO11 (Thy–1.2) spleens were i.v. injected in equal proportion (15 × 106 total cells) into Thy–1.1 Balb/c mice. After 24 h, mice were treated intrarectally with OVA (500 μg) and CT (10 μg) mixture. At indicated time, mice were i.p. injected with FTY720 or vehicle (PBS) and their tissues collected. Stock solutions of FTY720 (Sigma–Aldrich; 10 mg/mL ethanol) were diluted with PBS to 100 μg/mL, and i.p. injected at 1 mg/kg.
Comparative sequence analyses
We analyzed a highly conserved non–coding region downstream of mouse, chimpanzee, monkey, rat and human GPR15. The conserved region is located ~23kb and 15kb 3′ of GPR15 in human and mouse, respectively. The human sequence is within a region (chr3:98,272,711–98,276,110 in Hg19) highlighted in the ENCODE/BROAD histone modification track (UCSC genome browser) as an active promoter region in the GPR15–expressing B lymphoblastoid cell line GM12878. DNAase hypersensitivity marks from human TH2 cells, as well as histone modification comparisons with GM12878 and non GPR15–expressing H1–hESC embryonic stem cells, HepG2 hepatocyte line, HUVEC umbilical vein endothelial cells, HMEC microvascular endothelial cells, HSMM smooth muscle myoblasts, NHEK epidermal keratinocytes and NHLF lung fibroblasts were from the UCSC genome browser and the ENCODE project consortium. To verify the A–to–G mutation in the mouse GATA–3 consensus sequence adjacent to the conserved STAT6 binding motif, we surveyed mouse SNP databases (http://www.sanger.ac.uk/sanger/Mouse_SnpViewer/rel-1303) and found no variation in this nucleotide, indicating that it is a species– (not strain–) specific variant.
Foxp3 ChIP–Seq in mouse iTreg cells
CD4+ T cells from spleens and lymph nodes of 4– to 10–wk–old mice were purified by CD4 positive selection (Miltenyi Biotec) followed by sorting of naive CD4+, CD25−, CD62Lhi, CD44lo cells using a FACSAria II (BD Biosciences). Cells were activated by plate–bound anti–CD3 and anti–CD28 (both 10 μg/ml; clones 145–2C11 and 37.51, respectively; Bio X Cell). iTreg cells were generated by culturing in recombinant human TGF–β1 (33 ng/ml) and IL–2 (20 ng/ml; R&D Systems) for 7 days. Following activation, cross–linking, and sonication30, Foxp3–bound genomic DNA was isolated from whole cell lysate using a mix two antibodies (5 μg each); Santa Cruz S–31738 and eBioscience FJK–16. Libraries were constructed and sequenced as described30.
Statistical analysis
Prism software (GraphPad Software) was used for statistical analyses and statistical tests employed are indicated in the figure legends, with p values of <0.05 considered as significant.
Supplementary Material
1
2
3
Acknowledgments
This work was supported by National Institute of Health (NIH) grant R37 AI047822 to ECB, NIH grants K08 DK069385 and R03 DK085426 to AHa, the FACS Core facility of the Stanford Digestive Disease center (NIH Digestive Disease Center grants DK56339) and the VA Palo Alto Health Care System. Research was also supported by the Wellcome Trust (RGJ & GML, grant number 091009), the Medical Research Council (GML grant number G0802068), the National Institute for Health Research (NIHR) University College London Hospitals Biomedical Research Centre and the NIHR Biomedical Research Centre based at Guy’s and St Thomas’ National Health Service (NHS) Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NIH, NHS, the NIHR, or the Department of Health. LPN, HH, and TTD were recipients of fellowships under NIH Training Grant T32 AI07290. LPN was also a recipient of NIH postdoctoral fellowship Grant 1F32 AI082924. TTD is supported by a Cardiovascular Institute Fellowship under NIH T32 HL098049. HH is funded by an NIH grant AI109452A and was a recipient of an Investigator Career Award from the Arthritis Foundation and was supported in part by a CIRM grant. We thank J. Xue (Stanford University) for assistance with the RT–PCR experiments, J. Chen (Stanford University) for technical assistance with the animal work, and the Stanford Tissue Bank for providing the human colon specimens. We thank D. Littman (NYU) for recovering his cryopreserved Gpr15gfp mouse embryos for us in response to our request for collaboration to test our hypothesis that GPR15 mediates colon lymphocyte homing.
