| PMC full text: |
|
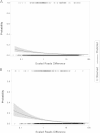
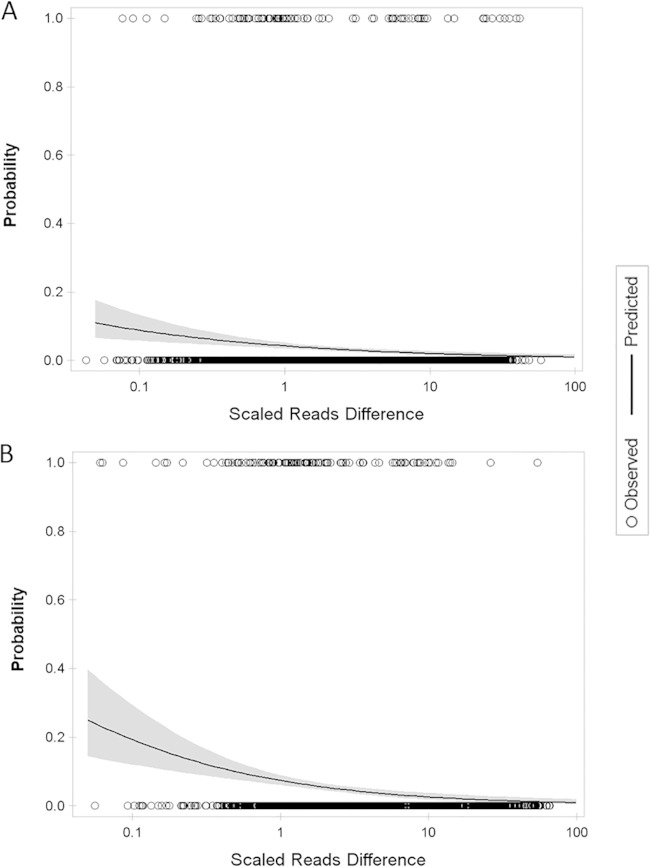
FIG 3
Predicted incorrect H antigen identification using reads mapping with 95% confidence limits. (A) Prediction for fliC identification. (B) Prediction for fljB identification. Logistic regression was used to estimate the probability of making an incorrect identification given the size of the mapped reads difference scaled by total number of reads sequenced from a genome. The GenomeTrakr data set selected for SeqSero validation was used for this analysis. Observed correct and incorrect antigens calls were based on the first round of reads mapping.