Abstract
Free full text

Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity
Associated Data
Abstract
Earlier studies reported allelic deletion of the essential autophagy regulator BECN1 in breast cancers implicating BECN1 loss, and likely defective autophagy, in tumorigenesis. Recent studies have questioned the tumor suppressive role of autophagy, as autophagy-related gene (Atg) defects generally suppress tumorigenesis in well-characterized mouse tumor models. We now report that, while it delays or does not alter mammary tumorigenesis driven by Palb2 loss or ERBB2 and PyMT overexpression, monoallelic Becn1 loss promotes mammary tumor development in 2 specific contexts, namely following parity and in association with wingless-type MMTV integration site family, member 1 (WNT1) activation. Our studies demonstrate that Becn1 heterozygosity, which results in immature mammary epithelial cell expansion and aberrant TNFRSF11A/TNR11/RANK (tumor necrosis factor receptor superfamily, member 11a, NFKB activator) signaling, promotes mammary tumorigenesis in multiparous FVB/N mice and in cooperation with the progenitor cell-transforming WNT1 oncogene. Similar to our Becn1+/−;MMTV-Wnt1 mouse model, low BECN1 expression and an activated WNT pathway gene signature correlate with the triple-negative subtype, TNFRSF11A axis activation and poor prognosis in human breast cancers. Our results suggest that BECN1 may have nonautophagy-related roles in mammary development, provide insight in the seemingly paradoxical roles of BECN1 in tumorigenesis, and constitute the basis for further studies on the pathophysiology and treatment of clinically aggressive triple negative breast cancers (TNBCs).
Introduction
Macroautophagy (hereforth referred to as autophagy) is a catabolic process whereby protein aggregates and damaged organelles are constitutively degraded in lysosomes.1 Cells also activate autophagy to survive under stress by recycling cytoplasmic material for energy production and biomolecular synthesis.1 Defective autophagy was first implicated in tumorigenesis with the report that the essential autophagy regulator BECN1/Beclin1 is allelically deleted in human cancers, including breast cancer,2 restoration of BECN1 expression in MCF7 breast carcinoma cells compromises xenograft tumor growth,3 and Becn1+/− mice develop spontaneous lung and liver carcinomas, lymphomas, and mammary hyperplasias.4,5 Furthermore, apoptosis-defective Becn1+/− immortalized mouse mammary epithelial cells (iMMECs) are more tumorigenic in nude mice, accumulate DNA damage and are more genomically unstable than their wild-type counterparts.6 Recently, work from Rosenfeldt and colleagues eloquently describes synergy between defective autophagy and tumor protein 53 (TP53/Trp53/p53, note that the mouse nomenclature is Trp53, but we use the TP53 acronym hereafter to refer to both the human and mouse genes/proteins for simplicity) loss in KRAS (Kirsten rat sarcoma viral oncogene homolog)-driven pancreatic cancer,7 while studies investigating the role of functional autophagy in ERBB2 (v-erb-b2 avian erythoblastic leukemia viral oncogene homolog 2)-positive and EGFR/ERBB1 (epidermal growth factor receptor)-expressing cells also implicate autophagy suppression in cell transformation driven by these oncogenes in mammary8 and lung9 epithelium, respectively.
Seemingly contradicting the results described above, several studies have reported that functional autophagy is required for tumorigenesis and tumor progression in some contexts.7,10-14 Recent publications shed light on the role of BECN1, and autophagy in general, in mammary physiology, as ATG proteins have been found important for breast cancer stem cell maintenance10 and dead cell clearance in mammary involution.15 The role of BECN1 in mammary tumorigenesis is likely complex, as mammary glands (MGs) from Becn1+/− mice exhibit increased proliferation in puberty and hyperplasias with age, but no mammary tumors.4 Furthermore, monoallelic Becn1 loss does not have an impact on ERBB2- or PyMT-driven mammary tumorigenesis,8 whereas it delays tumor formation due to mammary gland-specific biallelic Palb2 deletion in a wild-type Tp53 background.13 The tumor-suppressive role of BECN1 in tumorigenesis has also recently been challenged by the finding that in human breast and ovarian cancers, large genomic deletions encompassing both BRCA1 and BECN1, and deletions of only BRCA1 but not BECN1, are observed.16
Our earlier study demonstrates that phosphorylated keratin 8 (p-KRT8) accumulates in autophagy-deficient iMMECs, tissues, and tumors, implicating autophagy in keratin homeostasis.17 Building on this work, we now report that Becn1+/−, compared with Becn1+/+, iMMECs and iMMEC-generated tumors also exhibited KRT6 (keratin 6) upregulation. KRT6 is not only a basal cell marker in skin and mammary tissue, but also a marker for bipotent mammary progenitor cells and for cells with increased tumorigenic potential.18,19 We also report that KRT6-positive cells, likely representing bipotent mammary progenitor cells, are aberrantly present in MGs from postpubertal Becn1+/− mice. Furthermore, we demonstrate that monoallelic Becn1 loss results in augmented mammary stem and progenitor cell activities and TNFRSF11A-NFKB axis upregulation; it also promotes mammary tumor development following parity and accelerates WNT1-driven mammary tumorigenesis, which itself involves progenitor cell transformation.18,20 Thus, our studies elucidate 2 physiologically relevant contexts in which monoallelic Becn1 loss promotes mammary tumorigenesis, giving rise to tumors with basal-like characteristics.
