Abstract
Free full text

The end of the message: multiple protein–RNA interactions define the mRNA polyadenylation site
Abstract
The key RNA sequence elements and protein factors necessary for 3′ processing of polyadenylated mRNA precursors are well known. Recent studies, however, have significantly reshaped current models for the protein–RNA interactions involved in poly(A) site recognition, painting a picture more complex than previously envisioned and also providing new insights into regulation of this important step in gene expression. Here we review the recent advances in this area and provide a perspective for future studies.
Almost all eukaryotic mRNAs and primary microRNA transcripts and many long intergenic noncoding RNAs are polyadenylated at their 3′ ends. This involves a well-studied two-step reaction consisting of an endonucleolytic cleavage followed by the synthesis of the poly(A) tail (Colgan and Manley 1997; Zhao et al. 1999; Chan et al. 2011; Proudfoot 2011). Significantly, recent global analyses have revealed that ~70% of eukaryotic genes produce multiple mRNA isoforms with distinct 3′ ends through the process of alternative polyadenylation (APA) (Di Giammartino et al. 2011; Shi 2012; Elkon et al. 2013; Tian and Manley 2013). APA isoforms from the same gene may encode different proteins and/or contain different 3′ untranslated regions (UTRs). As longer 3′ UTRs often harbor more binding sites for microRNAs and/or RNA-binding proteins than shorter 3′ UTRs, APA isoforms may acquire different stability, translation efficiency, and/or intracellular localization. APA is dynamically regulated in development and in response to environmental stimuli (Flavell et al. 2008; Sandberg et al. 2008; Ji et al. 2009; Shepard et al. 2011; Graber et al. 2013), and deregulation of APA has been associated with a number of human diseases (Mayr and Bartel 2009; Jenal et al. 2012; Masamha et al. 2014). Therefore, it is critical to understand how mRNA 3′ processing sites, often referred to as the poly(A) site (PAS), are recognized and how PAS selection is regulated.
Mammalian PASs are generally AU-rich and have a highly conserved nucleotide composition profile (Shi 2012; Tian and Graber 2012). A number of key cis elements have been identified in the mammalian PAS, including the AAUAAA (or its close variant AUUAAA) located 10~30 nucleotides (nt) upstream of the cleavage site, which is frequently defined by a CA dinucleotide immediately 5′ to the site of endonucleolytic cleavage; a U/GU-rich downstream element (DSE) located within ~40 nt downstream from the cleavage sites; and U-rich upstream auxiliary elements (USEs) and upstream sequences conforming to the consensus UGUA. PAS sequences are divergent, and many PASs lack one or more of these cis elements. For example, ~30% of human PASs lack an A(A/U)UAAA hexamer (Beaudoing et al. 2000), and ~20% of human PASs do not possess a U- or GU-rich DSE (Zarudnaya et al. 2003). Unlike splicing, which relies on RNA base-pairing for splice site recognition (Wahl et al. 2009), PAS recognition is accomplished solely by protein–RNA interactions (Colgan and Manley 1997; Chan et al. 2011). The mRNA 3′ processing factors CPSF (cleavage and polyadenylation specificity factor) and CstF synergistically bind to the AAUAAA hexamer and the DSE, respectively, while the CFI complex binds to the UGUA motifs (Hu et al. 2005). CPSF, CstF, and CFI directly bind to RNAs to form a core complex and in turn recruit other factors, including CFII, the scaffolding protein Symplekin, and the poly(A) polymerase (PAP), to assemble the active mRNA 3′ processing complex (Shi et al. 2009; Chan et al. 2011). The assembly of these factors on PASs occurs cotranscriptionally and is facilitated by the C-terminal domain of the RNA polymerase II (RNAP II) large subunit (Hirose and Manley 2000; Proudfoot et al. 2002; Bentley 2005).
A detailed understanding of these key interactions is critical for delineating the mechanisms for PAS recognition. Additionally, an increasing number of regulatory factors have been shown to modulate PAS selection. There have been a number of recent reviews on APA and its physiological impact (Di Giammartino et al. 2011; Shi 2012; Elkon et al. 2013; Mueller et al. 2013; Tian and Manley 2013). Here we focus on the protein–RNA interactions that are responsible for PAS recognition and its regulation. An interesting and useful analogy to consider is the similarities between the core PAS and the core RNAP II promoter, specifically how multiple conserved sequence motifs are organized and recognized by multisubunit protein complexes, which in both cases function to recruit otherwise sequence-nonspecific polymerases.
CPSF and AAUAAA recognition
CPSF is a multisubunit protein complex consisting of CPSF160, Wdr33, CPSF100, CPSF73 (the cleavage endonuclease), Fip1, and CPSF30. CPSF specifically recognizes the most important cis element in the mammalian PAS, the AAUAAA hexamer (Fig. 1A; Bienroth et al. 1991; Murthy and Manley 1992; Shi et al. 2009). CPSF160 has long been implicated in directly binding to AAUAAA based on several lines of evidence. First, two CPSF subunits of ~160 and 30 kDa, presumed to be CPSF160 and CPSF30, could be UV cross-linked specifically to AAUAAA-containing RNA oligos (Keller et al. 1991). Second, Yhh1/Cft1, the yeast homolog of CPSF160, binds to PAS-containing RNA near the cleavage site via an evolutionarily conserved central domain (Dichtl et al. 2002). Third, recombinant CPSF160 possesses RNA-binding activity and shows preference for AAUAAA-containing sequences (Murthy and Manley 1995). However, the affinity and specificity of the CPSF160–RNA interaction is significantly weaker than that of the CPSF complex (Murthy and Manley 1995), and more recent global mapping of CPSF160–RNA interactions failed to detect specific enrichment of its binding signals at or near the AAUAAA hexamers (Martin et al. 2012). Therefore, >20 years after the isolation of CPSF, the identities of its subunits that directly bind to AAUAAA remained to be determined.
