Abstract
Free full text

The Roles of Chromatin-Remodelers and Epigenetic Modifiers in Kidney Cancer
Abstract
Clear cell renal cell carcinoma (ccRCC) is the major subtype of kidney cancer that is characterized by frequent inactivation of von Hippel-Lindau (VHL) gene in 80–90% of the tumors. Recent reports using massive parallel sequencing technologies have discovered more cancer driver genes. PBRM1 was found to be mutated in about 40% of ccRCC tumors, while BAP1 and SETD2 were mutated in about 10–15% of ccRCC tumors each. JARID1C and UTX, two histone H3 demethylases, were also found to harbor mutations in ccRCC, albeit at lower rates. ccRCC tumors display high degree of intra-tumoral heterogeneity, with some mutations present in everyone cancer cells (ubiquitous) while some other subclonal. The VHL mutations were always ubiquitous in the tumors, while PBRM1 mutations were also ubiquitous but to a lesser extent. On the contrary, the mutations in BAP1, SETD2, JARID1C, and UTX were all subclonal, meaning that they were present in a subset of cancer cells in a tumor. The prognosis value of PBRM1 mutations in ccRCC is still controversial, while BAP1 mutations were tightly linked to worse clinical outcome in multiple studies. The molecular functions of these newly identified cancer driver genes were discussed, and they were known reader, writer, or eraser of histone marks on histone H2 and H3 tails that are very close to each other, suggesting that these factors might functionally interact and affect common pathways. The studies on these newly identified tumor suppressors will shed light on ccRCC tumorigenesis and development, and will likely lead to development of novel therapeutic interventions for ccRCC patients.
VHL is the most frequently mutated gene in ccRCC
Inactivation of the VHL tumor suppressor gene is the causal event in the pathogenesis of clear cell renal cell carcinoma (ccRCC). Approximately 75% of the RCCs are of the clear cell type. Among them, 70–80% of ccRCC tumors harbor biallelic inactivation of VHL through mutation, deletion, or hypermethylation of its promoter that results in loss of expression [1]. In hereditary kidney cancer patients, a germline VHL mutation predisposes individuals to earlier onset bilateral kidney cancer. Because one allele is already defective, only the remaining copy of VHL must be altered for biallelic inactivation to occur. pVHL, the protein product of the VHL tumor suppressor gene, encodes the substrate recognition unit of an E3 ubiquitin ligase complex, which also includes Cul2 and Rbx1. This complex targets the α subunits of the heterodimeric transcription factor hypoxia-inducible factor (HIF) for poly-ubiquitylation and proteasomal destruction. pVHL recognizes HIFα only when HIFα is hydroxylated on either of two prolyl residues by members of the EglN family (also called PHDs or HPHs) (Fig. 1). When pVHL is inactivated, HIFα proteins are synthesized, accumulate and form a complex with HIF1β protein. The complex then activates transcriptional response to hypoxia in the nucleus. The constitutively active HIF activity subsequently drives tumorigenesis and growth of ccRCC tumors [2]. Interestingly, not all HIF-induced genes are tumor-promoting [3,4]. pVHL also has HIF-independent functions, but their relevance to tumor suppression remains unclear[5–7].
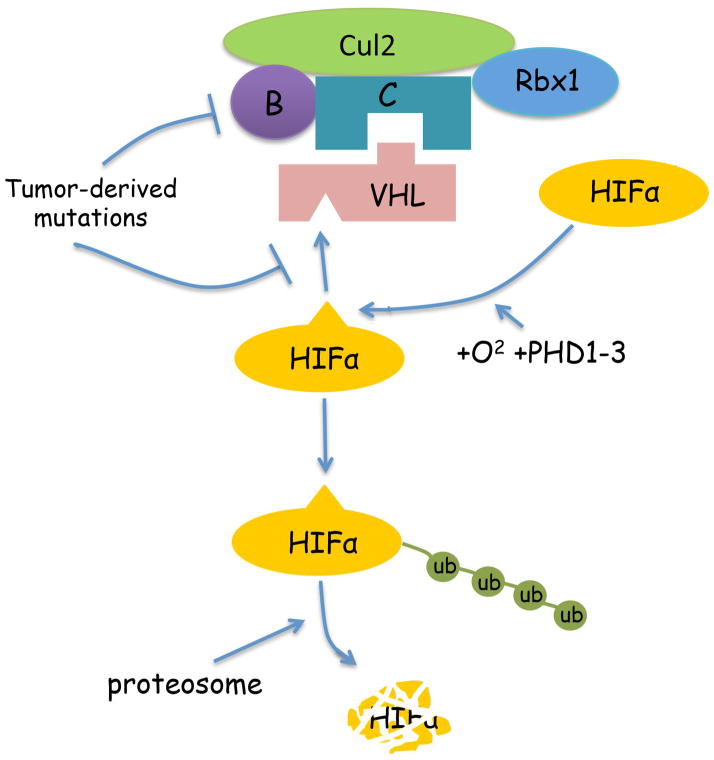
Schematic representation of HIF regulation by pVHL. pVHL is the substrate recognition subunit of an E3 ubiquitin ligase complex that contains Elongin B, C, Cul2 and Rbx1. Hypoxia-inducible factor α subunits (HIFα) are constantly translated. In the presence of oxygen, prolyl hydroxylases promote proline hydroxylation on HIFα proteins, which leads to recognition by pVHL and proteasome-mediated degradation. Tumor derived VHL mutations would either destroy the domain that mediates its interaction with the hydroxyl-prolines on HIFα, or disrupt the domain that is responsible for Elongin C binding, leading to HIFα stabilization and constitutive activation of HIF. This in turn leads to ccRCC initiation and development.