Results
Monoallelic Becn1 loss results in KRT6 upregulation in mammary epithelial cells and tissues
Apoptosis-defective Becn1+/− iMMECs have previously been reported to be more tumorigenic in nude mice than their Becn1+/+ counterparts.6 To investigate Becn1 genotype-specific differences in iMMECs and iMMEC-generated mammary tumors in nude mice, we performed gene expression analysis, which revealed that KRT6 was upregulated in Becn1+/− iMMECs (Fig. 1A and B) and, to an even higher and significant degree, in Becn1+/− iMMEC-generated allograft tumors (Fig. 1A and C). These tumors also displayed increased KRT14 expression compared with tumors resulting from orthotopic implantation of Becn1+/+ iMMECs (Fig. 1C), further indicating an association between monoallelic Becn1 loss and basal keratin expression in mammary tumor cells. This finding is in agreement with our recently published study, which reports that low BECN1 mRNA levels correlate with the basal-like and ERBB2, but not luminal, breast cancer subtypes.8
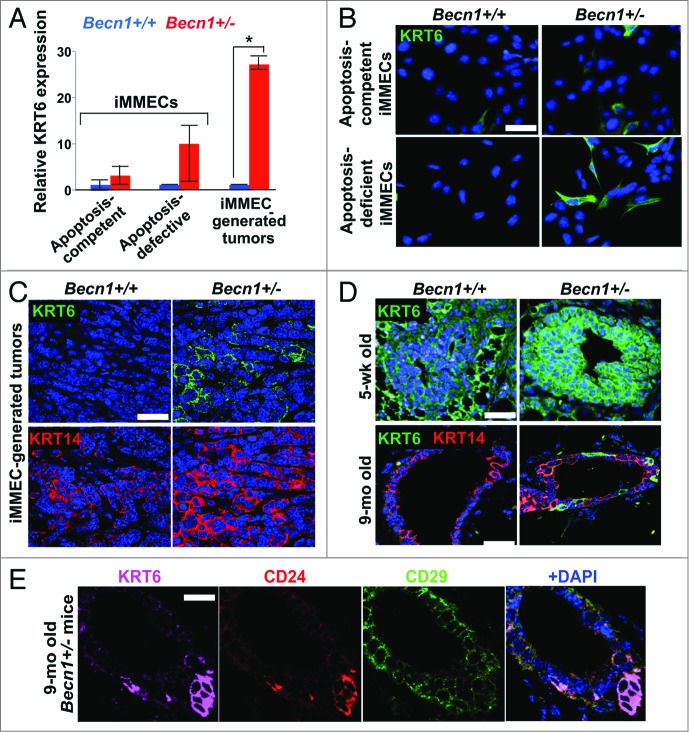
Monoallelic Becn1 loss results in KRT6 upregulation in mammary epithelial cells and tissues. (A) Microarray analysis was performed on samples (three each) of Becn1+/+ and Becn1+/− apoptosis-competent and apoptosis-defective iMMECS, as well as mammary tumors derived from orthotopic implantation of apoptosis-defective iMMECs in nude mice. KRT6 expression relative to that in apoptosis-competent Becn1+/+ iMMECs is presented as means ± SDs (n = 3). (B–D) Becn1+/+ (left) and Becn1+/− (right) with DAPI (blue) by IF: (B) More KRT6-expressing cells are seen in Becn1+/− apoptosis-competent iMMECs (top) and Becn1+/− apoptosis-defective iMMECs (bottom) under normal growth conditions. (C) More KRT6-expressing (green, top) and KRT14-expressing (red, bottom) cells are seen in allograft mammary tumors generated by Becn1+/− apoptosis-defective iMMECs. (D) MGs from 5-wk-old (top) and 9-mo-old (bottom) Becn1+/− mice reveals increased number of KRT6-positive (green) cells that are not KRT8-positive (red). (E) Costaining for KRT6 (purple), CD24 (red), CD29 (green) in MGs from 9-mo-old Becn1+/− mice reveals that KRT6-positve cells colocalize with mammary progenitor cell markers. Scale bar: (B and D) 30 μm; (C and E) 37.5 μm.
To exclude the possibility that KRT6 upregulation in Becn1+/− iMMECs and iMMEC-generated tumors resulted from the process of mammary epithelial cell (MEC) immortalization,6 we investigated KRT6 expression in MGs from Becn1+/+ and Becn1+/− mice. As previously reported,21 KRT6 was detected in MGs from pubertal wild-type mice, whereas hardly any KRT6-positive cells were seen in MGs from aged wild-type mice (Fig. 1D). Mammary epithelium from pubertal Becn1+/− mice exhibited uniformly robust KRT6 expression, whereas KRT6-positive, not costaining with the luminal cytokeratin KRT8, cells were observed in MGs from aged Becn1+/− mice (Fig. 1D).
Since bipotent mammary progenitor cells express KRT6,19 we examined whether KRT6-positive cells observed in MGs from older Becn1+/− mice also expressed mammary stem cell (MaSC) and progenitor cell markers.22,23 We indeed found that these cells costained with the MaSC and progenitor cell marker CD24 (cluster differentiation 24) and expressed low levels of the MaSC marker CD29/integrin-β1 (cluster differentiation 29) (Fig. 1E), thus suggesting that monoallelic Becn1 loss leads to accumulation of KRT6-positive bipotent mammary progenitor cells.
Monoallelic Becn1 loss results in accelerated mammary fat pad filling and excessive ductal side-branching
In agreement with earlier results using a different Becn1+/− transgenic line,4 we also detected increased proliferation in mammary ducts and terminal end buds (TEBs) from 5-wk-old Becn1+/− mice5 (Fig. S1A and S1B); apoptosis rates were similar (Fig. S1C and S1D). Consistent with the partially defective autophagic response observed in Becn1+/− iMMECs under metabolic stress,6 lower levels of MAP1LC3B/LC3B (microtubule-associated protein 1 light chain 3 β) and accumulation of SQSTM1/p62 (sequestosome 1) were observed in MGs from Becn1+/− mice (Fig. S1E and S1F).