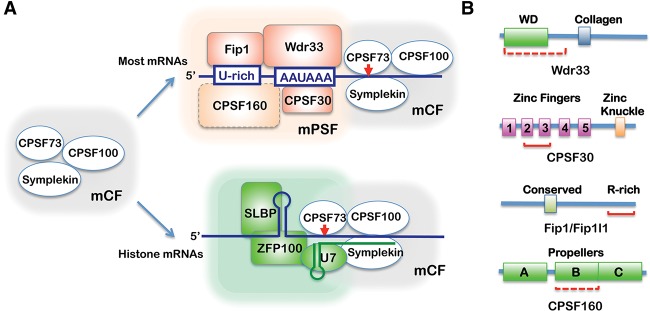
(A) Schematic models showing distinct modules within CPSF. CPSF73, CPSF100, and Symplekin form a module (which we refer to as mCF) that contains the endonuclease activity. mCF may cooperate with different RNA-binding modules, such as mammalian polyadenylation specificity factor (mPSF) (Wdr33, CPSF30, Fip1, and CPSF160), for cleavage/polyadenylation of most mRNAs or with SLBP and U7 snRNP for the cleavage of histone mRNAs. (B, bottom panel) Domain structures of the CPSF subunits involved or implicated in RNA binding. The known or putative RNA-binding regions are marked with red solid or dotted underlines.
Recent studies have provided unexpected new insight into AAUAAA recognition. Using intact CPSF complexes immunopurified from mammalian cell extracts or a CPSF subcomplex reconstituted with recombinant subunits, two recent studies provided direct evidence that Wdr33 and CPSF30 specifically recognize the AAUAAA hexamer in vitro (Chan et al. 2014; Schonemann et al. 2014). In vivo mapping of protein–RNA interactions using photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) detected specific binding of both proteins at the AAUAAA regions of active PASs at the global level (Chan et al. 2014; Schonemann et al. 2014). Based on these data, a new model for CPSF–RNA interactions was proposed in which Wdr33 and CPSF30 simultaneously and synergistically recognize AAUAAA (Fig. 1A). The RNA-binding activity of CPSF30, which consists of five C3H zinc finger repeats and a putative RNA-binding zinc knuckle motif at the C terminus (Barabino et al. 1997), is mediated primarily by its zinc fingers 2 and 3 (Fig. 1B; Chan et al. 2014). Wdr33, which is a large protein similar in size to CPSF160, interacts with RNA at least in part via its N-terminal region, which includes a highly conserved WD40 repeat domain (Fig. 1B; Schonemann et al. 2014). Both zinc finger and WD40 repeat domains are known to mediate sequence-specific RNA interactions (Hudson et al. 2004; Lau et al. 2009). It remains to be determined, however, whether Wdr33, CPSF30, or both are required to confer sequence specificity to CPSF–RNA interactions. A combination of biochemical and structural approaches will be needed to delineate the specific contribution of Wdr33 and CPSF30 to CPSF–RNA interactions as well as determine the precise role of CPSF160 in PAS recognition. It is noteworthy that both CPSF30 (Nemeroff et al. 1998; Twu et al. 2006) and Wdr33 (Brass et al. 2009) have been implicated in host cell defense against influenza virus infection, likely reflecting a pathway for viral inhibition of host mRNA production.
In addition to Wdr33 and CPSF30, and perhaps CPSF160, Fip1 also contributes CPSF–RNA interactions (Fig. 1A). Recombinant Fip1 binds to U-rich RNAs via its arginine-rich C-terminal region (Kaufmann et al. 2004). Within the CPSF complex, Fip1 binds to RNA sequences upstream of the AAUAAA hexamer in vitro (Chan et al. 2014). Consistent with this, in vivo mapping of Fip1–RNA interactions revealed that Fip1 binds to U-rich sequences in the AAUAAA hexamer region (Fig. 1B; Martin et al. 2012; Lackford et al. 2014). Fip1 was recently identified as a regulator of APA in embryonic stem cells (ESCs), and Fip1-mediated APA regulation is required for ESC self-renewal, further suggesting that Fip1–RNA interactions play an important role in PAS selection (Lackford et al. 2014). Finally, Fip1 and CPSF160 function directly in recruiting PAP to the mRNA 3′ processing site (Murthy and Manley 1995; Kaufmann et al. 2004).
What, then, is the function of CPSF160 in PAS recognition? Most surprisingly, CPSF160–RNA interactions were not detected in the two recent studies (Chan et al. 2014; Schonemann et al. 2014). Previous evidence for CPSF160–RNA interactions were mostly based on experiments using purified recombinant CPSF160 or its yeast homolog, Cft1 (Murthy and Manley 1995; Dichtl et al. 2002). In contrast, both recent studies used CPSF complexes (Chan et al. 2014; Schonemann et al. 2014). So it is possible that the RNA-binding activity of CPSF160 is inhibited within the CPSF complex. However, given the conserved nature of the CPSF160 RNA-binding activity, a more likely possibility is that CPSF160 participates in RNA interactions at a specific stage during mRNA 3′ processing or, intriguingly, at a specific subset of PASs.
Similarly complex protein–RNA interactions have been observed in yeast PAS recognition. The yeast mRNA 3′ processing factor CPF contains Mpe1 (Rbbp6 homolog) (see below), Pap1, Pta1 (Symplekin), and the homologs of all six subunits of the mammalian CPSF complex, including Cft1 (CPSF160), Cft2 (CPSF100), Ysh1/Brr5 (CPSF73), Fip1 (Fip1), Yth1 (CPSF30), and Pfs2 (Wdr33) (Zhao et al. 1999). Similar to CPSF, multiple CPF subunits have been shown to bind RNA, including Cft1, Cft2, Fip1, Yth1, and Mpe1 (Barabino et al. 1997; Zhao et al. 1997; Dichtl et al. 2002; Lee and Moore 2014).
