Abstract
Free full text

Rescue of an In Vitro Neuron Phenotype Identified in Niemann-Pick Disease, Type C1 Induced Pluripotent Stem Cell-Derived Neurons by Modulating the WNT Pathway and Calcium Signaling
Abstract
Niemann-Pick disease, type C1 (NPC1) is a familial disorder that has devastating consequences on postnatal development with multisystem effects, including neurodegeneration. There is no Food and Drug Administration-approved treatment option for NPC1; however, several potentially therapeutic compounds have been identified in assays using yeast, rodent models, and NPC1 human fibroblasts. Although these discoveries were made in fibroblasts from NPC1 subjects and were in some instances validated in animal models of the disease, testing these drugs on a cell type more relevant for NPC1 neurological disease would greatly facilitate both study of the disease and identification of more relevant therapeutic compounds. Toward this goal, we have generated an induced pluripotent stem cell line from a subject homozygous for the most frequent NPC1 mutation (p.I1061T) and subsequently created a stable line of neural stem cells (NSCs). These NSCs were then used to create neurons as an appropriate disease model. NPC1 neurons display a premature cell death phenotype, and gene expression analysis of these cells suggests dysfunction of important signaling pathways, including calcium and WNT. The clear readout from these cells makes them ideal candidates for high-throughput screening and will be a valuable tool to better understand the development of NPC1 in neural cells, as well as to develop better therapeutic options for NPC1.
Introduction
Niemann-Pick disease, type C1 (NPC1) is a lysosomal storage disorder that results in a neurodegenerative phenotype. Approximately half of the cases of NPC1 present in children under 10 years of age, and age of onset seems directly related to life span. The majority of cases (95%) are caused by a mutation in the NPC1 gene, whereas the remaining cases are caused by a mutation in the NPC2 gene [1]. Studies have shown that these two proteins interact closely and are involved with cholesterol binding and transfer [2, 3]. Disruptions in the NPC1 or NPC2 protein results in the accumulation of cholesterol and glycolipids within cells, and it is not clear which compound’s accumulation within neural cells is responsible for neurodegeneration [4]. However, this accumulation is correlated to progressive neurological disease with varying severity in each case. Additionally, a progressive loss of cerebellar Purkinje neurons occurs in mouse models of NPC1, indicating that the damage and loss of this cell type is likely responsible for the neurological symptoms seen in those affected by NPC1 [5, 6].
There is currently no Food and Drug Administration-approved drug for treating NPC1. However, work with animal models and screens using subject fibroblasts have identified several compounds that reduce abnormal lysosomal accumulations. These include δ-tocophorol (vitamin E) and histone deacetylase inhibitors and glycosphingolipids synthesis inhibitors such as migulstat [7, 8]. Additionally, cyclodextrin was found to increase life span in Npc1 and Npc2 mutant mice [9, 10]. Additional studies have identified curcumin (a calcium/calmodulin kinase II inhibitor) and TRPML1 channel agonists as compounds that could have therapeutic potential for NPC1 [11, 12]; however, subsequent studies determined that curcumin may not have as robust an effect on neurodegeneration in a mouse model of NPC1 [13]. The mechanisms underlying the effects of many of these compounds appear to involve lysosomal calcium signaling [12, 14, 15].
We generated an induced pluripotent stem cell (iPSC) line from an NPC1 subject fibroblast sample homozygous for the most frequent NPC1 mutation (p.I1061T) and subsequently differentiated the iPSCs to neural stem cells (NSCs), neurons, and astrocytes to more directly assess the effects of loss of NPC1 function on these cell types. We then compared their gene expression profiles to healthy and disease control lines to identify specific differences attributable to NPC1 genetic abnormalities. Few differences were seen at the NSC stage in cell growth, marker expression, and by whole transcriptome array analysis. However, Niemann-Pick disease, type C1 iPSC-derived neurons exhibited increased cell death and changes in calcium signaling, suggesting that alterations in calcium signaling may underlie the phenotype. Further examination of the gene expression profiles of iPSC-derived neurons identified WNT signaling and ryanodine receptor expression as significantly altered. Our data suggest that altered WNT or calcium signaling is an important event in the etiology of the disease and implicate the modulation of these signaling pathways as possible therapeutic targets at early stages of NPC1.
Materials and Methods
iPSC Generation
iPSCs were generated from fibroblasts obtained from a subject with Niemann-Pick disease, type C1 with a mutation status as I1061T/I1061T under an National Institute of Child Health and Human Development Institutional Review Board-approved protocol and with informed consent. These fibroblasts were reprogrammed using the STEMCCA lentivirus system to create iPSCs on a mouse fibroblast feeder culture, in which they were maintained in stem cell medium consisting of knockout Dulbecco’s modified Eagle’s medium (DMEM)/F-12, 20% knockout serum replacement, GlutaMAX (2 mM), 1× nonessential amino acids (NEAA), β-mercaptoethanol, and 10 ng/ml basic fibroblast growth factor (bFGF) (all from Life Technologies, Carlsbad, CA, http://www.lifetech.com). Medium was changed daily. iPSCs were then adapted to feeder-free culture on Matrigel (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com) and maintained in E8 medium (Life Technologies). NPC1 iPSCs expressed the pluripotency markers OCT4, NANOG, and TRA-1-60 through immunocytochemistry (described below).
Neural Stem Cell Culture
NSC lines were generated from the episomally derived NCRM-1 iPSC line, patient-derived NPC1 iPSC line, and patient-derived amyotropic lateral sclerosis (ALS) iPSC line via an embryoid body intermediate method by XCell Sciences [16] (Novato, CA, http://www.xcell2.com). Karyotype analysis at passage 2 revealed a normal karyotype and no clonal abnormalities (supplemental online Fig. 1B). NSCs were maintained on culture dishes coated with Geltrex (Life Technologies) in NSC medium. The NSC medium is comprised of neurobasal medium with GlutaMAX (2mM) and 1× NEAA, 1× B27 supplement, and 10 ng/ml of bFGF (all from Life Technologies). Cells were passaged at 80% confluence in a 1:4 ratio, and cells in experiments were used between passages 4 and 12.
Neural Differentiation and Culture
For neuronal differentiation, NSCs between passages 4 and 12 were first cultured on a Geltrex-coated plate and then passaged to 1 million cells per well of a 6-well plate. Culture medium was changed 1 day later to neuronal differentiation medium (NDM), containing DMEM/F-12 supplemented with GlutaMAX, 1.8% bovine serum albumin, 1× StemPro hESC Supplement (all from Life Technologies), 10 ng/ml brain-derived neurotrophic factor, and glial-derived neurotophic factor [17] (R&D Systems, Minneapolis, MN, http://www.rndsystems.com). After 4 days in NDM, cells were passaged to a 6-well or 96-well plate coated with 0.002% poly-l-ornithine (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) and 15 μg/ml laminin (Life Technologies) or Geltrex. NDM was refreshed by removing 30% of the medium and adding 50% fresh NDM.