Systematic examination of MGs from cohoused Becn1+/+ and Becn1+/− littermates revealed that 6.5-wk-old Becn1+/− mice exhibited accelerated mammary fat-pad filling and increased ductal side-branching (Fig. 2A and B). The latter phenotype persisted and was further accentuated in MGs from 6-mo-old nulliparous Becn1+/− mice (Fig. 2C, upper panel); at that age, Becn1+/− mice also exhibited mammary hyperplasias (Fig. 2C, lower panel). The difference in ductal side-branching in MGs from Becn1+/+ and Becn1+/− mice became less pronounced by 12 mo of age (Fig. 2D), possibly indicating that increased ductal side-branching may be a hormonally regulated phenotype.
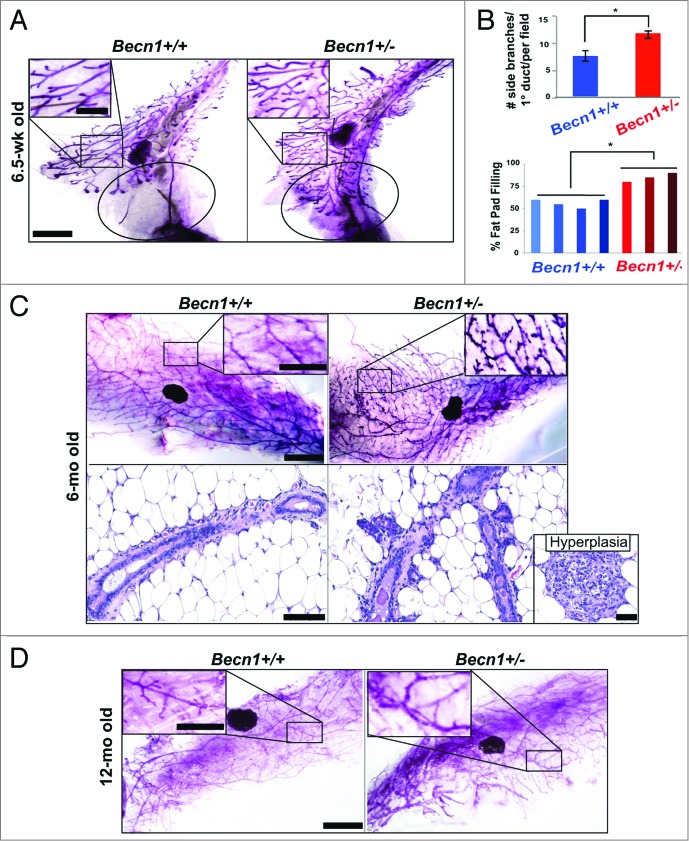
Monoallelic Becn1 loss results in accelerated mammary fat pad filling and excessive ductal side-branching. (A–D) Analysis of mouse mammary epithelium from Becn1+/+ (left) and Becn1+/− (right) mice (n = 3 to 5): (A) MG whole mounts from 6.5-wk-old mice, with circle identifying an unfilled (left) vs. filled (right) fat pad and (B) (top) quantification of number of side-branches per primary branch per field and (bottom) quantification of percentage of mammary fat pad 4 filled with epithelium. Three MG specimen per genotype from cohoused Becn1+/+ and Becn1+/− littermates were evaluated. Results are presented as means ± SDs. (C) MG whole mounts (top) and hematoxylin and eosin staining of MG sections (bottom) from 6-mo-old mice, with mammary hyperplasia in separate insert. (D) MG whole mounts from 12-mo-old mice. Scale bar: (A, C,and D) 5 mm for whole mounts; (A, C, and D) 2 mm for whole mount enlargements. Scale bar: (C) 100 μm for H&Es; 200 μm for hyperplasia. *P < 0.05 by a 2-tailed Student t test.
Monoallelic Becn1 loss results in functionally enriched mammary stem and progenitor cell populations
Faster mammary fat pad filling and excessive ductal side-branching result from increased mammary stem and progenitor cell populations/activity, respectively.22 We utilized fluorescence-activated cell sorting (FACS) to isolate PI− LIN− CD24+ CD29hi, i.e. mammary stem cell (MaSC)-containing, and PI− LIN− CD24+ CD29lo, i.e. mammary progenitor cell (MaPC)-containing, populations22 from cohoused Becn1+/+ and Becn1+/− littermates. Although the relative proportions of MaSC-, MaPC-, and CD61 (cluster differentiation 61)+ luminal progenitor cell-containing populations22-24 were similar between the 2 genotypes (Fig. S2A and S2B), MGs from Becn1+/− mice consistently demonstrated higher MaSC and MaPC activities by in vitro and in vivo functional assays (Fig. 3; Fig. S2C and S2D). Specifically, CD24+ CD29hi Becn1+/− MECs exhibited increased MaSC activity, as measured by primary and limiting dilution mammosphere (Fig. 3A, left and right panel, respectively) and 3D colony-formation (Fig. 3B) assays.23,25 Furthermore, CD24+ CD29lo Becn1+/− MECs exhibited increased MaPC activity (Fig. 3C), as measured by number of colonies formed in 2D-matrigel assays.23,25
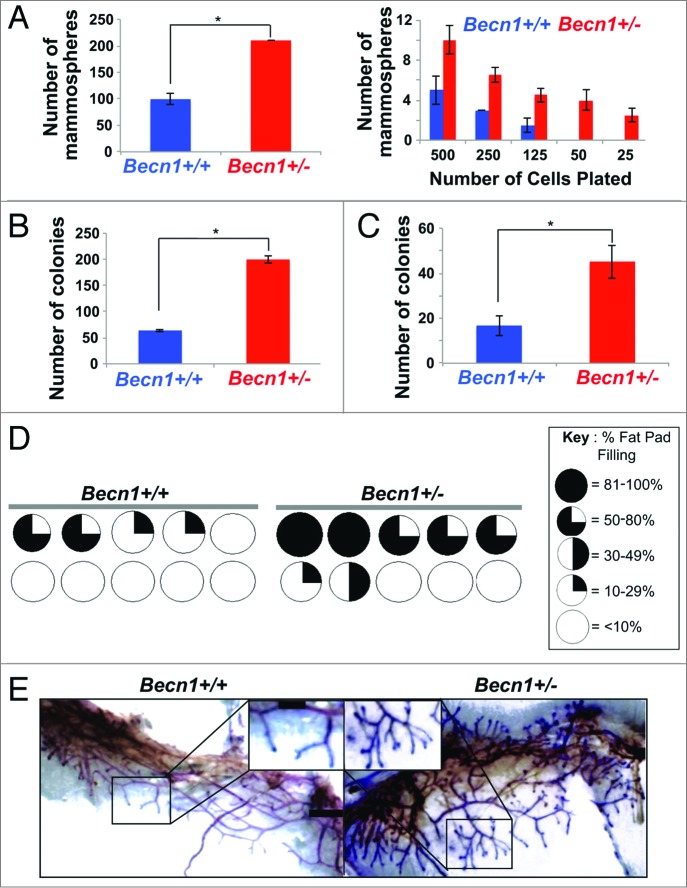