We envision several scenarios to explain why such complex protein–RNA interactions are necessary. First, given the generally lower specificity of RNA–protein interactions compared with DNA–protein interactions or RNA base-pairing, combinatorial interactions may be necessary to achieve the RNA-binding specificity necessary for accurate PAS recognition. Second, due to the dynamic nature of mRNA 3′ processing (Chan et al. 2011), it is possible that different factors participate in PAS binding at different stages of 3′ processing. For example, Yth1 binds to RNA and Fip1 in a mutually exclusive manner, and it was proposed that Yth1 binds to PAS RNA during the cleavage step but releases RNA to associate with Fip1 during the poly(A) synthesis step (Tacahashi et al. 2003). To determine whether CPSF160 participates in RNA binding at specific stages of mRNA 3′ processing, it will be critical to characterize better the dynamics of the cleavage/polyadenylation machinery. Third, an attractive idea is that there may be distinct CPSF subcomplexes or alternative complexes that bind to PAS sequences through distinct protein–RNA interactions. As mentioned earlier, mammalian PASs are highly diverse and may require multiple alternative factors for their recognition.
An intriguing similarity exists between CPSF and the general RNAP II transcription factor TFIID (Cler et al. 2009). Both are multisubunit complexes that not only recognize key promoter/PAS sequences, which are strikingly similar (TATAAA vs. AAUAAA), but also contain subunits that bind additional promoter/PAS elements. TFIID exists as tissue-specific and cell type-specific subcomplexes and can also contain distinct isoforms of different subunits (Muller et al. 2010). As we shall see, a similar picture of CPSF is beginning to emerge.
Several lines of evidence point to the existence of heterogeneous CPSF complexes. For example, Schonemann et al. (2014) showed that a relatively stable complex, called mammalian polyadenylation specificity factor (mPSF), can be reconstituted with Wdr33, CPSF30, Fip1, and CPSF160 and is active, with PAP, in AAUAAA-dependent poly(A) synthesis (Fig. 1A). A distinct subcomplex consisting of CPSF100, Symplekin, and CPSF73 functions in 3′ processing of nonpolyadenylated histone mRNA precursors (Sullivan et al. 2009). This factor, which we refer to as mCF, can either partner with SLBP, the RNA-binding protein that functions in recognition of the histone mRNA 3′ processing site, to carry out histone mRNA cleavage (Marzluff et al. 2008) or, we suggest, associate with mPSF to form CPSF (Fig. 1A). This modular design seems to be evolutionarily conserved, as mPSF shares most of its components with the yeast PFI, and mCF shares most of its components with CFII (Zhao et al. 1999). Another more speculative idea is that distinct CPSF complexes exist and may function at different PASs. For example, CPSF160 and Wdr33 are similar in size and domain structure, and perhaps both function in different CPSF complexes. Although there is currently no evidence in support of this, it is noteworthy that early size estimates of native CPSF were in the 200- to 300-kDa range (Christofori and Keller 1988; Takagaki et al. 1989), too small to include both of these ~160-kDa proteins. Finally, another possibility is that distinct CPSF complexes can contain different subunit isoforms. For example, two splice isoforms of CPSF30 exist and can be found in CPSF complexes (Chan et al. 2014).
CstF and the DSE
CstF is a trimeric complex consisting of CstF77, CstF50, and CstF64 or its paralog, CstF64τ (Takagaki et al. 1990; Gilmartin and Nevins 1991; Wallace et al. 1999). CstF is believed to form a homodimer, as both CstF77 and CstF50 self-associate (Fig. 2A; Takagaki and Manley 2000; Bai et al. 2007; Moreno-Morcillo et al. 2011). CstF specifically recognizes the DSEs, which are generally characterized as U/GU-rich, and this interaction is mediated by CstF64 or CstF64τ (MacDonald et al. 1994; Takagaki and Manley 1997). It was originally suggested that CstF64τ is a testis-specific factor that has more distinct RNA-binding specificity than CstF64 and that CstF64τ may play a role in mediating testis-specific PAS selection (Wallace et al. 1999; Monarez et al. 2007). However, recent studies provided compelling evidence that CstF64τ, similar to CstF64, is widely expressed in mammalian tissues (Yao et al. 2013). Furthermore, in vitro and in vivo analyses demonstrated that CstF64 and CstF64τ have nearly identical RNA-binding specificities (Takagaki and Manley 1997; Yao et al. 2013). Finally, individual knockdowns of CstF64 or CstF64τ had little effect on global PAS selection, but double knockdowns led to significant APA changes (Yao et al. 2012). Together, these recent studies strongly suggest that CstF64 and CstF64τ play largely redundant roles in mediating CstF–RNA interactions. Another CstF64 isoform, called βCstF64, was detected predominantly in the CNS (Shankarling et al. 2009), but its role in PAS recognition has yet to be characterized.
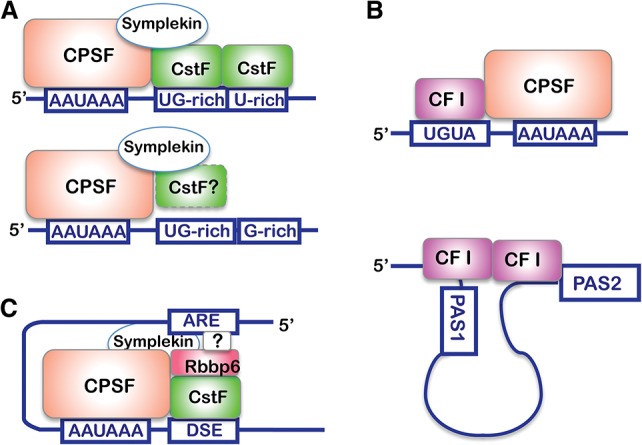
(A) CstF-dependent and potentially CstF-independent PAS recognition. In the CstF-independent model, CstF is shown in a dotted boundary to indicate that CstF may or may not be present in this complex. The scaffolding protein Symplekin plays a role in bridging CPSF–CstF complexes. (B) CFI–RNA interactions. (Top panel) CFI binds to UGUA motifs often found upstream of the AAUAAA hexamer. (Bottom panel) The CFI dimer may bind to two UGUA sequences at different PASs, thus looping out the proximal PASs. (C) The role of Rbbp6 in PAS recognition. The question mark denotes the fact that Rbbp6 may or may not directly interact with upstream AU-rich elements (AREs) or may rely on an unknown factor.