Restoration of pVHL expression and function in VHL-defective kidney cancer cells suppresses their ability to form tumors in nude mice[8], while stabilization of HIF2α in VHL-proficient cancer cells drives tumor growth [9,10]. Conversely, reduction of high-level endogenous HIF2α expression in VHL-defective kidney cancer cells severely blocks their ability to form tumors in a xenograft model [11,12]. Consistent with the notion that an abnormally activated HIF pathway is critical to tumor growth and maintenance, sunitinib, a small molecule antagonist against VEGF and PDGF receptors (which are activated by two HIF-responsive genes), generates a 30% to 40% response rate and a five-month increase in overall survival (OS) of kidney cancer patients [13]. Other anti-angiogenesis drugs such as sorafenib, pazopanib, and axitinib hit the same targets and have similar effects. However, only a third of the patients respond strongly to these drugs, and almost all the tumors will develop drug resistance over time. Given that so many drugs are directed at the same molecular targets and that a large portion of ccRCC patients are not benefiting from them, other drug target(s) are urgently needed.
PBRM1 is another key tumor suppressor in kidney cancer
While VHL is frequently mutated in ccRCC and is has been shown to be essential to development in this disease, it is a well-known fact that most types of cancer harbor multiple driver mutations, sometimes dozens of them, which collectively establish the hallmarks of cancer that are too numerous for one mutation to achieve [14,15]. Large-scale sequencing efforts have been undertaken to uncover genes mutated at a significant level in addition to VHL mutations in ccRCC. Varela et al. reported that 41% of ccRCC tumors harbor inactivating mutations in a SWI/SNF chromatin remodeling complex gene known as PBRM1 [16]. Interestingly, PBRM1 is also mutated in Bladder carcinoma, Chromophobe and Papillary Renal Cell Carcinoma (Table 1), although at lower frequencies. The high rate of PBRM1 mutations in ccRCC have been confirmed by multiple studies, along with mutations in several other tumor suppressor genes, including BAP1, SETD2, JARID1C and PTEN, as well as the histone H3 lysine 27 demethylase gene UTX (KMD6A)[17–21]. However, the mutations of these other genes occur at much lower rates than that of PBRM1.
Table 1
The cancer alteration rates for the described genes in clear cell Renal Cell Carcinoma, Chromophobe Renal cell Carcinoma, Papillary Renal Cell Carcinoma, Prostate Adenocarcinoma, and Urothelial Bladder Carcinoma were obtained from the indicated publications or from cBioportal website. The cancer alteration includes mutation, deletion, amplification and multiple alterations.