Astrocytes were generated in a similar fashion in astrocyte differentiation medium containing DMEM/F-12 supplemented with GlutaMAX, 1× B27 supplement, 8 ng/ml bFGF, and 10 ng/ml heregulin [18, 19]. Astrocytes were passaged every 4–5 days as they became confluent, and immunocytochemistry was performed at day 55 of astrocyte differentiation.
Immunocytochemistry
Cells were washed once with warmed base medium (DMEM/F-12 or neurobasal medium) and fixed with 4% paraformaldehyde in DMEM/F-12 for 20 minutes at room temperature or at 4°C overnight. Fixed cells were then washed three times with phosphate-buffered saline (PBS) and blocked using a buffer containing 10% normal goat serum (NGS) for 1 hour at room temperature. Primary antibody was diluted in blocking buffer containing 0.1% Triton X-100 in PBS and 1% NGS and incubation was performed at 4°C overnight. Cells were then washed again with PBS three times, and secondary antibody was diluted in blocking buffer. Secondary antibody incubation was performed for 1 hour at room temperature. Cells were then washed twice with PBS and used for imaging. Appropriate secondary antibodies were used for single and double labeling, and all secondary antibodies were tested for specificity. The primary antibodies used here are as follows: anti-GFAP antibody at 1:2,000 dilution (Dako, Glostrup, Denmark, http://www.dako.com), anti-βIII tubulin at 1:1,000 dilution (Millipore, Billerica, MA, http://www.millipore.com), and anti-MAP2 at 1:1,000 dilution (Millipore). Secondary antibodies used are Alexa Fluor 555 goat anti-mouse and Alexa Fluor 488 goat anti-rabbit (Life Technologies). Hoechst 33342 (H3570; Molecular Probes, Eugene, OR, http://probes.invitrogen.com) at 1:2,000 dilution was used for nuclei staining. Images were captured using a Leica fluorescence microscope.
Filipin Staining and Confocal Spinning Disk Images
For filipin/TUJ1 (β-III tubulin)/TOPRO-3I stain, medium on cells plated to Geltrex-coated multiwall slides was removed. Cells stained for TUJ1 were processed as described above and processed as described here after secondary antibody. Otherwise, cells were fixed in 4% formaldehyde for 20 minutes, incubated with 1 μM TOPRO-3I (Life Technologies) for 40 minutes, and then incubated in 50 μg/ml filipin (Sigma-Aldrich). The filipin step and all following steps were protected from light. The cells were coverslipped with Prolong Gold antifade (Life Technologies). All steps were at room temperature; all solutions diluted into PBS, with one PBS wash step between each solution change. Spinning-disk microscope illumination greatly reduced filipin photobleaching. Wide-field images were collected using a Zeiss Yokogawa spinning disk system (Carl Zeiss Inc., Thornwood, NY, http://www.zeiss.com) mounted on an inverted Zeiss Axio Observer Z1 microscope with an oil immersion Plan-Apochromat ×63/1.4 differential interference contrast lens. All images were acquired using a 16-bit Photometrics Evolve EMCCD camera (Photometrics, Tucson, AZ, http://www.photometrics.com/) with exposure times ranging from 150 to 1,000 milliseconds. Excitation wavelengths of 405 nm (65%), 561 nm (9%), and 639 nm (1.5%) were used for detection of the filipin-stained free cholesterol, TUJ1, and TOPRO-3I-stained nuclei, respectively. Fluorescent emissions were collected in a band pass (BP) 450/50-nm filter, a BP 629/62-nm filter, and a BP 690/50-nm filter, respectively. Zeiss ZEN blue 2012 software package was used for collection and postprocessing of the images.
Gene Expression Analysis
Gene expression analysis was performed on iPSC-derived NSC samples and day 14 neuron samples derived from those NSCs. An additional NSC sample from the healthy control line NCRM-5 is also included. The passage numbers of NSCs used in gene expression analysis are as follows: NPC1, passage 3; NCRM-1, passage 5; ALS, passage 5; and NCRM-5, passage 6. Samples were collected on different days and processed together. Total RNA was isolated and prepared using the RNeasy mini kit according to the manufacturer’s instructions (Qiagen, Valencia, CA, http://www.qiagen.com). RNA samples were hybridized to Illumina Human HT-12 BeadChip by Qiagen. Data processing was performed using the algorithms included with the gene expression module of the Illumina BeadStudio software with the background method used for normalization. The maximum expression value of gene for the probe set was used as the expression value of the gene. The data were exported to Microsoft Excel for generation of gene lists for assessing presence or absence of cell specific markers. Microarray data were deposited in Gene Expression Omnibus (GSE55379).
Drug Treatment and MTT Assay
Cells undergoing neuronal differentiation at day 4 were passaged on to 96-well plates coated in Geltrex to a density of 1 million cells per plate. On day 6, cells were treated with 10 μM of curcumin, dantrolene, BIO, TNP-470, or CCT036477 (all from Tocris, Bristol, U.K., http://www.tocris.com). Drugs were diluted to a concentration of 10 μM in dimethyl sulfoxide (DMSO). At day 14, an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed to assess cell viability with absorbance measured at 590 nm. All treatments were performed in triplicate, and the results are reported as relative absorbance in medium with DMSO vehicle only.
Results
Disease-Line iPSCs Can Be Generated From Niemann-Pick Disease, Type C1 Fibroblasts
Fibroblast samples from an individual diagnosed with Niemann-Pick disease, type C1 homozygous for the p.I1061T mutation were isolated, cultured, and reprogrammed to iPSCs using the STEMCCA lentivirus system. Immunocytochemistry was performed on NPC1 iPSCs to identify markers routinely used to determine pluripotency and establish iPSC identity [20]. NPC1 iPSCs expressed the pluripotency markers OCT4 and NANOG, as well as the stem cell-specific surface marker TRA-1-60 (Fig. 1A). The NPC1 iPSC line showed no overt phenotypic defects and could be maintained in both feeder and feeder-free cultures. These iPSCs were then differentiated for use in subsequent experiments (supplemental online Fig. 1A).