The DSEs of mammalian PASs are highly variable, and thus an important question is how CstF64/τ recognize such diverse sequences. While properties of the CstF64 RNA recognition motif (RRM) allow it to recognize diverse GU-rich sequences (Perez Canadillas and Varani 2003), recent in vivo and in vitro analyses of CstF64/τ–RNA interactions provided evidence that these “general” 3′ processing factors only bind to a subset of PASs, which are characterized by GU-rich sequences in the first 20 nt downstream from the cleavage site followed by U-rich sequences (Fig. 2A; Yao et al. 2012, 2013). In contrast, PASs that are not stably bound by CstF64/τ contain more G-rich DSEs (Fig. 2A). These results raised the possibility that there may be distinct types of PASs that require different combinations of factors for their recognition. This conclusion is consistent with earlier studies. For example, it has been shown that CstF and mRNA 3′ processing activity is transiently inhibited following DNA damage due to an interaction between CstF50 and the BARD1–BRCA1 complex (Kleiman and Manley 2001). In the meantime, DNA damage response genes must be expressed, which would require mRNA 3′ processing activity (Gomes et al. 2006). Providing an answer to this paradox, a recent study revealed that the PAS of p53, a DNA damage-induced gene, contains a G-rich DSE. A unique G-quadruplex structure is formed within this DSE region and is recognized by hnRNP F/H to allow 3′ end formation of p53 mRNAs after DNA damage (Decorsiere et al. 2011). Together, these studies provide evidence for CstF-independent PAS recognition, but additional work is needed to delineate such mechanisms fully.
CFI–upstream UGUA interactions
CFI binds to UGUA motifs, which are typically located upstream of the AAUAAA hexamer (Fig. 2B; Brown and Gilmartin 2003; Hu et al. 2005). CFI consists of CFI25 and one of two related subunits, CFI59 or CFI68 (Ruegsegger et al. 1996), and appears to exist as a tetramer of two CFI25 subunits and two large subunits (Yang et al. 2011). All three CFI subunits participate in RNA binding, as they can be UV cross-linked to RNA (Ruegsegger et al. 1996). The small subunit, CFI25, contains a Nudix domain, which, in its canonical form, possesses pyrophosphohydrolase activities. The CFI25 Nudix domain, however, lacks such enzymatic activity due to substitutions at key residues and instead is involved in specific interactions with the UGUA motif (Yang et al. 2010b). Similar to CstF subunits, CFI25 forms a dimer and thus can bind and recognize two UGUA sequences simultaneously (Fig. 2B, bottom panel), a property that has been proposed to regulate PAS selection (Yang et al. 2010a,b). Extending these results, PAR-CLIP analyses detected robust CFI binding to UGUA motifs at the global level (Martin et al. 2012). Both CFI59 and CFI68 contain an N-terminal RRM, a proline-rich central region, and a C-terminal region rich in arginine–serine (RS) repeats (Ruegsegger et al. 1998). CFI59/68 share domain structures similar to SR proteins, which are well-known splicing regulators (Tacke and Manley 1996). Like SR proteins, CFI subunits have been detected in purified spliceosomes (Rappsilber et al. 2002; Zhou et al. 2002), implicating CFI in splicing itself or in mediating cross-talk between splicing and 3′ processing. In fact, CFI has been shown to interact with U2AF65, which is involved in the early steps of splicing, and this interaction helps to stimulate mRNA 3′ processing (Millevoi et al. 2002, 2006).
CFI plays an important role in PAS recognition. Initial studies established that, like CstF, it functions early in the process, functioning to help stabilize CPSF binding to the PAS (Ruegsegger et al. 1996, 1998). This again suggests an analogy with core RNAP II promoter recognition, in which two general factors, TFIIA and TFIIB, function in part by stabilizing TFIID binding to promoter DNA (Thomas and Chiang 2006). Given the importance of CFI in not only PAS recognition but also APA (Gruber et al. 2012; Martin et al. 2012; Masamha et al. 2014), it will be important to delineate the detailed mechanisms by which CFI–RNA interactions contribute to PAS recognition.
Rbbp6 and PAS recognition
Rbbp6 is a newly identified 3′ processing factor that also functions in part by contacting RNA. Rbbp6 was originally identified in a screen for Rb-binding proteins (Saijo et al. 1995; Sakai et al. 1995), hence the name, and was later identified as a p53-binding protein as well (Simons et al. 1997). Although Rbbp6 was suggested to be the putative mammalian homolog of the yeast 3′ processing factor Mpe1 based on limited sequence similarity (Vo et al. 2001), it was not functionally implicated in mRNA 3′ processing until a proteomic analysis detected Rbbp6 in the purified mRNA 3′ processing complex (Shi et al. 2009), and direct evidence for its involvement in 3′ processing was provided only recently (Di Giammartino et al. 2014). Rbbp6 was found to associate with CstF via its evolutionarily conserved N-terminal regions, which contain a DWNN (domain with no name)/ubiquitin-like domain, a zinc knuckle, and a RING finger. This N-terminal region, which is the region of similarity with its yeast homolog, Mpe1, is sufficient for 3′ processing activity in vitro. In both proteins, the zinc knuckle and RING domains function in RNA binding, although in neither case does binding appear sequence-specific (Di Giammartino et al. 2014; Lee and Moore 2014). Interestingly, knockdown of Rbbp6 in mammalian cells preferentially inhibits the 3′ processing of mRNAs that contain AU-rich elements (AREs) in their 3′ UTRs (Di Giammartino et al. 2014). It remains unknown, however, whether Rbbp6 directly interacts with AREs or whether additional factors are involved in determining this apparent specificity (Fig. 2C).