| Gene | Cancer Alteration rates in clear cell Renal Cell Carcinoma (TCGA) [20] | Cancer Alteration rates in Chromophobe Renal Cell Carcinoma [81] | Cancer Alteration rates in Papillary Renal Cell Carcinoma (TCGA) | Cancer Alteration rates in Prostate Adenocarcinoma | Cancer Alteration rates in Urothelial Bladder Carcinoma (TCGA) [82] |
|---|---|---|---|---|---|
| PBRM1 | 34.8% | 4.2% | 3.7% (provisional) | 0% (TCGA) | 10% |
| SETD2 | 13.7% | 3% | 6.8% (provisional) | 3.6% [83](Broad/Cornell) | 7.7% |
| BAP1 | 10.8% | 0% | 3.1% (provisional) | 0.9% [83](Broad/Cornell) | 4.6% |
| JARID1C /KDM5C | 6.7% | 3% | 1.2% (provisional) | 9.8% (Amplification) [84] (Michigan) | 1.5% |
| UTX /KDM6A | 1.2% | 0% | 4.3% (provisional) | 9.8% (Michigan) [84] | 26.1% |
PBRM1 is likely a tumor suppressor as the reported mutations are mostly inactivating ones; the main PBRM1 mutations that have been observed are missense, frameshift truncation, frameshift insertion, and nonsense mutations, all of which are inactivating in nature. Knockdown of PBRM1 has been shown to significantly increase cell proliferation, cell migration, and colony formation in soft agar, indicating that PBRM1 loss leads to a transformed phenotype. Transcriptionally, PBRM1 suppression led to altered pathways governing chromosome instability and cell proliferation [16]. The importance of PBRM1 mutations to ccRCC development is further supported by the following discovery: PBRM1 mutations, like those of VHL, occur early in ccRCC tumorigenesis, with mutations being present in all cancer cells within a tumor in many cases. Notably, however, it was discovered through multi-region exome sequencing that ccRCCs display profound intra-tumor heterogeneity [22–24]. The majority of the driver mutations, including those of BAP1, SETD2 and JARID1C, are subclonal, meaning that they occur later during tumor development and are present only in a subpopulation of the cancer cells. Based on the mutations identified from different regions of a tumor, phylogenetic trees of mutations were constructed and the driver mutations were mapped onto them. VHL mutations were ubiquitous and mapped to the trunk of the mutational phylogenetic trees in the all cases, suggesting that it is the fundamental event of ccRCC development. PBRM1 mutations were mapped onto the trunk in ~50% of cases when mutated, suggesting that in many cases PBRM1 mutations happen very early as well. The mutations of all the other genes mentioned above were mapped onto the branches, suggesting that they occur later during tumor development [23]. Collectively, these lines of evidence strongly suggest that, like VHL, PBRM1 is a key tumor suppressor in ccRCC.
To date, the contribution of PBRM1 mutations to the clinical outcome of ccRCC patients has been controversial. Some groups reported that PBRM1 mutations occurred at a similar rate in different tumor stages, and these mutations did not seem to correlate with adverse patient survival[25,26]. However, other groups have reported that PBRM1 mutations are positively linked to tumor invasiveness[27] and, based on IHC findings, loss of PBRM1 protein was associated with advanced tumor stage, high Fuhrman grade and poor overall survival[28,29]. Our own studies of PBRM1 protein loss in ccRCC tissue microarrays also indicate that defects in PBRM1 are strongly linked to higher tumor stages (Jiang, et al., unpublished). Thus, it is possible that PBRM1 protein deficiency itself is strongly tumor promoting, but the dampening effects of other concurrent mutations may mask its effect.
With the high percentage of ccRCC tumors carrying PBRM1 mutations and deficiency, understanding its molecular functions, its contribution to clinical disease progression and outcome, its interaction of other mutated genes in ccRCC, and how to suppress or kill PBRM1-deficient ccRCC cancer cells therapeutically are important areas of renal cancer research. It is worth mentioning that expression of ARID1A, another targeting subunit of the SWI/SNF complex that competes with PBRM1 for binding to core subunits, is absent or downregulated in more than half of ccRCC tumors [30,31]. Since its mutation rate in ccRCC is low[16,20], the widespread loss of protein expression likely arises through other mechanisms such as promoter methylation and/or silencing by microRNA.
PBRM1’s molecular and biological functions
PBRM1 was originally discovered as BAF180, which is a component of a SWI/SNF-B complex that resembles the yeast Rsc chromatin remodeling complex [32]. This complex also contains BRD7 and BAF200 as the specificity subunits [33]. This complex is called PBAF to distinguish it from the BAF complex that has ARID1A or ARID1B as specificity subunits. Both PBAF and BAF complexes share the core subunits such as BAF155/BAF170, BAF57, BAF47, ACTL6, BAF60, and the enzymatic subunits BRG1. Both complexes possess ATP-dependent mono-nucleosome disruption activity that can slide or eject nucleosomes on DNA [32]. PBRM1 deficiency in mice leads to embryonic lethality[34], thought to result from a failure in cardiac chamber maturation. It was later discovered that PBRM1-deficient mice more likely have a failure in coronary vessel development [35]. Deletion of PBRM1 in mouse T cells enhanced Th2 differentiation and significantly elevated IL-10 expression [36]. PBRM1 is mutated in some human breast cancers, and its re-expression inhibits cell proliferation and induces p21 expression [37]. Furthermore, PBRM1 and BRD7 are found to be critical for p53 to induce replicative senescence [38]. Interestingly, genes that are regulated by PBRM1 are highly dependent on the cell state, as its repression targets are very different in resting and stimulated Th2 cells [36]. PBRM1 was also found to be required for retinoic acid-dependent gene activation [34]. Recently, PBRM1 was found to be required for centromeric cohesion and prevention of genomic instability [39], and it is also reported to help repress transcription and repair at DNA damage sites [40]. Some tumor-derived PBRM1 mutants failed to perform these functions, suggesting that they might contribute to PBRM1’s tumor suppressor functions [39,40].