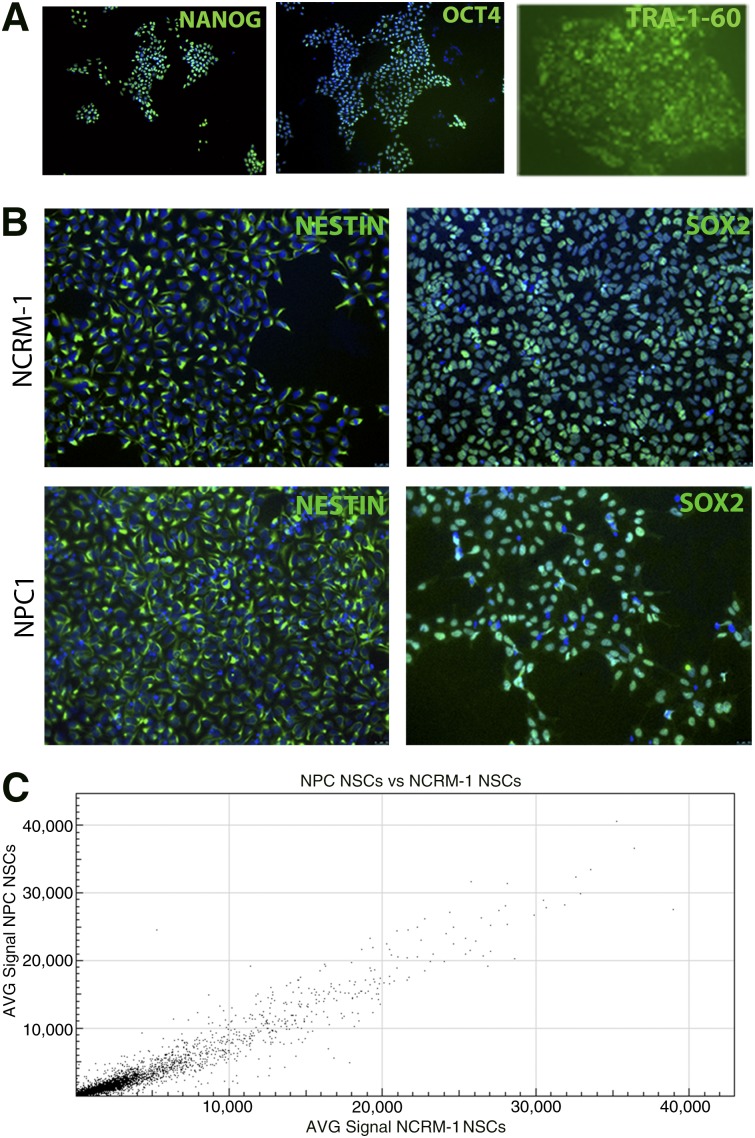
NPC1 induced pluripotent stem cell (iPSC)-derived NSCs do not display major disease-specific differences from control line NSCs. (A): NPC1 subject iPSCs express the stem cell-specific markers OCT4, TRA-1-60, and NANOG, shown in green (images at ×10). (B): NPC1 and NCRM-1 NSCs express NSC markers NESTIN and SOX2, shown in green (images at ×20). (C): Gene signal intensity for NPC1 and NCRM-1 NSCs displays an R2 value of 0.96, suggesting high overlap in gene expression between these two cell types. Abbreviations: AVG, average; NPC1, Niemann-Pick disease, type C1; NSC, neural stem cell.
NPC1 NSCs Display Minimal Disease-Specific Morphological and Functional Differences
iPSCs were then directed to form multipotent NSCs by a rosette-based method [16]. NSCs are a stable intermediate cell type that express some features of neural cells and have the capacity to differentiate to both neurons and glia [16]. All NPC1 NSC-expressed NSC markers including NESTIN and SOX2 (Fig. 1B) were karyotypically normal (supplemental online Fig. 1B) and did not display any observable morphological differences compared with a healthy control NSC line NCRM-1. The similarities between the disease NPC1 NSC line and the NCRM-1 NSC line were further confirmed by gene expression analysis of NSCs using microarray, which indicated no major differences between these lines. A scatterplot generated from these data indicated an R2 value of 0.96, indicating high levels of similarity in the gene expression patterns of the two cell types. (Fig. 1C). Because these cell types showed many similarities, we sought to explore whether there were any overt differences between these two cell types after neural differentiation.
NPC1 Neurons Display an Early Death Phenotype That Is Not Observed in Astrocytes
iPSC-derived NSCs from the NPC1 line were directed to differentiate to neurons using a previously published protocol (supplemental online Fig. 1A) [17]. However, NPC1 neurons began to show morphological differences as early as day 6 and an obvious difference in cell survival at day 10 (Fig. 2A), which could be mitigated by frequent medium changes but ultimately culminated in early neuronal cell death compared with the NCRM-1 control cell line. Daily medium changes and culture at high density allowed for cell survival to neuronal differentiation day 14, when cells were stained for neuronal and glial markers and collected for whole genome analysis by microarray. The majority of NPC1 cells at this stage expressed the neuronal markers TUJ1 and MAP2, indicating successful neuronal differentiation. A small number of cells also expressed the mature astrocyte marker GFAP, indicating that some cells underwent spontaneous astrocyte differentiation despite being in neuronal differentiation conditions (Fig. 2B).
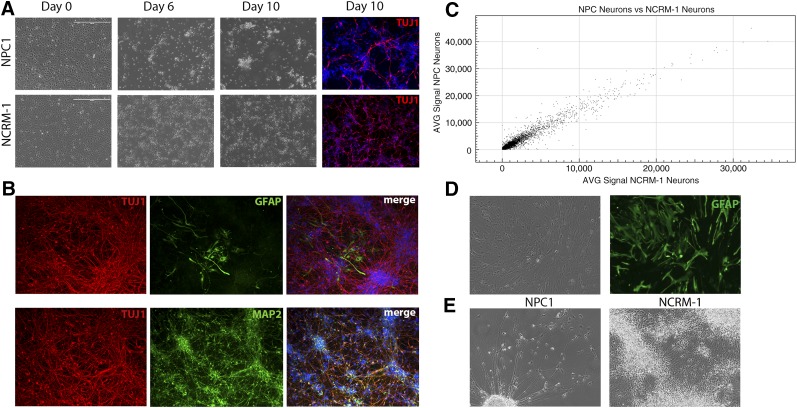
NPC1 induced pluripotent stem cell-derived neurons display an early death phenotype. (A): NPC1 and NCRM-1 neural stem cells (NSCs) display morphological changes during neuronal differentiation and can be differentiated to neurons by day 10 as indicated by TUJ1 expression (red). Scale bar = 400 μm. (B): NPC1 neurons at day 14 of neuronal differentiation display neuronal markers TUJ1 and MAP2. NPC1 cells also reveal some spontaneous differentiation to astrocytes, as indicated by expression of GFAP (images at ×20). (C): Comparison of gene signal intensity for NPC1 and NCRM-1 neurons at day 14 reveals an R2 value of 0.94, suggesting high overlap in gene expression but more variability than what is seen in NSCs. (D): NPC1 NSCs can be differentiated to GFAP (green) expressing astrocytes and maintained in culture for up to 55 days (images at ×20). (E): Representative phase image illustrating the morphological differences and high numbers of cell death seen in NPC1 neurons at 14 days with medium changes every 2 days compared with NCRM-1 neurons under the same conditions (images at ×20). Abbreviations: AVG, average; NPC1, Niemann-Pick disease, type C1.