Rbbp6 has several other intriguing properties. For example, Mpe1 has been implicated in regulating ubiquitination of PAP and potentially other factors (Lee and Moore 2014). Rbbp6 has also been shown to possess E3 ligase activity and function as an activator of Mdm2-mediated ubiquitination of p53 (Li et al. 2007; Miotto et al. 2014). Finally, a splice isoform of Rbbp6, called iso3 and consisting solely of the DWNN, competes with the full-length protein to modulate cleavage efficiency (Di Giammartino et al. 2014). As with a number of 3′ processing factors, levels of Rbbp6 are elevated in many types of cancers (Chen et al. 2013). Coupled with the fact that iso3 levels are reduced (Mbita et al. 2012), this likely contributes to the changes in processing efficiency that modulate APA in cancer cells (Mayr and Bartel 2009). It will be important to determine how all of these properties of Rbbp6 function in modulating 3′ processing and in tumorigenesis.
Regulation of PAS recognition
A growing number of factors have been shown to modulate PAS recognition by the core 3′ processing machinery. In many cases, the effect of these regulatory factors, most of which were initially described as functioning in pre-mRNA splicing, depends on the positions of their binding sites relative to a PAS (Fig. 3). For example, U1 snRNP suppresses polyadenylation at downstream PAS by U1A-70K-mediated and U1-70K-mediated inhibition of PAP activity (Gunderson et al. 1997, 1998). More recent global analyses revealed that U1 snRNP plays a critical role in protecting pre-mRNAs from premature cleavage and polyadenylation (Kaida et al. 2010). Additionally, U1 snRNP levels relative to the cellular demand has been proposed to control mRNA length and isoform expression by modulating PAS selection (Berg et al. 2012). On the other hand, U2 snRNP promotes the usage of downstream PASs through multiple protein–protein interactions (Fig. 3; Kyburz et al. 2006; Millevoi et al. 2006). In addition to splicing factors, the m7G cap at the 5′ ends of mRNAs can stimulate mRNA 3′ processing (Cooke and Alwine 1996; Flaherty et al. 1997), possibly through interactions between the cap-binding complex and 3′ processing factors (Yang et al. 2011).
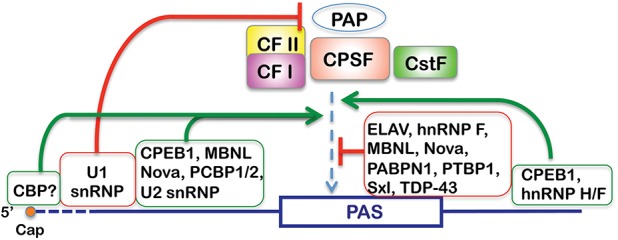
Context-dependent regulation of PAS recognition. Regulatory factors bound at different locations relative to the core PAS sequence have different effects on PAS recognition by the mRNA 3′ processing factors. Positive effects are indicated by an arrow, and negative effects are indicated by a vertical line.
Many RNA-binding proteins, also initially described as splicing regulators, have been implicated in control of PAS recognition (Fig. 3). Some of these factors directly compete with core 3′ processing factors to block PAS recognition (Fig. 3). For example, PTBP1, TDP-43, sex-lethal (Sxl), and ELAV/HuR have all been shown to directly compete with CstF for binding to DSEs and in turn inhibit PAS usage (Castelo-Branco et al. 2004; Gawande et al. 2006; Zhu et al. 2007; Avendano-Vazquez et al. 2012). PABPN1 was suggested to compete with CPSF for binding to the AAUAAA hexamer region, especially at many proximal PASs (Jenal et al. 2012).
Notably, the effects of some regulatory factors on PAS recognition are position-dependent. For example, NOVA proteins, which are known splicing regulators in the brain (Jensen et al. 2000), were shown to regulate APA by binding to sites in 3′ UTRs and repress or enhance PAS usage in a position-dependent manner (Licatalosi et al. 2008). Binding near the PASs was found to be repressive, likely by interfering with core factor binding, while binding to more distant sites tended to result in activation of PAS utilization by currently unknown mechanisms. Likewise, when muscleblind (Mbnl) proteins bind to core PAS sequences, especially between the cleavage sites and the DSEs, they tend to inhibit PAS recognition, presumably through steric hindrance (Batra et al. 2014). In contrast, when they bind to sequences upstream of the core PASs, Mbnl proteins stimulate PAS usage. CPEB1 has been suggested to promote PAS recognition when it binds to either upstream or downstream sequences (Bava et al. 2013). Similar position-specific effects of RNA-binding proteins on splice site recognition have also been reported (Wang et al. 2012; Erkelenz et al. 2013; Fu and Ares 2014). It remains an important question for future studies as to how RNA-binding proteins bound to neighboring sequences regulate the interactions between the core mRNA 3′ processing factors and the PASs. Furthermore, recent studies have provided evidence that the effect of some RNA-binding proteins, such as ELAV, on PAS selection is linked to the promoter and RNAP II pausing (Oktaba et al. 2015).
Conclusions and future perspectives
Recent studies suggest that the protein–RNA interaction network involved in PAS recognition is more complex than previously thought, which raises many important questions for future studies. For example, which factors—Wdr33, CPSF30, or both—provide specificity for the CPSF–AAUAAA interaction? Does CPSF160 participate in PAS recognition? If so, when and where? How does RNA binding by other CPSF subunits contribute to PAS definition? How do alternative protein isoforms and distinct core complexes contribute to PAS recognition? How do regulatory proteins modulate PAS usage in a position-dependent manner? Solving these important problems will undoubtedly bring our understanding of mRNA 3′ processing and its regulation to a new height.
Acknowledgments
Work in our laboratories was supported by the following grants: National Institutes of Health (NIH) GM090056 and CA177651 and American Cancer Society RSG-12-186 to Y.S., and NIH GM28983 to J.L.M.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.261974.115.