The bromodomain 2 of PBRM1 has high affinity for acetylated K14 on histone H3 (H3K14Ac)
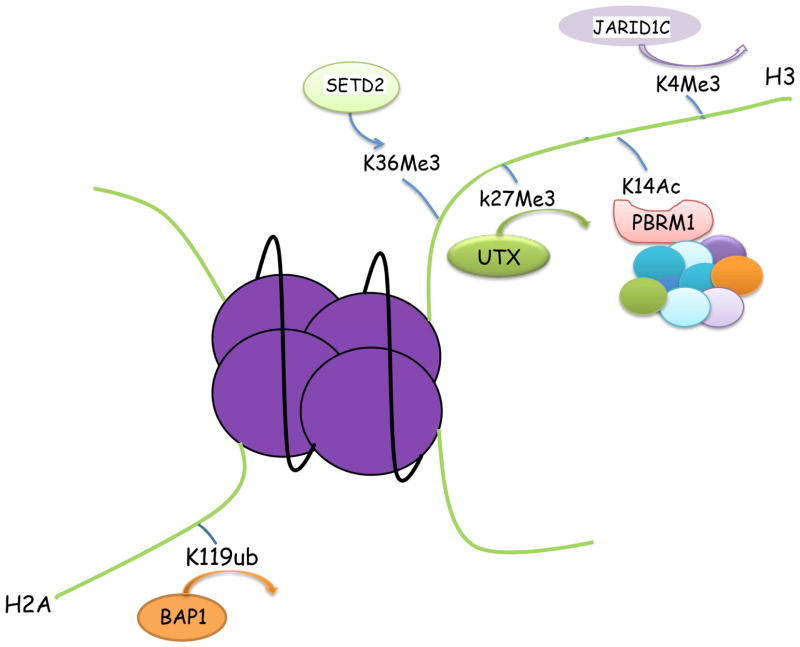
The PBRM1 protein contains six bromodomains, two bromo-adjacent homology (BAH) domains, and one high-mobility-group (HMG) motif (Fig. 2). The bromodomains are well-characterized motifs that bind to acetylated lysine [41]. The BAH domains are proposed to be involved in protein-protein interactions, and the HMG motif has been shown to bind to DNA [42]. It was hypothesized that PBRM1’s ability to regulate gene expression is dependent upon its bromodomain-mediated interaction with lysine-acetylated histones. In one investigation, individual bromodomains of PBRM1 displayed high affinity for different acetylation sites on histone H3 tails, with the dissociation constants (Kd) in the sub-micromolar range [43]. Another report suggested that the bromodomain 2 of PBRM1 preferentially recognizes the acetylated lysine 14 on histone H3 (H3K14Ac) (Fig. 3), with a Kd of about 500μM [44]. Because of the many discrepancies among these reports, it will be important to characterize the specificity of full-length PBRM1 toward acetylated-lysines on histone H3. It is highly probable that PBRM1, through its multiple bromodomains, would interact with other lysine-acetylated tumor suppressors or oncogenes to exert its tumor suppressor functions.