Gene expression analysis at this stage confirmed the purity of neuronal cultures at day 14 of differentiation (supplemental online Tables 1–12). Neurons expressed both early and mature neuronal markers, including synaptic vesicle genes, channels and transporters, and intermediate and late neurofilaments. We also detected neurotransmitter synthesizing enzymes and markers for both excitatory and inhibitory neurons. The cells also expressed few to no glial markers, including astrocyte markers and oligodendrocyte markers, and low levels of NSC markers (supplemental online Tables 1–12).
To determine whether early cell death was NPC1-specific and not a result of culture conditions, we also differentiated two other iPSC-derived NSC lines to neurons using the same protocol. These included the normal control line (NCRM-1) and a disease line from a subject with ALS. Neither of these lines displayed the same early death phenotype, suggesting that it was specific to the NPC1 line. Comparison of gene expression profiles of NPC1 neurons with NCRM-1 neurons revealed R2 values of 0.94, indicating that there may be more variability in gene expression at this stage as compared with the NSCs (Fig. 2C).
We then sought to determine whether NPC1 cell death was limited to neurons or whether other differentiated cell types were also affected. We have previously shown that iPSC-derived NSCs can be directed to form astrocytes through the addition of heregulin to the cell culture medium [18, 19]. Cell death did not occur in NPC1 NSCs directed to differentiate to astrocytes under this protocol, and we were able to continue culturing these cells for up to 55 days (Fig. 2D). We therefore decided to focus our attention on neurons, because they exhibited an obvious phenotype.
NPC1 NSCs and Neurons Do Not Display Evident Differences in Cholesterol Accumulation
Because fibroblasts from NPC1 lines are commonly used source for diagnosing NPC1, as well as for screening potential drugs [21], we sought to perform similar tests on our disease-line cells. We performed a filipin staining assay to determine whether NPC1 NSCs showed a significant increase in free unesterified cholesterol accumulation. NPC1 NSCs did not show a discernible difference in the accumulation of filipin staining as compared with NCRM-1 NSCs (Fig. 3A).
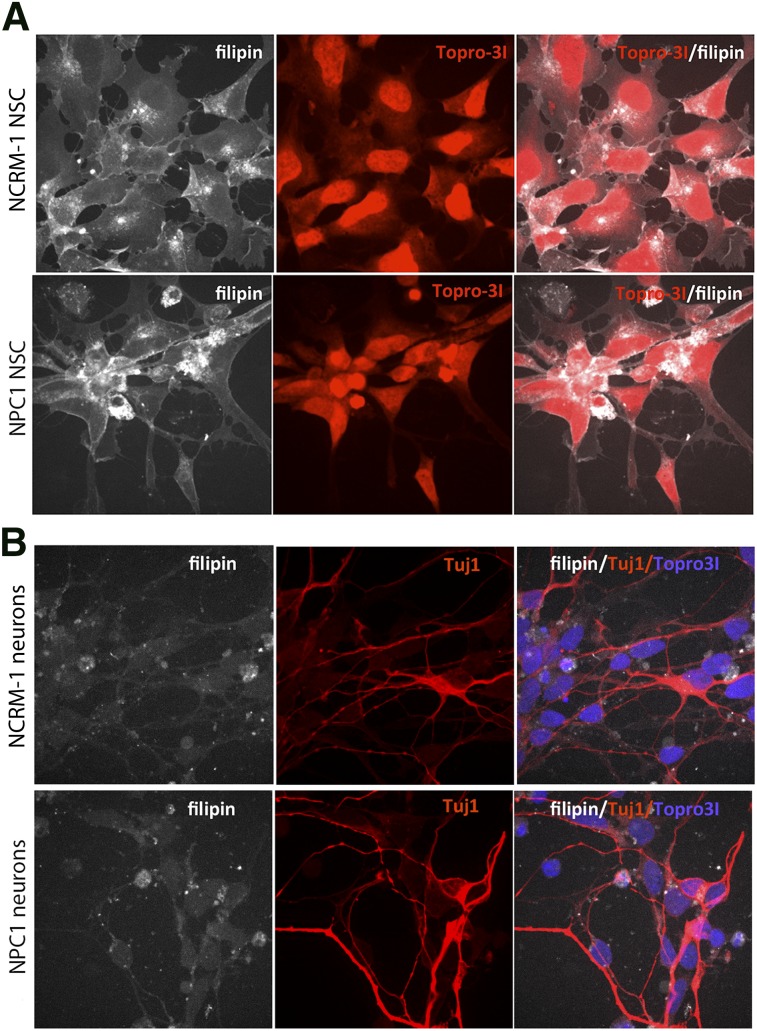
Filipin staining on induced pluripotent stem cell-derived NSCs and neurons does not reveal any evident disease-specific differences. (A): Representative images of filipin (white) and Topro31 nuclear stain (red) on NPC1 and NCRM-1 NSCs reveal similar patterns of filipin accumulation. (B): Filipin (white), TUJ1 (red), and Topro31 (blue) staining on NPC1 and NCRM-1 NSC-derived neurons does not reveal any evident differences in filipin accumulation between healthy and disease neurons. All images are at ×63. Abbreviations: NPC1, Niemann-Pick disease, type C1; NSC, neural stem cell.
It has been previously established that filipin staining can also be performed on iPSC-derived neurons to investigate cholesterol accumulation in these cells [22]. We also performed filipin staining on both NPC1 and NCRM-1 neurons to determine whether there were any overt differences in filipin accumulation between these two cell types but again did not notice a discernible difference (Fig. 3B). Therefore, we chose to determine whether the neuronal death phenotype was a good functional readout for use in a screening assay.