References
- Avendano-Vazquez SE, Dhir A, Bembich S, Buratti E, Proudfoot N, Baralle FE. 2012. Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev 26: 1679–1684. [Europe PMC free article] [Abstract] [Google Scholar]
- Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. 2007. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol Cell 25: 863–875. [Abstract] [Google Scholar]
- Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev 11: 1703–1716. [Abstract] [Google Scholar]
- Batra R, Charizanis K, Manchanda M, Mohan A, Li M, Finn DJ, Goodwin M, Zhang C, Sobczak K, Thornton CA, et al.2014. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell 56: 311–322. [Europe PMC free article] [Abstract] [Google Scholar]
- Bava FA, Eliscovich C, Ferreira PG, Minana B, Ben-Dov C, Guigo R, Valcarcel J, Mendez R. 2013. CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature 495: 121–125. [Abstract] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res 10: 1001–1010. [Europe PMC free article] [Abstract] [Google Scholar]
- Bentley DL. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256. [Abstract] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al.2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64. [Europe PMC free article] [Abstract] [Google Scholar]
- Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. 1991. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J Biol Chem 266: 19768–19776. [Abstract] [Google Scholar]
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, et al.2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139: 1243–1254. [Europe PMC free article] [Abstract] [Google Scholar]
- Brown KM, Gilmartin GM. 2003. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell 12: 1467–1476. [Abstract] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N. 2004. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol 24: 4174–4183. [Europe PMC free article] [Abstract] [Google Scholar]
- Chan S, Choi EA, Shi Y. 2011. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA 2: 321–335. [Europe PMC free article] [Abstract] [Google Scholar]
- Chan S, Huppertz I, Yao C, Weng L, Moresco J, Yates JR, Ule J, Manley J, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 28: 2370–2380. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen J, Tang H, Wu Z, Zhou C, Jiang T, Xue Y, Huang G, Yan D, Peng Z. 2013. Overexpression of RBBP6, alone or combined with mutant TP53, is predictive of poor prognosis in colon cancer. PLoS One 8: e66524. [Europe PMC free article] [Abstract] [Google Scholar]
- Christofori G, Keller W. 1988. 3′ cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell 54: 875–889. [Abstract] [Google Scholar]
- Cler E, Papai G, Schultz P, Davidson I. 2009. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci 66: 2123–2134. [Abstract] [Google Scholar]
- Colgan DF, Manley JL. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766. [Abstract] [Google Scholar]
- Cooke C, Alwine JC. 1996. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol 16: 2579–2584. [Europe PMC free article] [Abstract] [Google Scholar]
- Decorsiere A, Cayrel A, Vagner S, Millevoi S. 2011. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev 25: 220–225. [Europe PMC free article] [Abstract] [Google Scholar]
- Dichtl B, Blank D, Sadowski M, Hubner W, Weiser S, Keller W. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J 21: 4125–4135. [Europe PMC free article] [Abstract] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866. [Europe PMC free article] [Abstract] [Google Scholar]
- Di Giammartino DC, Li W, Ogami K, Yashinskie JJ, Hoque M, Tian B, Manley JL. 2014. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3′ UTRs. Genes Dev 28: 2248–2260. [Europe PMC free article] [Abstract] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506. [Abstract] [Google Scholar]
- Erkelenz S, Mueller WF, Evans MS, Busch A, Schoneweis K, Hertel KJ, Schaal H. 2013. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA 19: 96–102. [Europe PMC free article] [Abstract] [Google Scholar]
- Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci 94: 11893–11898. [Europe PMC free article] [Abstract] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038. [Europe PMC free article] [Abstract] [Google Scholar]
- Fu XD, Ares M Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15: 689–701. [Europe PMC free article] [Abstract] [Google Scholar]
- Gawande B, Robida MD, Rahn A, Singh R. 2006. Drosophila sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J 25: 1263–1272. [Europe PMC free article] [Abstract] [Google Scholar]
- Gilmartin GM, Nevins JR. 1991. Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol Cell Biol 11: 2432–2438. [Europe PMC free article] [Abstract] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 20: 601–612. [Europe PMC free article] [Abstract] [Google Scholar]
- Graber JH, Nazeer FI, Yeh PC, Kuehner JN, Borikar S, Hoskinson D, Moore CL. 2013. DNA damage induces targeted, genome-wide variation of poly(A) sites in budding yeast. Genome Res 23: 1690–1703. [Europe PMC free article] [Abstract] [Google Scholar]
- Gruber AR, Martin G, Keller W, Zavolan M. 2012. Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol 9: 1405–1412. [Abstract] [Google Scholar]
- Gunderson SI, Vagner S, Polycarpou-Schwarz M, Mattaj IW. 1997. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev 11: 761–773. [Abstract] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1: 255–264. [Abstract] [Google Scholar]
- Hirose Y, Manley JL. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429. [Abstract] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11: 1485–1493. [Europe PMC free article] [Abstract] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. 2004. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257–264. [Abstract] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, et al.2012. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149: 538–553. [Abstract] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. 2000. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25: 359–371. [Abstract] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. [Europe PMC free article] [Abstract] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23: 616–626. [Europe PMC free article] [Abstract] [Google Scholar]
- Keller W, Bienroth S, Lang KM, Christofori G. 1991. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J 10: 4241–4249. [Europe PMC free article] [Abstract] [Google Scholar]