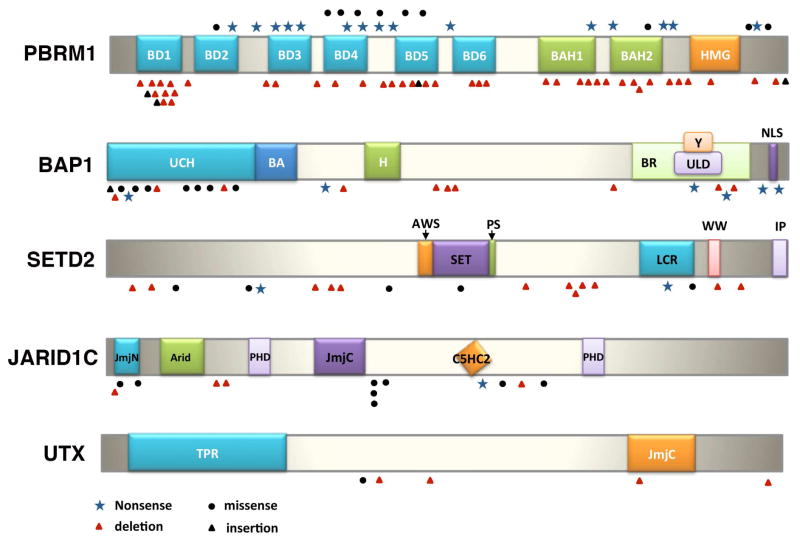
The tumor-derived somatic mutations on the newly discovered cancer genes in ccRCC. PBRM1 has six bromodomains (known to bind to acetyl lysines), two BAH domains (postulated for protein-protein interaction) and one HMG domain (known to bind to DNA), and the mutations are inactivating ones [16]. BAP1 has an UCH domain that possesses deubiquitinase activity, and an H domain that mediates interaction with HCF-1. There are numerous mutations affecting the UCH domain, including many missense mutations[19]. The SET domain in SETD2 is required for its histone demethylase activity, and the mutations seem to be evenly distributed[27,85]. The Jumonji domains in JARID1C are important for its histone demethylase activity. Many identified mutations are either inside or near the Jumonji domains[27]. The Jumonji domain is also critical to the histone demethylase activity of UTX[79].
BAP1 is a key tumor suppressor in ccRCC and other cancers
Inactivating mutations of the BRCA1 associated protein-1 gene (BAP1) gene have been reported in 10–15% of ccRCC[18,19] (Fig. 2 and Table 1). In other urological malignancies such as Bladder carcinoma and Papillary Renal Cell Carcinoma, BAP1 mutations are also found at lower frequencies (Table 1). BAP1 was initially discovered as a BRCA1-interacting protein in a yeast two-hybrid screen[45], and BAP1 protein inhibited the E3 ubiquitin ligase activity of BRCA1/BARD1 complex [46]. However, this interaction was not easy to detect in some systems, suggesting that they might function together under specific conditions. Notably, the BAP1 gene is located on the short (p) arm of chromosome 3, at band location 3p21, very near (<140,000 base pairs away) from the PBRM1 gene. SETD2 and VHL are also located in chromosome 3p, and allelic losses of all four of these tumor suppressor genes are frequently observed in ccRCC tumors due to chromosomal deletions of large segments of 3p. As with VHL, PBRM1 and SETD2, biallelic inactivation of BAP1 via mutation and allelic loss of the remaining wild type allele fit a two-hit tumor suppressor profile.
Germline mutations of BAP1 predispose patients to ccRCC and other type of cancers
Somatic mutations of BAP1 have been reported in nearly 85% of metastasizing uveal melanomas and in 20–25% of malignant pleural mesotheliomas [47–49]. In 2011, Testa et al. reported germline mutations of BAP1 in two families with multiple mesothelioma and various other malignant tumors[49]. One of the two families also exhibited two uveal melanomas and two skin cancers, whereas several BAP1 mutation carriers in the second family were afflicted with carcinomas of the kidney, breast or ovary. In a concurrent report, Wiesner et al. described germline BAP1 mutations in two families with high incidence of benign atypical melanocytic tumors as well as several uveal melanomas and cutaneous melanomas[50]. Collectively, the observations in these high-risk cancer families have led investigators to propose the existence a BAP1 cancer susceptibility syndrome characterized by a high incidence of malignant mesothelioma, uveal and other melanocytic neoplasms, and most likely in other cancers[51]. The latter possibility is now certain, based on the subsequent identification of multiple BAP1 families with one or more ccRCCs[51–53]. Other studies suggest that the clinical phenotype of the BAP1 syndrome may be expanded to other cancers such as lung adenocarcinoma, meningioma, breast carcinoma and paraganglioma[54–57].
In vivo studies have revealed that systemic or hematopoietic-restricted homozygous deletion of Bap1 in adult mice recapitulates features of human myelodysplastic syndrome[58]. Bap1 deletion was found to result in decreased levels of host cell factor-1 (HCF-1) and O-linked N-acetylglucosamine transferase, indicating a critical role for BAP1 in stabilizing these epigenetic regulators. To address experimentally the question of why mesothelioma is the predominate malignancy in some BAP1 syndrome families but not in others, Xu et al. exposed heterozygous Bap1 knockout mice to asbestos[59]. They found that germline mutation of Bap1 accelerates the rate of development and progression of asbestos-induced mesothelioma, although no spontaneous mesotheliomas were seen in unexposed Bap1 knockout mice followed for up to 21 months of age. Drawing parallels to the human disease counterpart, these in vivo genetic findings indicate that BAP1 mutation carriers are predisposed to the tumorigenic effects of asbestos and suggest that high penetrance of mesothelioma requires such environmental exposure.