Monoallelic Becn1 loss results in functionally enriched mammary stem and progenitor cell populations. (A–C) Freshly isolated Becn1+/+ (blue) and Becn1+/− (red) MECs were used, experiments were performed 3 independent times and outcomes are presented as means ± SDs. (A) Primary mammosphere assays performed using Pi− LIN(CD31,CD45,LY76)− CD24+ CD29hi MECs plated at 20,000 cells per well (left) and in limiting dilution of 500 to 25 CD24+ CD29hi MECs plated (right) reveal increased MaSC activity in Becn1+/− MECs. (B) 3D colony-formation by CD24+ CD29hi Becn1+/− compared with Becn1+/+ MECs plated at 20,000 cells per well shows increased MaSC activity. (C) Colony formation by CD24+ CD29lo Becn1+/− compared with Becn1+/+ MECs plated at 1,000 cells per well reveals increased colony forming ability in 2D-matrigel conditions. (D) Increased repopulation frequency and mammary fat pad filling are seen following transplantation of 250 CD24+ CD29hi Becn1+/− MECs isolated from 5-wk-old mice. Becn1+/+ (left) and Becn1+/− (right), contralateral transplantations in wild-type recipient mouse. Circles represent fat pads and the black color represents the percentage of the mammary fat pad that is filled in. (E) Representative whole mount images of outgrowths from contralateral CD24+ CD29hi Becn1+/+ and Becn1+/− MEC transplantation are shown and reveal increased ductal side branching in Becn1+/− samples. *P < 0.05 by a 2-tailed Student t test. Scale bar: (E) 5 mm for whole mounts; 2 mm for whole mount enlargements.
The in vivo stem/progenitor cell activity of CD24+ CD29hi Becn1+/+ and Becn1+/− MECs was evaluated by contralateral transplantation of these cells into cleared 4th mammary fat pads of wild-type mice.22 CD24+ CD29hi Becn1+/− MECs consistently resulted in higher number of successful (i.e. giving rise to mammary outgrowths) transplantations (Fig. 3D; Fig. S2C and S2D). Six weeks post-transplantation, comparison of similarly filled fat pads revealed that CD24+ CD29hi Becn1+/− MECs gave rise to outgrowths with more extensive side-branching than CD24+ CD29hi Becn1+/+ MECs (Fig. 3E). This phenotype was similar to that observed in MGs from Becn1+/− mice (Fig. 2), suggesting that transplantation of MaSC-containing Becn1+/− MECs in wild-type mice sufficiently reproduced the side-branching phenotype observed in native MGs from Becn1+/− mice, where all cell types (MECs and stroma) are missing one Becn1 allele.
Since transplantations of CD24+ CD29hi Becn1+/+ and Becn1+/− MECs were performed in wild-type recipient mice, we examined the autophagy status of resulting outgrowths and found that similar to MGs from Becn1+/− mice (Fig. S1E and S1F), CD24+ CD29hi Becn1+/− MEC-generated outgrowths exhibited LC3B expression decrease and SQSTM1 accumulation (Fig. S2E and S2F). Furthermore, similar to MGs from Becn1+/− mice (Fig. S1A and S1B), proliferation rates in TEBs and mammary ducts were significantly higher in outgrowths derived from CD24+ CD29hi Becn1+/− MEC transplantation (Fig. 4A and B). In contrast to similar apoptosis rates observed in native glands (Fig. S1C and S1D), in CD24+ CD29hi Becn1+/− MEC-generated outgrowths, apoptosis was suppressed (Fig. 4C and D), possibly due to enrichment in more apoptosis-resistant MaSCs and MaPCs.