NPC1 Neurons Display Aberrant Calcium and WNT Signaling, Along With an Upregulation of Glial Markers
We hypothesized that major signaling pathways were likely disrupted in NPC1 neurons and that this dysregulation was contributing to early neuronal death. Therefore, we analyzed our microarray data to determine differences in global expression patterns between the NPC1 neurons, NCRM-1 neurons, and ALS neurons. The data revealed three major differences that were unique to our NPC1 sample as compared with the NCRM-1 and ALS samples: (a) NPC1 neurons showing increased gene expression of genes involved in calcium signaling, (b) disruption of genes involved in WNT signaling, and (c) an upregulation of glial-associated gene markers (Tables 1, ,2).2). This increase in glial marker expression could be due to NPC1 neuronal death, after which a high number of glial cells remained, or it is possible that these cells are predisposed to a glial fate. The latter may be the case, because the levels of mature astrocyte markers such as GFAP and NFIX were much higher than those seen in NCRM-1 and ALS neuronal samples, and the presence of glial markers in day 14 neurons was confirmed using immunocytochemistry (Fig. 2B). However, changes in glial marker expression do not reflect larger pathway dysfunction that is easily targetable or adaptable to our screening platform. The data collected by our microarray analysis confirm previously published literature indicating that the calcium signaling pathway is disrupted in mouse and human fibroblast models of NPC1 [12, 14, 23–26]. Conscious of the fact that we only have one cell line, we therefore chose to concentrate on the disruption of calcium and WNT signaling.
Table 1.
Microarray analysis indicating high gene expression
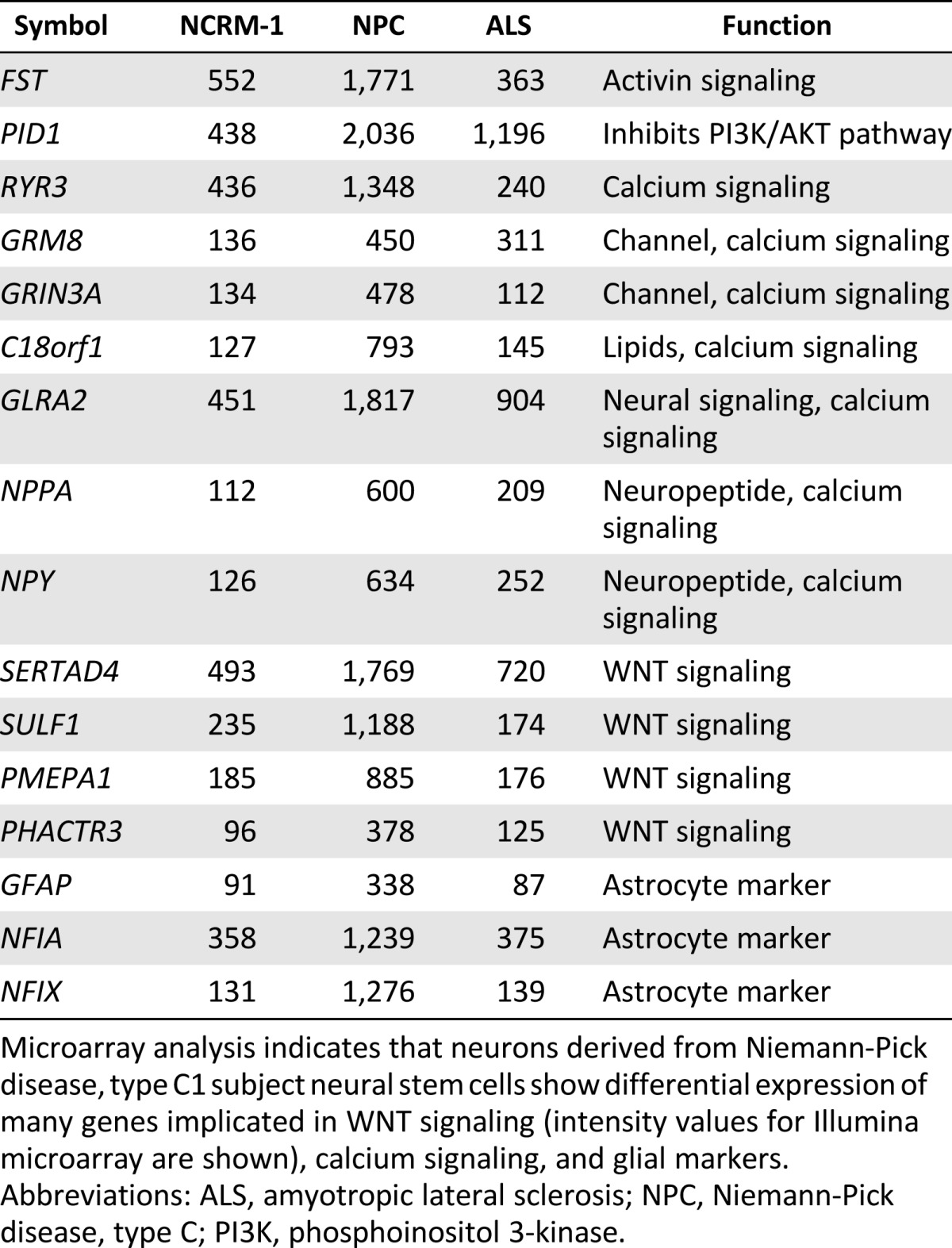
Microarray analysis indicates that neurons derived from Niemann-Pick disease, type C1 subject neural stem cells show differential expression of many genes implicated in WNT signaling (intensity values for Illumina microarray are shown), calcium signaling, and glial markers.
Abbreviations: ALS, amyotropic lateral sclerosis; NPC, Niemann-Pick disease, type C; PI3K, phosphoinositol 3-kinase.
Table 2.