The molecular functions of BAP1 that might be important to its tumor suppressor functions
BAP1 is a histone deubiquitinase, with the ubiquitin C-terminal hydrolase (UCH) domain located at its N-terminal domain (Fig. 2). It also harbors a BARD1 interaction domain (BA), and its interaction with HCF-1 needs the NHNY sequence (H). The C-terminus of BAP1 also contains a UCH37-like domain (ULD), a YY1-bindng domain (Y), and two nuclear localization signals (NLS) (Fig. 2). BAP1 was found to deubiquitinate the mono-ubiquitinated K119 residue on histone 2A (Fig. 3), and possibly other proteins as well[58,60]. However, its ability to repress cell growth did not seem to be associated with its ability to deubiquitinate H2A. As BAP1 can both promote and suppress cell proliferation based on the cell types, its ability to repress cellular growth in vitro does not necessarily correlate with its major tumor suppressor function in vivo[61]. Indeed, even though BAP1’s deubiquitinase activity is not required to suppress cell growth in vitro, its N-terminal UCH region is often targeted by tumor-derived mutations, suggesting that this activity is important to it to be a tumor suppressor.
In addition to its deubiquitinase activity, BAP1 also has a HBM motif that mediates the interaction with HCF-1, which could serve as a chromatin scaffold for a few chromatin-remodeling complexes[62–64]. Interestingly, BAP1 with a mutation in the HBM motif that failed to interact with HCF-1 also failed to repress cell growth in RCC cells[19]. It remains to be determined whether its deubiquitinase activity and/or its ability to interact with HCF-1 is required for its tumor suppressor function.
In addition to a potentially critical role for BAP1 inactivation in epigenetic deregulation in ccRCC, BAP1 has been found to promote DNA double-strand break repair by homologous recombination, thereby adding to its tumor suppressor function[24]. In other work, germline mutations in BAP1 were found to impair its function in DNA double-strand break repair[65]. BAP1 was shown to mediate rapid poly(ADP-ribose)-dependent recruitment of the polycomb deubiquitylase complex PR-DUB to sites of DNA damage and was identified as a phosphorylation target for the ATM, further suggesting a DNA repair response mechanism that also contributes to the tumor-suppressive function of BAP1.
BAP1 mutations are not ubiquitous in ccRCC but do associate with tumor aggressiveness and poor outcome
The mutations rate of BAP1 was 10–15% compared to PBRM1’s 30–40%. The mutations of BAP1 are mostly inactivating mutations (Fig. 2). Multiple studies have revealed that in a substantial portion of ccRCC tumors with BAP1 mutations, the mutations are subclonal and the protein losses are focal[21–24,66]. Such tumor heterogeneity suggests that BAP1 alterations occur late in disease progression. Interestingly, in ccRCC tumors, the mutations of BAP1 and PBRM1 are largely mutually exclusive[19,25]. Such a reciprocal pattern usually occurs when two genetic mutations affect the same pathway, such that there is no selection pressure to mutate the second gene once the first is inactivated. However, this does not seem to be the case for BAP1 and PBRM1, as both can occur in the same tumor.
Using whole-genome and exome sequencing, Peña-Llopis et al. identified inactivating mutations of BAP1 in 15% of ccRCCs, and BAP1 loss was associated with high tumor grade[19]. In a retrospective analysis of 145 patients with primary ccRCC, Kapur et al. reported that median overall survival was significantly shorter for patients with BAP1-mutant tumors than for patients with PBRM1-mutant tumors[25]. A second independent cohort from The Cancer Genome Atlas (TCGA) was used for validation, with similar differences between these two patient groups. Similarly, a study of 609 patients with ccRCC from two distinct cohorts revealed that mutations of 3p21 epigenetic regulators BAP1 and SETD2 were associated with worse cancer-specific survival (CSS), suggesting that they play key roles in disease progression[26]. In an independent European study of ccRCC, BAP1-mutated tumors were associated with metastatic disease at presentation, advanced clinical stage, and a trend towards shorter recurrence free survival when compared with tumors exclusively mutated for PBRM1[67]. These results provide further support indicating a potential role for BAP1 and PBRM1 mutations in the risk stratification of ccRCCs. Based on their roles as ccRCC driver genes and the function of their gene products, investigators have proposed ways in which mutations in these genes may be exploited therapeutically[68].