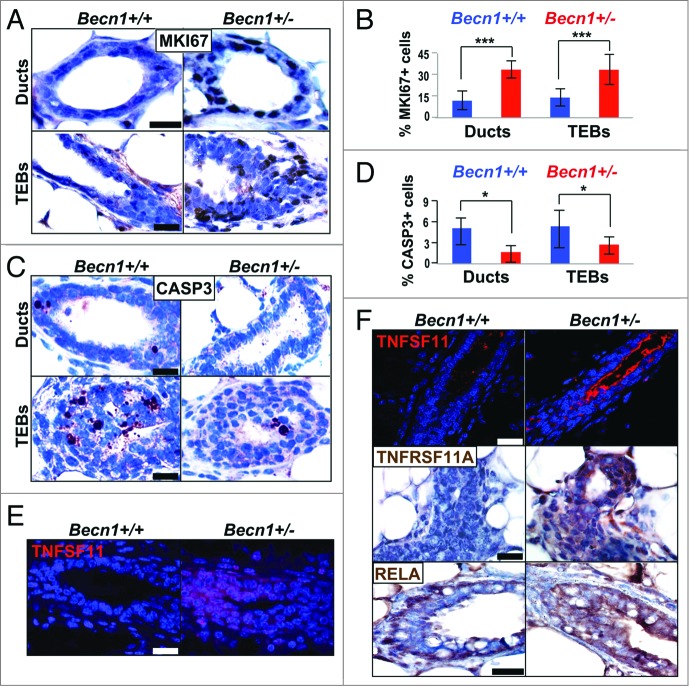
Outgrowths from CD24+ CD29hi Becn1+/− MEC transplantation display increased proliferation and TNFSF-NFKB pathway activation. (A–D, and F) Examination of outgrowths from contralateral transplantations of CD24+ CD29hi Becn1+/+ and Becn1+/− MECs, and (E) native MGs from 6.5-wk-old cohoused Becn1+/+ and Becn1+/− littermates. (A) Representative images show more MKI67+ cells in ducts (top) and TEBs (bottom) of outgrowths from CD24+ CD29hi Becn1+/− MEC transplantation; (B) quantification of MKI67+ cells in mammary ducts and TEBs is shown as means ± SDs (n = 3 mammary gland specimens per genotype). (C) Representative images show fewer cleaved CASP3-staining cells in ducts (top) and TEBs (bottom) of outgrowths from CD24+ CD29hi Becn1+/− MEC transplantation; (D) quantification of cleaved CASP3+ cells in mammary ducts and TEBs is shown as means ± SDs (n = 3 mammary gland specimens per genotype). Increased TNFSF11 (red) expression is seen in (E) native MGs from 6.5-wk-old Becn1+/− mice and (F) outgrowths from contralateral CD24+ CD29hi Becn1+/− MEC transplantation (top panel). Increased TNFRSF11A (middle panel) and RELA (bottom panel) levels are also seen in outgrowths from CD24+ CD29hi Becn1+/− MEC transplantation. Scale bar: (A, C, E, and F, middle and bottom panels) 30 μm ; (F, top panel) 40 μm.
MEC subpopulations exhibit different levels of autophagic activity
Given that monoallelic Becn1 loss resulted in deregulation of the mammary hierarchy, thus implicating BECN1, and possibly autophagy in general, in mammary differentiation, we isolated MEC populations from wild-type mice, as described above, and examined basal autophagy in these populations by electron microscopy (EM)17 (Fig. S3). The CD24+ CD29hi population exhibited paucity in mitochondria, autophagosomes, and other cytoplasmic organelles (Fig. S3, left panel), whereas the CD24+ CD29lo population showed high autophagic activity, as illustrated by the presence of numerous autophagosomes (Fig. S3, middle panel). Interestingly, the autophagosomal cargo was different between CD24+ CD29hi and CD24+ CD29lo MECs; in the former (MaSC-containing) population, autophagosomes appeared largely carrying mitochondria, while in the latter (progenitor cell-containing) population, diverse cargo was observed in autophagosomes. Lastly, CD24− MECs contained fewer autophagosomes than the CD24+ CD29lo population, but more than the CD24+ CD29hi compartment, and more mitochondria than both other MEC populations examined (Fig. S3, right panel). High autophagy induction in CD24+ CD29lo MECs, though not a proof, suggests that autophagy may play a significant role in mammary differentiation, which is also accompanied by enrichment in mitochondria.
Krt14-driven biallelic Atg7 deletion delays mammary fat pad filling, without altering ductal side-branching
In an effort to investigate whether the increased ductal side-branching and enriched MaSC and progenitor cell activities observed in MGs from Becn1+/− mice were secondary to defective autophagy associated with monoallelic Becn1 loss or to a nonautophagy-related BECN1 function, we generated mice with conditional biallelic deletion of the essential autophagy regulator Atg726 utilizing Krt14 promoter-driven expression of CRE recombinase.27 In agreement with prior studies showing that Atg7 is not happloinsufficient for autophagy deficiency or other phenotypes,26 we found that MGs from Atg7+/+ and Atg7F/+;Krt14-Cre mice displayed similar morphological characteristics (data not shown).
In MGs from atg7F/F;Krt14-Cre mice, Atg7 deletion and defective autophagy status were confirmed (Fig. S4A and S4B). In contrast to our observations in developing MGs from Becn1+/− mice (Fig. 2A and B), whole MG mounts revealed no difference in ductal side-branching between Atg7+/+ and atg7F/F;Krt14-Cre mice and, more impressively, showed delayed—rather than accelerated—fat pad filling in 6.5-wk-old atg7F/F;Krt14-Cre mice (Fig. S4C and S4D). While proliferation rates were similar, increased apoptosis was detected in MGs from atg7F/F;Krt14-Cre compared with Atg7+/+ mice (Fig. S4E and S4F). Furthermore, KRT6, KRT8, and KRT14 levels were comparable in MGs from atg7F/F;Krt14-Cre and Atg7+/+ mice (Fig. S4G and S4H). Although functional in vitro and in vivo studies for MaSC and progenitor cell activities in Atg7+/+ and Atg7-null CD24+ CD29hi and CD24+ CD29lo MECs were not performed, decreased mammary fat pad filling in pubertal MGs from atg7F/F;Krt14-Cre mice suggests, at least partial, impairment of MaSC or progenitor cell function in association with MEC-specific biallelic Atg7 deletion, and points to a nonautophagy-related role of BECN1 in mammary differentiation. Alternatively, and together with the evidence for high autophagic activity during mammary epithelial differentiation (Fig. S3) and the mammary phenotypes observed in MGs from Becn1+/− mice (Figs. 1, ,2,2, and and3),3), it raises the possibility of a dose-dependent requirement for functional autophagy in MaSC and progenitor cell maintenance. If this is the case, partial autophagy deficiency may affect mammary cell hierarchy differently than a more severe autophagy defect. Further investigation of these 2 not necessarily mutually exclusive, mechanistic possibilities will be needed.