Microarray analysis indicating low gene expression
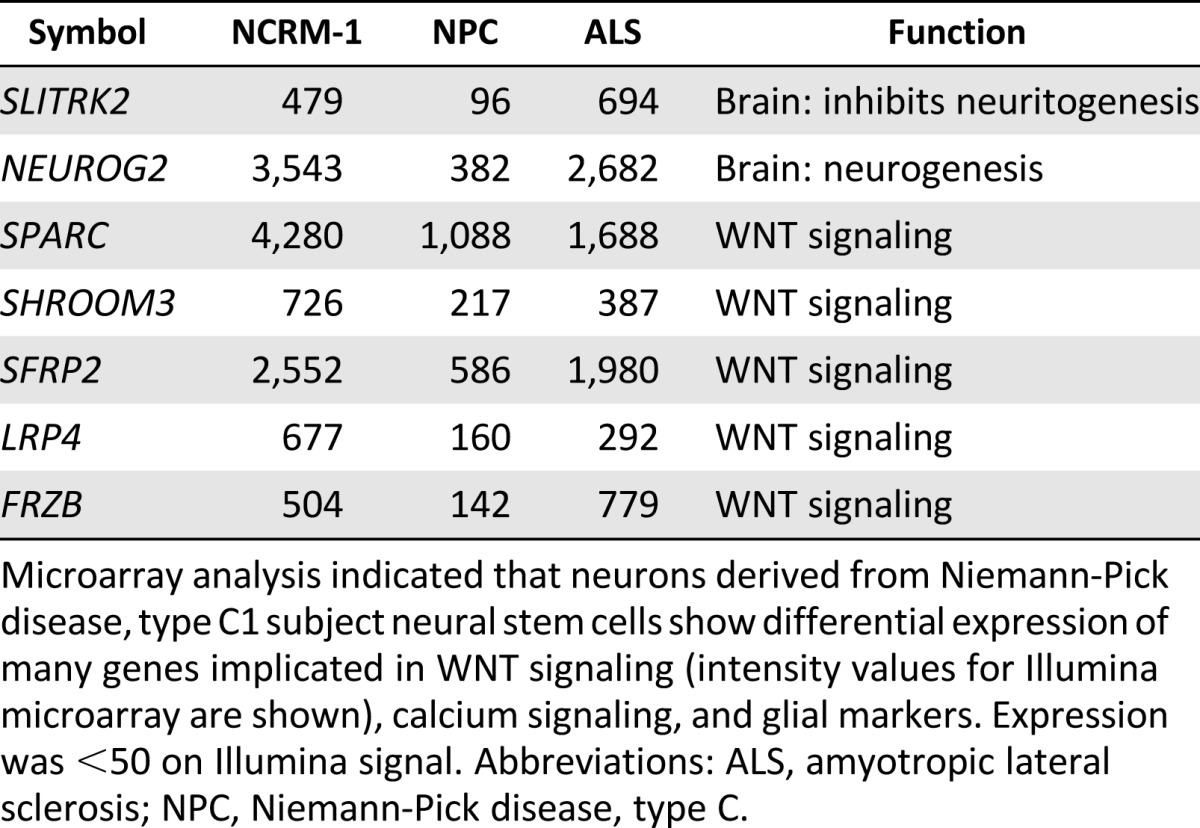
Microarray analysis indicated that neurons derived from Niemann-Pick disease, type C1 subject neural stem cells show differential expression of many genes implicated in WNT signaling (intensity values for Illumina microarray are shown), calcium signaling, and glial markers. Expression was <50 on Illumina signal. Abbreviations: ALS, amyotropic lateral sclerosis; NPC, Niemann-Pick disease, type C.
NPC1 Neurons Can Be Rescued by Modulation of Calcium Signaling or the WNT Pathway
NPC1 neuronal cell death in our system provided an obvious functional readout based on intrinsic cellular traits, thereby allowing us to monitor the health and viability of our disease-line cells. We adapted these cells to our previously described high-throughput screening system, thus allowing us to perform viability screens on the NPC1 neurons [17]. Because the microarray analysis indicated disturbances in calcium and WNT signaling, we tested the calcium inhibitors curcumin and dantrolene, which have been tested for treatment in human fibroblast and animal models of NPC1, to determine whether they could rescue the NPC1 neurons from premature cell death.
NPC1 and NCRM-1 cells at day 4 of neuronal differentiation were passaged to 96-well plates for cell viability and drug screening. Curcumin and dantrolene were both tested on all cells in triplicate at a concentration of 10 μM at day 6 of neuronal differentiation, preceding any observable cell death. Cells were treated with drug only at this time point, but fresh medium was replenished every 2 days for seven additional days, after which cell viability was determined using an MTT assay. Addition of the calcium modulators curcumin or dantrolene was sufficient to promote NPC1 neuronal survival (Fig. 4).
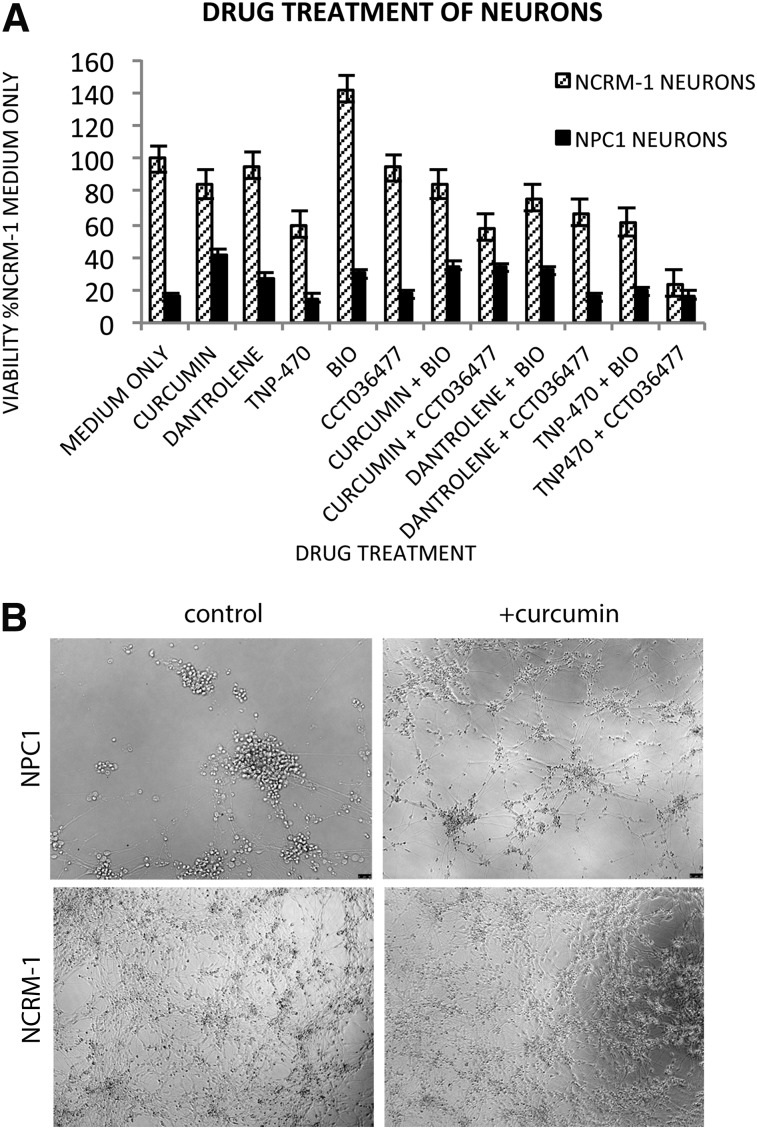
Drug treatment on NPC1 and NCRM-1 neurons supports roles of calcium and WNT signaling in NPC1 neuronal cell death. (A): NPC1 neuronal cell death can be rescued by the addition of curcumin, dantrolene, and WNT agonists. Percentages were calculated in relation to treatment with medium without drug. (B): Representative images of NPC1 and NCRM-1 neuronal survival with or without curcumin at day 14 reveal NPC1 neuronal death can be rescued by single early dose of drug. All images are at ×10. Abbreviation: NPC1, Niemann-Pick disease, type C1.