SETD2 gene
SETD2 protein is a histone H3 Lysine 36 trimethylating (H3K36Me3) enzyme (Fig. 3), and it is mutated in 10–15% of ccRCC tumors[17,20,21,26] (Fig. 2 and Table 1). In other urological malignancies SETD2 mutations are found as well (Table 1). As mentioned before it is also located on chromosome 3p and it is a two-hit tumor suppressor. Mutational analysis indicated that SETD2 mutations tended to be subclonal[22–24], and sampling analysis found that different SETD2 mutations could exist in different regions of the same tumor[22], suggesting high selection pressure for SETD2 mutations. It was suggested that SETD2 mutations were associated with worse cancer specific survival[26]. Furthermore, SETD2 mutations were found to occur much more frequently when PBRM1 was mutated, suggesting that these mutations were cooperating with each other[69].
It is not clear how SETD2 mutations lead to ccRCC. Without SETD2, the H3K36Me3 mark on histone is significantly down-regulated. This mark is usually associated with active transcription, although it is also reported to be associated with alternative splicing, transcriptional repression[70]. Indeed, it was reported that SETD2 mutations the ccRCC tumors associated with changes in chromatin accessibility and DNA methylation[71], and another group reported that SETD2 mutations were linked to variation in chromatin accessibility and widespread RNA processing defects[72]. Multiple reported also revealed that SETD2 played an important role in DNA damage repair through promoting homologous recombination, mismatch repair, and activation of p53-mediated checkpoint[73–75]. It remains to be seen which function of SETD2 plays the most significant role in ccRCC tumor suppression, and whether it is worthwhile to target the dysfunctional pathway associated with SETD2 inactivation in ccRCC.
JARID1C/KDM5C
JARID1C/KDM5A is a histone demethylase that takes off methyl group from lysine 4 on histone H3 (H3K4Me3) (Fig. 3). Its gene was inactivated with mutations in 3–7% of ccRCC tumors[16,17,20,21] (Fig. 2 and Table 1). Interestingly, its mutations were mostly subclonal, and if multiple regions of ccRCC tumors were sampled, 25% of them would harbor JARID1C mutations[24]. JARID1C/KDM5A mutations or amplification are also identified in other urological malignancies (Table 1). Its prognosis value is unknown, but JARID1C was found to be a HIF target[3]. Without JARID1C, the overall level of H3K4Me3 in VHL-defective kidney cancer cells would go up, and these cancer cells would form much larger tumors in a xenograft model, suggesting that HIF-induced JARID1C expression and function constituted a negative feedback loop in regard to tumor growth[3]. H3K4Me3 is a histone mark that is tightly linked to actively transcribing gene[76] as it provides the targeting site for TFIID[77]. Recently it was found that H3K4Me3 breadth is linked to cell identity, and the broadest H3K4Me3 domains exhibited enhanced transcriptional consistency rather than transcriptional levels[78].
UTX/KDM6A
UTX/KDM6A is a histone H3 lysine 27 demethylase (Fig. 3). It was mutated at a low frequency in ccRCC tumors [17,79] (Fig. 2 and Table 1). Strikingly, its mutation rate in Bladder cancer is quite high (26.1%), and its mutations are also discovered in Prostate adenocarcinoma and Papillary Renal Cell Carcinoma (Table 1). Very little is known how UTX mutations could impact ccRCC. However, it is worth noting that H3K27Me3 is the most prominent histone mark that is associated with repressed transcription[76], and the enzymes that modify this site such as EZH2 and UTX were frequently mutated and/or altered in human cancers[79,80]. How UTX inactivation contribute to ccRCC tumor biology remains to be determined.