Monoallelic Becn1 loss results in TNFRSF11A-NFKB pathway activation in mammary tissues
TNFSF11/TNF11/RANKL (tumor necrosis factor [ligand] superfamily, member 11) has been implicated in progesterone-induced MaSC expansion28 and mice overexpressing TNFSF11 or its receptor, TNFRSF11A/RANK (tumor necrosis factor receptor superfamily, member 11a, NFKB activator), under the mouse mammary tumor virus (MMTV) promoter,29,30 exhibit mammary phenotypes similar to Becn1+/− mice, including excessive ductal side-branching (Fig. 2), increased proliferation (Fig. S1A and S1B), and mammary hyperplasias (Fig. 2C), but no mammary tumors in nulliparous mice.4,15 We hypothesized that ectopic TNFRSF11A-TNFSF11 signaling may be mediating the increased mammary stem and progenitor cell activities in MGs from Becn1+/− mice. TNFSF11 was indeed detected in MGs from pubertal Becn1+/− mice (Fig. 4E) and robustly in outgrowths from CD24+ CD29hi Becn1+/− MEC transplantations (Fig. 4F). Consistent with aberrant TNFRSF11A pathway activation, outgrowths from CD24+ CD29hi Becn1+/− MEC transplantation also displayed increased TNFRSF11A expression (Fig. 4F) and likely activation of the downstream effector NFKB axis,31 as indicated by increased expression, though not apparent nuclear localization, of RELA (Fig. 4F). In agreement with previous studies,28-30,32-34 this data suggests that aberrant TNFRSF11A pathway activation may play a role in the MaSC and progenitor cell expansion observed in association with monoallelic Becn1 loss. However, further studies will need to be conducted to determine whether TNFRSF11A signaling is responsible for the mammary phenotypes observed in Becn1+/− mice.
Locally increased progesterone signaling in MGs from Becn1+/− mice
Given that progesterone is required for mammary ductal side-branching35 and TNFSF11 is a mediator of both progesterone signaling28,32 and progestin-induced mammary tumorigenesis,34 we examined MGs from cohoused Becn1+/+ and Becn1+/− littermate mice for evidence of progesterone signaling. As shown (Fig. 5A and B), monoallelic Becn1 loss resulted in higher proportion of PGR (progesterone receptor)-positive MECs. Transplantation of CD24+ CD29hi Becn1+/+ and Becn1+/− MECs into contralateral fat pads of a wild-type recipient resulted in mammary outgrowths of both Becn1 genotypes in the same mouse; similar to native MGs, and despite uniform systemic hormone levels, the proportion of nuclear PGR-positive cells was again higher in outgrowths generated by Becn1+/− MECs (Fig. 5C and D), indicating that monoallelic Becn1 loss results in locally-regulated increased progesterone signaling.
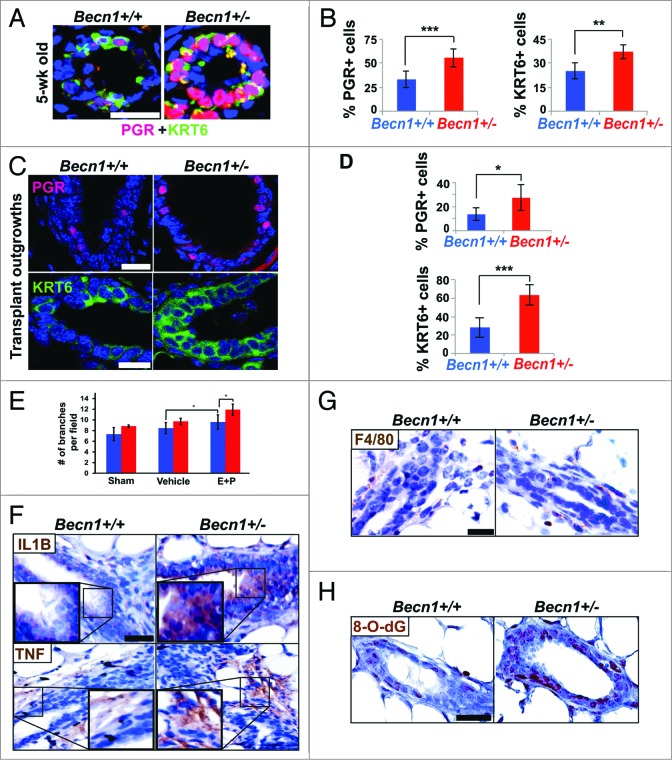
Augmented progesterone signaling, cytokines, and oxidative stress detected upon monoallelic Becn1 loss. (A and C) Nuclear PGR (red) and KRT6 (green) cell staining in native MGs from cohoused Becn1+/+ and Becn1+/− MEC and outgrowths from contralateral CD24+ CD29hi Becn1+/+ and Becn1+/− MEC transplantation. (A) Representative images of PGR+ and KRT6+ cells and (B) quantification of nuclear PGR+ cells (left) and KRT6+ cells (right) in MGs from 5-wk old mice reveals increased numbers of PGR+ and KRT6+ cells in Becn1+/− mice. (C) Representative images of nuclear PGR+ cells (top panel) and KRT6+ cells (bottom panel) in outgrowths from contralateral CD24+ CD29hi Becn1+/+ and Becn1+/− MEC transplantation with (D) quantification of PGR+ cells (top panel) and KRT6+ cells (bottom panel). Results are presented as means ± SDs (n = 3 mammary gland specimens per genotype). (E) Quantification of ductal side-branching in MGs from hormonally stimulated following ovariectomization, where Sham = ovariectomized, Vehicle = ovariectomized + 100 μL sesame oil daily, and E + P = ovariectomized + 5 μg 17B-estradiol + 5 mg progesterone in 100 μL of sesame oil daily. (F–H) Examination of outgrowths from contralateral transplantations of CD24+ CD29hi Becn1+/+ and Becn1+/− MECs. (F) Increased IL1B (top panel) and TNF (bottom panel) seen in outgrowths from Becn1+/− MEC transplantation. (G) Similar F4/80 staining detected in outgrowths from Becn1+/+ and Becn1+/− MEC transplantation. (H) Increased 8-O-dG is seen in outgrowths from Becn1+/− MEC transplantation. *P < 0.05, **P < 0.01, and ***P < 0.001 by a 2-tailed Student t test. Scale bar: (A, C, F, G, and H) 30 μm; (E) 50 μm.