- Kleiman FE, Manley JL. 2001. The BARD1–CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104: 743–753. [Abstract] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W. 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 23: 195–205. [Abstract] [Google Scholar]
- Lackford B, Yao C, Charles GM, Weng L, Zheng X, Choi EA, Xie X, Wan J, Xing Y, Freudenberg JM, et al.2014. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J 33: 878–889. [Europe PMC free article] [Abstract] [Google Scholar]
- Lau CK, Bachorik JL, Dreyfuss G. 2009. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat Struct Mol Biol 16: 486–491. [Abstract] [Google Scholar]
- Lee SD, Moore CL. 2014. Efficient mRNA polyadenylation requires a ubiquitin-like domain, a zinc knuckle, and a RING finger domain, all contained in the Mpe1 protein. Mol Cell Biol 34: 3955–3967. [Europe PMC free article] [Abstract] [Google Scholar]
- Li L, Deng B, Xing G, Teng Y, Tian C, Cheng X, Yin X, Yang J, Gao X, Zhu Y, et al.2007. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc Natl Acad Sci 104: 7951–7956. [Europe PMC free article] [Abstract] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469. [Europe PMC free article] [Abstract] [Google Scholar]
- MacDonald CC, Wilusz J, Shenk T. 1994. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol 14: 6647–6654. [Europe PMC free article] [Abstract] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep 1: 753–763. [Abstract] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854. [Europe PMC free article] [Abstract] [Google Scholar]
- Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ. 2014. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510: 412–416. [Europe PMC free article] [Abstract] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [Europe PMC free article] [Abstract] [Google Scholar]
- Mbita Z, Meyer M, Skepu A, Hosie M, Rees J, Dlamini Z. 2012. De-regulation of the RBBP6 isoform 3/DWNN in human cancers. Mol Cell Biochem 362: 249–262. [Abstract] [Google Scholar]
- Millevoi S, Geraghty F, Idowu B, Tam JL, Antoniou M, Vagner S. 2002. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep 3: 869–874. [Europe PMC free article] [Abstract] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. 2006. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J 25: 4854–4864. [Europe PMC free article] [Abstract] [Google Scholar]
- Miotto B, Chibi M, Xie P, Koundrioukoff S, Moolman-Smook H, Pugh D, Debatisse M, He F, Zhang L, Defossez PA. 2014. The RBBP6/ZBTB38/MCM10 axis regulates DNA replication and common fragile site stability. Cell Rep 7: 575–587. [Abstract] [Google Scholar]
- Monarez RR, MacDonald CC, Dass B. 2007. Polyadenylation proteins CstF-64 and τCstF-64 exhibit differential binding affinities for RNA polymers. Biochem J 401: 651–658. [Europe PMC free article] [Abstract] [Google Scholar]
- Moreno-Morcillo M, Minvielle-Sebastia L, Mackereth C, Fribourg S. 2011. Hexameric architecture of CstF supported by CstF-50 homodimerization domain structure. RNA 17: 412–418. [Europe PMC free article] [Abstract] [Google Scholar]
- Mueller AA, Cheung TH, Rando TA. 2013. All's well that ends well: alternative polyadenylation and its implications for stem cell biology. Curr Opin Cell Biol 25: 222–232. [Europe PMC free article] [Abstract] [Google Scholar]
- Muller F, Zaucker A, Tora L. 2010. Developmental regulation of transcription initiation: more than just changing the actors. Curr Opin Genet Dev 20: 533–540. [Abstract] [Google Scholar]
- Murthy KG, Manley JL. 1992. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem 267: 14804–14811. [Abstract] [Google Scholar]
- Murthy KG, Manley JL. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev 9: 2672–2683. [Abstract] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell 1: 991–1000. [Abstract] [Google Scholar]
- Oktaba K, Zhang W, Lotz TS, Jun DJ, Lemke SB, Ng SP, Esposito E, Levine M, Hilgers V. 2015. ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol Cell 57: 341–348. [Europe PMC free article] [Abstract] [Google Scholar]
- Perez Canadillas JM, Varani G. 2003. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J 22: 2821–2830. [Europe PMC free article] [Abstract] [Google Scholar]
- Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev 25: 1770–1782. [Europe PMC free article] [Abstract] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [Abstract] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245. [Europe PMC free article] [Abstract] [Google Scholar]
- Ruegsegger U, Beyer K, Keller W. 1996. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 271: 6107–6113. [Abstract] [Google Scholar]
- Ruegsegger U, Blank D, Keller W. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell 1: 243–253. [Abstract] [Google Scholar]
- Saijo M, Sakai Y, Kishino T, Niikawa N, Matsuura Y, Morino K, Tamai K, Taya Y. 1995. Molecular cloning of a human protein that binds to the retinoblastoma protein and chromosomal mapping. Genomics 27: 511–519. [Abstract] [Google Scholar]
- Sakai Y, Saijo M, Coelho K, Kishino T, Niikawa N, Taya Y. 1995. cDNA sequence and chromosomal localization of a novel human protein, RBQ-1 (RBBP6), that binds to the retinoblastoma gene product. Genomics 30: 98–101. [Abstract] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [Europe PMC free article] [Abstract] [Google Scholar]
- Schonemann L, Kuhn U, Martin G, Schafer P, Gruber AR, Keller W, Zavolan M, Wahle E. 2014. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev 28: 2381–2393. [Europe PMC free article] [Abstract] [Google Scholar]
- Shankarling GS, Coates PW, Dass B, Macdonald CC. 2009. A family of splice variants of CstF-64 expressed in vertebrate nervous systems. BMC Mol Biol 10: 22. [Europe PMC free article] [Abstract] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772. [Europe PMC free article] [Abstract] [Google Scholar]
- Shi Y. 2012. Alternative polyadenylation: new insights from global analyses. RNA 18: 2105–2117. [Europe PMC free article] [Abstract] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376. [Europe PMC free article] [Abstract] [Google Scholar]