Conclusion
The field of kidney cancer research had focused on VHL-HIF axis for two decades. It produced tremendous amount of insights and led to the development of anti-angiogenesis therapies against ccRCC, which are partially and transiently effective. The complete sequencing of the kidney cancer genomes in the last few years revealed that many chromatin regulators such as histone code reader (PBRM1), writer (SETD2), and erasers (BAP1, JARID1C and UTX) were also mutated in ccRCC to various extents, suggesting that these factors could contribute to ccRCC initiation, development, and evolvement into more aggressive states. It is also worth noting that the same genes are also significantly altered in other subtypes of kidney cancer and other urological malignancies (Table 1), indicating the pan-cancer significance of these genes, and suggesting that the lessons learned from ccRCC might be applicable to other cancers as well. Interestingly, even though the chromatin landscape is vast, these proteins all targeted toward a crowded area of histone H2 and H3 tails. Some of these sites were only a few amino acids apart (Fig. 3), suggesting that these proteins might be functionally linked. Currently their studies in the cancer biology are still in early stage, so it is hard to predict or speculate their therapeutic relevance. As they are tumor suppressor genes, direct inhibition of their activities is unlikely to generate positive outcome. However, as the story of VHL/HIF indicates, the loss of an important tumor suppressor will lead to the activation of major oncoprotein(s), whose inhibition could be therapeutically beneficial. Undoubtedly the intense studies of these factors in ccRCC will provide significant insights into how their losses contribute, either alone or in cooperation with HIF and/or each other, to cancer biology of ccRCC tumors. These studies will likely lead to the identification of new target(s) and development of new therapeutics against ccRCC tumors that are sorely needed.
Acknowledgments
Work by J.R.T. involving BAP1 is supported in part by NCI Grant R01 CA175691. J.R.T. also received support from Fox Chase Cancer Center’s Personalized Kidney Cancer Therapy Keystone Program. Work by H. Y. involving JARID1C is partially supported by National Institute of Health/NCI grant R01 CA155015. We also thank Mitchell Cheung in Dr. Testa’s lab for his suggestion on BAP1-related literature, and we thank Dr. Frank J. Rauscher III for the clarification on BAP1-BRCA1 connection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cancergen.2015.02.008
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4494076?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1016/j.cancergen.2015.02.008
Article citations
AURKB promotes immunogenicity and immune infiltration in clear cell renal cell carcinoma.
Discov Oncol, 15(1):286, 16 Jul 2024
Cited by: 0 articles | PMID: 39014265 | PMCID: PMC11252114
Histone demethylase KDM6A coordinating with KMT2B regulates self-renewal and chemoresistance of non-small cell lung cancer stem cells.
Transl Oncol, 37:101778, 06 Sep 2023
Cited by: 2 articles | PMID: 37683307 | PMCID: PMC10493599
Rare germline heterozygous missense variants in BRCA1-associated protein 1, BAP1, cause a syndromic neurodevelopmental disorder.
Am J Hum Genet, 109(2):361-372, 19 Jan 2022
Cited by: 8 articles | PMID: 35051358 | PMCID: PMC8874225
Background, applications and challenges of radiogenomics in genitourinary tumor.
Am J Cancer Res, 11(5):1936-1945, 15 May 2021
Cited by: 3 articles | PMID: 34094662 | PMCID: PMC8167692
Review Free full text in Europe PMC
HIF-1-Independent Mechanisms Regulating Metabolic Adaptation in Hypoxic Cancer Cells.
Cells, 10(9):2371, 09 Sep 2021
Cited by: 47 articles | PMID: 34572020 | PMCID: PMC8472468
Review Free full text in Europe PMC
Go to all (38) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Clinical and pathological impact of VHL, PBRM1, BAP1, SETD2, KDM6A, and JARID1c in clear cell renal cell carcinoma.
Genes Chromosomes Cancer, 53(1):38-51, 29 Oct 2013
Cited by: 77 articles | PMID: 24166983
Aberrant promoter hypermethylation of PBRM1, BAP1, SETD2, KDM6A and other chromatin-modifying genes is absent or rare in clear cell RCC.
Epigenetics, 8(5):486-493, 01 May 2013
Cited by: 35 articles | PMID: 23644518 | PMCID: PMC3741218
PBRM1 and BAP1 as novel targets for renal cell carcinoma.
Cancer J, 19(4):324-332, 01 Jul 2013
Cited by: 66 articles | PMID: 23867514 | PMCID: PMC4222578
BAP1, PBRM1 and SETD2 in clear-cell renal cell carcinoma: molecular diagnostics and possible targets for personalized therapies.
Expert Rev Mol Diagn, 15(9):1201-1210, 11 Jul 2015
Cited by: 53 articles | PMID: 26166446
Review
Funding
Funders who supported this work.
Fox Chase Cancer Center's Personalized Kidney Cancer Therapy Keystone Program
NCI (1)
Grant ID: R01 CA155015
NCI NIH HHS (3)
Grant ID: P30 CA006927
Grant ID: R01 CA155015
Grant ID: R01 CA175691
NIEHS NIH HHS (1)
Grant ID: P30 ES013508
National Cancer Institute (1)
Grant ID: R01 CA175691