The rescue of NPC1 neuronal cell death using these calcium inhibitors validated the changes seen in our microarray analysis of the calcium signaling pathway. We then repeated these experiments using WNT signaling modulators. The WNT signaling modulator BIO also increased neuronal viability (Fig. 4). However, not all WNT signaling modulators rescued neuronal viability, because TNP-470 and CCT036477 were unable to partially rescue NPC1 neurons. It should be noted that neuronal viability was partial with addition of drugs and did not recover to levels seen in the NCRM-1 line. These results suggest that the iPSC-derived NPC1 neurons may be adapted to larger scale assays for the identification of novel therapeutic compounds for the treatment of this disease.
Discussion
The use of pluripotent stem cells has major potential for the modeling of neurodegenerative disease and for developing appropriate and beneficial screening assays. In this study, we describe a method by which fibroblasts from a subject with Niemann-Pick disease, type C1 can be reprogrammed to iPSCs and differentiated to develop a disease model of NPC1 within an appropriate cellular phenotype. Although other disease models do exist, including in vivo models in mouse and cat, these animal models are not amenable to high-throughput screens that may be used to identify therapeutic compounds. Furthermore, the only widely available human samples for NPC1 are fibroblasts, which are not physiologically relevant to disease pathology.
As stated, the majority of in vitro experiments on NPC1 have been performed on fibroblasts. Indeed, fibroblasts offer a cost-effective and less labor-intensive method to screen human cells to identify potentially therapeutic compounds. However, fibroblasts are functionally distinct/dissimilar from neural cells, and not all therapeutic candidates identified through screens of fibroblasts have proven to have utility in treating NPC1. It has previously been shown that the tests used on NPC1 fibroblasts, including drug testing and measurement of cholesterol and tracking of lysosomes, can be conducted on neurons [21]. We have taken this one step further in demonstrating that such experiments can be conducted on disease-line neurons derived from a subject with NPC1 in a scalable platform and that these disease-line cells can provide insight as to mechanisms of disease pathology and potential therapeutic targets.
NPC1 line iPSCs were generated from a human sample and differentiated to NSCs, a stable intermediate stage with expansion capacity and differentiation capability to neurons and astrocytes. NPC1 line iPSC-derived NSCs displayed no major differences with control line NCRM-1 NSCs, based on morphological and functional characteristics, including NSC-specific markers and the ability to generate both neurons and astrocytes. Filipin staining on NCRM-1 and NPC1 NSCs for cholesterol accumulation also showed no evident differences between neural stem cells from disease and control line.
It became apparent after neuronal differentiation of NPC1 NSCs that these cells were not able to survive under our standard neuronal culture conditions. Although NPC1 neuron cell death was consistent and reproducible, this phenotype was also influenced by external environmental factors including the frequency of medium changes and density of cell cultures. We attempted to control for our use of only one NPC1 iPSC line in this study by using additional controls, including an ALS iPSC line. Although ALS is also a neurodegenerative disorder, it primarily affects other cell types (astrocytes and motor neurons) through a different mechanism of action. We did find several commonalities while analyzing neuronal microarray data from these two samples. However, the main findings summarized in this study are unique to NPC1 cells, and we believe that other NPC1 lines are likely to show similar differences.
Because of the small sample size, we were careful to focus on findings that were similar or related to those already seen in fibroblast and animal models of the disease, including calcium signaling [23]. Despite the lack of previous findings indicating that the WNT pathway may be involved, this pathway is closely regulated with the calcium signaling pathway [27]. We therefore chose to test compounds affecting both pathways in an initial viability screen in an effort to identify compounds that could rescue the death phenotype displayed by NPC1 neurons.
We have previously described a system in which iPSC-derived neural stem cells can be differentiated to neurons in a 96-well format that is suitable for high-throughput screening [17]. We adapted this platform to use the cell death phenotype displayed by the NPC1 neurons as a functional readout for our assay. This assay, when combined with our gene expression data and a NPC1 neuronal death, confirmed the previously reported changes in calcium signaling, as well as revealing a role for WNT signaling in NPC1 disease pathology. Therefore, our assay indicates that iPSC-derived neurons may be a viable model for NPC1, because they can be used to validate several previous discoveries in NPC1 fibroblasts and animal models and show promise as a tool in future screening assays.
NPC1 iPSC-derived neurons show great utility in screens because they are a more physiologically relevant cell type to explore the neurodegenerative effects of the disease. This can be taken a step further when considering neuronal subtypes, because NPC1 is well documented to result in cerebellar Purkinje cell neurodegeneration [28–30]. Although protocols for the generation of Purkinje cells from mouse iPSCs are available, they have not yet been verified in human iPSCs [31]. We were therefore limited to differentiating our cells to cortical neurons but believe that as more neuronal differentiation protocols are developed NPC1 disease line, iPSCs can be differentiated to Pukinje cells to develop a better understanding of one of the major pathologies associated with this disease.
Despite being unable to look at Purkinje cells in vitro, we were able to see larger changes in the gene expression data of NPC1 neurons that hint at disrupted Purkinje cell development, including low levels of NEUROG2. NEUROG2 has previously been shown to be expressed in murine cerebellar Purkinje cell progenitors, and a mouse null for this gene results in mice that have defective Purkinje cell dendrites [32]. It is also well known that WNT signaling is essential to the proper development of the cerebellum and that high levels of WNT have been found in the Purkinje cells of mice [33]. Additionally, because GSK3 has been shown to negatively regulate NEUROG2-mediated neurogenesis, it is possible that Purkinje cell development or maturation may be regulated by the WNT pathway via NEUROG2 [34]. Further studies will be needed to determine whether these correlations are more strongly connected in our NPC1 model.