To investigate the role of hormonal signaling in the mammary hyper side-branching phenotype observed in association with monoallelic Becn1 loss, Becn1+/+ and Becn1+/− mice were ovariectomized at 3 to 4 wk of age. In the absence of endogenous hormone production, MGs from Becn1+/+ and Becn1+/− mice displayed similar ductal side-branching (Fig. 5E), whereas hormonal stimulation of ovariectomized mice with exogenous 17β-estradiol and progesterone32 rescued the hyper ductal side-branching phenotype in Becn1+/− vs. Becn1+/+ mice (Fig. 5E). This data implies that hormonal signaling is required for the increased ductal side-branching seen in MGs from Becn1+/− mice, and supports our observations that this phenotype is associated with progesterone axis activation (Fig. 2C and D) and is likely hormonally regulated.
In progesterone-driven TNFRSF11A signaling, exogenous progesterone activates PGR-positive MECs to secrete TNFSF11, which in turn induces proliferation of neighboring TNFRSF11A-expressing mammary stem and progenitor cells and subsequent generation of more PGR-positive MECs, thus creating a positive feedback loop.28,32 In macrophages, TNFSF11 production and activation of TNFRSF11A-NFKB signaling is driven by proinflammatory cytokines.36 In our studies, where contralaterally transplanted Becn1+/+ and Becn1+/− MECs are exposed to the same systemic hormones, outgrowths generated by CD24+ CD29hi Becn1+/− MECs displayed increased IL1B (interleukin 1, β) and TNF/TNFα (tumor necrosis factor) levels (Fig. 5F), which were not accompanied by enhanced macrophage infiltration (Fig. 5G). Together these studies indicate that IL1B and TNF were likely secreted by and acting autonomously on Becn1+/− MECs.
Autophagy-deficient tissues commonly harbor a proinflammatory microenvironment,37 mainly attributed to increased reactive oxygen species (ROS) due to impaired removal of damaged mitochondria.37 Consistent with their functional autophagy defect (Fig. S2E and S2F), CD24+ CD29hi Becn1+/− MEC-generated outgrowths exhibited increased oxidative stress, as evaluated by 8-oxo-7,8-dihydroguanine (8-O-dG) staining (Fig. 5H).
Becn1+/− mice develop spontaneous mammary tumors following parity
In earlier studies which reported mammary hyperplasias, but no mammary tumors, in Becn1+/− mice independent of age and parity,4,15 the mouse genetic background was C57BL/6, which is known to suppress mammary tumorigenesis.38 To ensure that mammary tumor studies in Becn1+/− mice were not confounded by the C57BL/6 status, we changed the genetic background of Becn1+/− mice5 to FVB/N, which is more amenable to mammary tumorigenesis studies.38
Nulliparous and multiparous (post 2 pregnancy/lactation/involution cycles) Becn1+/+ and Becn1+/− female mouse cohorts were generated, cohoused and monitored for mammary tumor formation. Similar to C57BL/6 mice,4,15 both nulliparous and multiparous Becn1+/− FVB/N mice developed lymphomas, lung and liver tumors, and mammary hyperplasias with age. Furthermore, mammary ducts from older (24-mo-old) nulliparous Becn1+/− FVB/N mice showed increased cellular density and irregular epithelial cell layering (Fig. 6A). Similar, but more pronounced, changes of increased epithelial density and disorganized ductal architecture were observed in MGs from multiparous Becn1+/− multiparous mice at a younger age (14 to 17 mo) (Fig. 6B). In contrast to C57BL/6 mice and nulliparous FVB/N mice, 4 out of 21 mice (19%) of multiparous Becn1+/− female mice developed mammary tumors between the ages of 14 and 24 mo (Fig. 6C). Tumor #1 displayed squamous and focal glandular differentiation, with keratinizing differentiated squamous and nonkeratinized poorly differentiated regions. Tumor #2 displayed well-differentiated keratinizing squamous differentiation with foci of basaloid squamous and glandular components; both tumors #1 and 2 showed pervasive KRT6- and KRT14-positivity, but no PGR staining. Tumor #3 had characteristics of glandular differentiation with small foci of well-differentiated keratinizing squamous component, and was positive for KRT6 and KRT14, but not PGR. Tumors #1–3 were characterized as invasive adenosquamous carcinomas, a type of metaplastic carcinoma seen in the mammary gland. Tumor #4 displayed a marked cribriform pattern and luminal eosinophilic cylinders resembling the invasive mammary tumor known as an adenoid cystic carcinoma, and was KRT6-negative, but PGR-positive. Despite diversity in histology likely due to tumor heterogeneity,39 most tumors were squamous, KRT6-positive, and basal-like (KRT14-positive) and all showed negative correlation between KRT6 and PGR expression.