Footnotes
Accession codes. GEO Accession codes for published ChIP–Seq data are provided in online Supplementary Table 2.
Competing interests
The authors declare no competing interests.
Literature Cited
Full text links
Read article at publisher's site: https://doi.org/10.1038/ni.3079
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4338558?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Triggers for eosinophilic esophagitis (EoE): The intersection of food allergy and EoE.
J Allergy Clin Immunol, 153(6):1500-1509, 01 Jun 2024
Cited by: 0 articles | PMID: 38849185
Review
Molecular recognition of the atypical chemokine-like peptide GPR15L by its cognate receptor GPR15.
Cell Discov, 10(1):69, 25 Jun 2024
Cited by: 0 articles | PMID: 38918398 | PMCID: PMC11199581
Developmental self-reactivity determines pathogenic Tc17 differentiation potential of naive CD8+ T cells in murine models of inflammation.
Nat Commun, 15(1):2919, 04 Apr 2024
Cited by: 0 articles | PMID: 38575593 | PMCID: PMC10994929
GPR15-mediated T cell recruitment during acute viral myocarditis facilitated virus elimination and improved outcome.
Nat Cardiovasc Res, 3(1):76-93, 27 Dec 2023
Cited by: 0 articles | PMID: 39195892 | PMCID: PMC11357984
The selection landscape and genetic legacy of ancient Eurasians.
Nature, 625(7994):312-320, 10 Jan 2024
Cited by: 17 articles | PMID: 38200293 | PMCID: PMC10781624
Go to all (99) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
IGSR: The International Genome Sample Resource
- (2 citations) IGSR/1000 Genomes - GM12878
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
GPR15-mediated homing controls immune homeostasis in the large intestine mucosa.
Science, 340(6139):1456-1459, 09 May 2013
Cited by: 177 articles | PMID: 23661644 | PMCID: PMC3762262
GPR15: a tale of two species.
Nat Immunol, 16(2):137-139, 01 Feb 2015
Cited by: 7 articles | PMID: 25594457
The aryl hydrocarbon receptor regulates expression of mucosal trafficking receptor GPR15.
Mucosal Immunol, 14(4):852-861, 05 Mar 2021
Cited by: 13 articles | PMID: 33674764 | PMCID: PMC7934811
Emerging roles of a chemoattractant receptor GPR15 and ligands in pathophysiology.
Front Immunol, 14:1179456, 30 Jun 2023
Cited by: 1 article | PMID: 37457732 | PMCID: PMC10348422
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Academy of Medical Sciences (1)
Grant ID: AMS-SGCL7-Gökmen
Biotechnology and Biological Sciences Research Council (1)
Grant ID: BB/L009277/1
Cancer Research UK (1)
Grant ID: G0802068
Medical Research Council (1)
Defining the cellular and molecular pathogenesis of ulcerative colitis
Professor Graham Lord, King's College London
Grant ID: G0802068
NHLBI NIH HHS (1)
Grant ID: T32 HL098049
NIAID NIH HHS (7)
Grant ID: R01 AI109452
Grant ID: 1F32 AI082924
Grant ID: T32 AI07290
Grant ID: F32 AI082924
Grant ID: R37 AI047822
Grant ID: T32 AI007290
Grant ID: AI109452A
NIDDK NIH HHS (4)
Grant ID: K08 DK069385
Grant ID: DK56339
Grant ID: R03 DK085426
Grant ID: P30 DK056339
Wellcome Trust (1)
Transcriptional regulation of human T cell differentiation.
Professor Graham Lord, King's College London
Grant ID: 091009