The changes in calcium signaling that we observed are similar to previously reported data obtained from investigations using NPC1 mutant fibroblasts and mouse models of NPC1 [14]. To confirm these results and to verify the utility of NPC1 neuronal cells, we decided to test two known calcium inhibitors that have been demonstrated to help protect against disease effects in other models of NPC1: curcumin and dantrolene. Curcumin is a calcium inhibitor [35] that shows promise in the treatment of neurological diseases [36]. Dantrolene is a calcium release inhibitor and vitamin D receptor ligand, which may work through the ryanodine receptor class of intracellular calcium channels [37]. One such receptor, RYR3, was expressed more than threefold more in NPC1 neurons as compared with NCRM-1 and ALS neurons in our study. Addition of curcumin and dantrolene at day 6 of neuronal differentiation, thereby preceding the observed cell death, was sufficient to suspend the consistent and specific loss of NPC1 neurons in culture. Importantly, both curcumin and dantrolene have been found to ameliorate the Niemann-Pick phenotype in patient fibroblasts [14, 38].
To confirm that the WNT signaling pathway may play a role in disease etiology as suggested by the microarray data, we also tested the WNT compounds BIO, TNP-470, and CCT036477. The WNT activator BIO was also able to partially rescue neuronal death in NPC1 neurons. However, TNP-470 and CCT036477 had no effect. The known existence of considerable crosstalk between calcium signaling and the WNT pathway and our array data suggest that both pathways are affected in NPC1 neurons and that the two pathways may be coordinately dysregulated in NPC1 neurons.
The compounds that are most effective at rescuing NPC1 neuronal cell death—curcumin, dantrolene, and BIO—have many potential mechanisms of action. Dantrolene broadly affects calcium signaling, which can alter multiple signaling pathways [39]. Various studies have shown that curcumin broadly affects many signal transduction pathways, including calcium signaling and the WNT pathway through GSK3-b [40, 41]. BIO is a GSK3-β inhibitor that activates WNT signaling, but GSK3-β is an enzyme that is a central node in several signaling pathways [42]. Because all three of these compounds share a role in calcium signaling and the WNT pathway, we believe that this is a good starting point for future investigation that matches up well with our microarray data. More effort will be needed to determine the precise mechanism through which these compounds rescue the observed neuronal death phenotype.
Conclusion
We have derived an iPSC line from an NPC1 subject fibroblast line that was then used to generate NSCs. Differentiation of these disease-line NSCs to neurons to create a human neuronal model for NPC1 displayed a cell type-specific premature death phenotype, although neurons could be partially rescued with several drugs that modulate calcium signaling known to be effective in treating NPC1 fibroblasts and also with a compound that affects WNT signaling. Our data suggest that targeting both pathways may be an effective means to treat the neuronal deficits associated with NPC1. As this manuscript was being prepared, several other groups described their generation of iPSC lines from human subjects with NPC1 [22, 43]. These studies also generated NSCs and neurons from NPC1 iPSC lines, although these cells did display cholesterol accumulation. This difference may reflect patient-specific differences or technical differences in how the NSCs and neurons were generated and highlights the need for the availability of iPSC lines from multiple patients for future studies to better understand the disease mechanisms of NPC1. We believe that the line described in this study can help in this process and will be especially helpful for drug-screening studies to identify novel compounds that can help in the treatment of NPC1.
Acknowledgments
This work was supported by the NIH Common Fund and the intramural research program of the National Institute of Child Health and Human Development.
Author Contributions
A.G.E. and N.M.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; J.S., W.J.P., S.W., and D.M.L.: collection and assembly of data; F.D.P.: provision of study material, clinical protocol; M.S.R.: conception and design, data analysis and interpretation, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
Articles from Stem Cells Translational Medicine are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.5966/sctm.2014-0127
Read article for free, from open access legal sources, via Unpaywall:
https://stemcellsjournals.onlinelibrary.wiley.com/doi/pdfdirect/10.5966/sctm.2014-0127
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.5966/sctm.2014-0127
Article citations
Upregulation of NPC1 and its association with poor prognosis in gastric cancer.
Clin Transl Oncol, 26(10):2665-2673, 02 May 2024
Cited by: 0 articles | PMID: 38698279
ApoE maintains neuronal integrity via microRNA and H3K27me3-mediated repression.
iScience, 27(3):109231, 15 Feb 2024
Cited by: 0 articles | PMID: 38439966 | PMCID: PMC10909902
Lithium ameliorates Niemann-Pick C1 disease phenotypes by impeding STING/SREBP2 activation.
iScience, 26(5):106613, 08 Apr 2023
Cited by: 2 articles | PMID: 37128603 | PMCID: PMC10148154
A neuropathological cell model derived from Niemann-Pick disease type C patient-specific iPSCs shows disruption of the p62/SQSTM1-KEAP1-NRF2 Axis and impaired formation of neuronal networks.
Mol Genet Metab Rep, 28:100784, 24 Jul 2021
Cited by: 2 articles | PMID: 34377675 | PMCID: PMC8327345
Deregulation of signalling in genetic conditions affecting the lysosomal metabolism of cholesterol and galactosyl-sphingolipids.
Neurobiol Dis, 146:105142, 17 Oct 2020
Cited by: 5 articles | PMID: 33080336 | PMCID: PMC8862610
Review Free full text in Europe PMC
Go to all (29) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE55379
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Diversity of glycosphingolipid GM2 and cholesterol accumulation in NPC1 patient-specific iPSC-derived neurons.
Brain Res, 1657:52-61, 05 Dec 2016
Cited by: 15 articles | PMID: 27923633
Patient-Specific iPSC-Derived Neural Differentiated and Hepatocyte-like Cells, Carrying the Compound Heterozygous Mutation p.V1023Sfs*15/p.G992R, Present the "Variant" Biochemical Phenotype of Niemann-Pick Type C1 Disease.
Int J Mol Sci, 22(22):12184, 10 Nov 2021
Cited by: 3 articles | PMID: 34830064 | PMCID: PMC8624182
Decreased calcium flux in Niemann-Pick type C1 patient-specific iPSC-derived neurons due to higher amount of calcium-impermeable AMPA receptors.
Mol Cell Neurosci, 83:27-36, 27 Jun 2017
Cited by: 17 articles | PMID: 28666962
Pluripotent Stem Cells for Disease Modeling and Drug Discovery in Niemann-Pick Type C1.
Int J Mol Sci, 22(2):E710, 12 Jan 2021
Cited by: 6 articles | PMID: 33445799 | PMCID: PMC7828283
Review Free full text in Europe PMC
Funding
Funders who supported this work.
 a
a