- Simons A, Melamed-Bessudo C, Wolkowicz R, Sperling J, Sperling R, Eisenbach L, Rotter V. 1997. PACT: cloning and characterization of a cellular p53 binding protein that interacts with Rb. Oncogene 14: 145–155. [Abstract] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF. 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34: 322–332. [Europe PMC free article] [Abstract] [Google Scholar]
- Tacahashi Y, Helmling S, Moore CL. 2003. Functional dissection of the zinc finger and flanking domains of the Yth1 cleavage/polyadenylation factor. Nucleic Acids Res 31: 1744–1752. [Europe PMC free article] [Abstract] [Google Scholar]
- Tacke R, Manley JL. 1996. SR proteins and splicing control. Genes Dev 10: 1569–1579. [Abstract] [Google Scholar]
- Takagaki Y, Manley JL. 1997. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol 17: 3907–3914. [Europe PMC free article] [Abstract] [Google Scholar]
- Takagaki Y, Manley JL. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525. [Europe PMC free article] [Abstract] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL. 1989. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev 3: 1711–1724. [Abstract] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev 4: 2112–2120. [Abstract] [Google Scholar]
- Thomas MC, Chiang CM. 2006. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178. [Abstract] [Google Scholar]
- Tian B, Graber JH. 2012. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA 3: 385–396. [Europe PMC free article] [Abstract] [Google Scholar]
- Tian B, Manley JL. 2013. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci 38: 312–320. [Europe PMC free article] [Abstract] [Google Scholar]
- Twu KY, Noah DL, Rao P, Kuo RL, Krug RM. 2006. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J Virol 80: 3957–3965. [Europe PMC free article] [Abstract] [Google Scholar]
- Vo LT, Minet M, Schmitter JM, Lacroute F, Wyers F. 2001. Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol Cell Biol 21: 8346–8356. [Europe PMC free article] [Abstract] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [Abstract] [Google Scholar]
- Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. 1999. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc Natl Acad Sci 96: 6763–6768. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang Y, Ma M, Xiao X, Wang Z. 2012. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat Struct Mol Biol 19: 1044–1052. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang Q, Coseno M, Gilmartin GM, Doublie S. 2010a. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure 19: 368–377. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang Q, Gilmartin GM, Doublie S. 2010b. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci 107: 10062–10067. [Europe PMC free article] [Abstract] [Google Scholar]
- Yang Q, Gilmartin GM, Doublie S. 2011. The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5′ capping and splicing. RNA Biol 8: 748–753. [Europe PMC free article] [Abstract] [Google Scholar]
- Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, Shi Y. 2012. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci 109: 18773–18778. [Europe PMC free article] [Abstract] [Google Scholar]
- Yao C, Choi EA, Weng L, Xie X, Wan J, Xing Y, Moresco JJ, Tu PG, Yates JR III, Shi Y. 2013. Overlapping and distinct functions of CstF64 and CstF64τ in mammalian mRNA 3′ processing. RNA 19: 1781–1790. [Europe PMC free article] [Abstract] [Google Scholar]
- Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. 2003. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res 31: 1375–1386. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhao J, Kessler MM, Moore CL. 1997. Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J Biol Chem 272: 10831–10838. [Abstract] [Google Scholar]
- Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. [Abstract] [Google Scholar]
- Zhu H, Zhou HL, Hasman RA, Lou H. 2007. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J Biol Chem 282: 2203–2210. [Abstract] [Google Scholar]
Articles from Genes & Development are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gad.261974.115
Read article for free, from open access legal sources, via Unpaywall:
http://genesdev.cshlp.org/content/29/9/889.full.pdf
Citations & impact
Impact metrics
Article citations
Alternative mRNA polyadenylation regulates macrophage hyperactivation via the autophagy pathway.
Cell Mol Immunol, 13 Nov 2024
Cited by: 0 articles | PMID: 39537902
Thr<sup>4</sup> phosphorylation on RNA Pol II occurs at early transcription regulating 3'-end processing.
Sci Adv, 10(36):eadq0350, 06 Sep 2024
Cited by: 0 articles | PMID: 39241064 | PMCID: PMC11378909
DEAD-box ATPase Dbp2 is the key enzyme in an mRNP assembly checkpoint at the 3'-end of genes and involved in the recycling of cleavage factors.
Nat Commun, 15(1):6829, 09 Aug 2024
Cited by: 0 articles | PMID: 39122693 | PMCID: PMC11315920
SnoRNAs: Exploring Their Implication in Human Diseases.
Int J Mol Sci, 25(13):7202, 29 Jun 2024
Cited by: 0 articles | PMID: 39000310
Review
Identifying human pre-mRNA cleavage and polyadenylation factors by genome-wide CRISPR screens using a dual fluorescence readthrough reporter.
Nucleic Acids Res, 52(8):4483-4501, 01 May 2024
Cited by: 1 article | PMID: 38587191 | PMCID: PMC11077057
Go to all (168) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Recent molecular insights into canonical pre-mRNA 3'-end processing.
Transcription, 11(2):83-96, 01 Apr 2020
Cited by: 42 articles | PMID: 32522085 | PMCID: PMC7549739
Review Free full text in Europe PMC
Biophysical characterizations of the recognition of the AAUAAA polyadenylation signal.
RNA, 25(12):1673-1680, 28 Aug 2019
Cited by: 12 articles | PMID: 31462423 | PMCID: PMC6859858
Understanding RNA Binding by the Nonclassical Zinc Finger Protein CPSF30, a Key Factor in Polyadenylation during Pre-mRNA Processing.
Biochemistry, 60(10):780-790, 22 Feb 2021
Cited by: 1 article | PMID: 33615774 | PMCID: PMC8575367
Reconstitution of 3' end processing of mammalian pre-mRNA reveals a central role of RBBP6.
Genes Dev, 36(3-4):195-209, 17 Feb 2022
Cited by: 22 articles | PMID: 35177537 | PMCID: PMC8887130
Funding
Funders who supported this work.
American Cancer Society (1)
Grant ID: RSG-12-186
HSRD VA (1)
Grant ID: CDP 12-186
NCI NIH HHS (2)
Grant ID: R01 CA177651
Grant ID: CA177651
NIGMS NIH HHS (4)
Grant ID: GM28983
Grant ID: R01 GM090056
Grant ID: R01 GM028983
Grant ID: GM090056
NIH (1)
Grant ID: GM28983
National Institutes of Health (2)
Grant ID: CA177651
Grant ID: GM090056

 1 and
1 and 