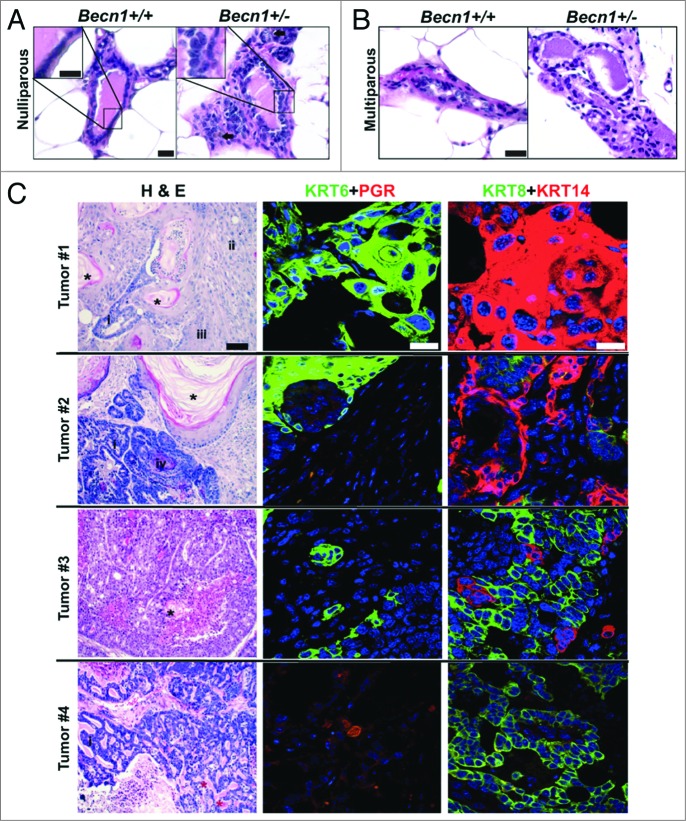
Spontaneously arising mammary tumors are detected in multiparous Becn1+/− FVB/N mice. (A and B) Hematoxylin and eosin staining (H&E) of MG sections from Becn1+/+ (left) and Becn1+/− (right) FVB/N mice: (A) Representative images from MGs of nulliparous 21- to 22-mo-old mice reveals increased epithelial cell density and inflammatory cell infiltration (brown, identified with arrow) in Becn1+/− mice. (B) Representative H&E images of MGs from parous 14- to 17-mo-old mice reveals increased epithelial cell density and disorganized cell layering in MGs from Becn1+/− mice. (C) Representative images from 4 spontaneously arising mammary tumors in parous Becn1+/− FVB/N mice (Tumors #1 to 4). Left panels are representative H&Es; middle panels, KRT6 (green) and nuclear PGR (red) staining; and right panels, KRT8 (green) and KRT14 (red) staining. (*) and (*) denote keratin pearls and basement membrane/eosinophilic hyaline materials, respectively. Tumor type denotations are identified by: (i) for glandular components, (ii) for poorly to moderately differentiated squamous cell carcinoma, (iii) for well-differentiated squamous cell carcinoma, and (iv) for basaloid squamous cell carcinoma. Scale bar: (A and B) 30 μm and KRT8 and KRT14 (C); (C) 200 μm for H&Es; (C) 50 μm for KRT6 and PGR.
Monoallelic Becn1 loss accelerates WNT1-driven mammary tumorigenesis
Given our findings that monoallelic Becn1 loss promotes KRT6-positive immature mammary cell accumulation (Fig. 1), we investigated whether Becn1 heterozygosity has an impact on mammary tumorigenesis in the context of oncogenic WNT1 activation, where KRT6-positive bipotent MaPCs are considered tumor-initiating cells.18,20
As shown (Fig. 7A; Fig. S5A), Becn1+/−;MMTV-Wnt1 mice displayed significantly shorter mammary tumor-free survival (4 vs. 7.2 mo; P 0.004) and overall survival (4.7 vs. 7.5 mo; P < 0.001) compared with Becn1+/+;MMTV-Wnt1 mice. Consistent with defective autophagy status, mammary tumors from Becn1+/−;MMTV-Wnt1 mice showed reduced LC3B levels and SQSTM1 aggregates (Fig. S5B). Tumors arising in Becn1+/+;MMTV-Wnt1 mice displayed known characteristics of MMTV-Wnt1 mammary tumors,40 as they were KRT6-positive adenosquamous carcinomas with high KRT8 and low KRT14 levels, and variable ESR (estrogen receptor) and PGR expression (Fig. 7B). Mammary tumors from Becn1+/−;MMTV-Wnt1 mice were also mostly adenosquamous carcinomas, but exhibited higher KRT6 levels, were more basal-like, as KRT14 expression was very prominent, and did not express ER or PGR (Fig. 7B). Furthermore, while proliferation was similar, apoptosis was suppressed in tumors from Becn1+/−;MMTV-Wnt1 mice (Fig. S5C and S5D).
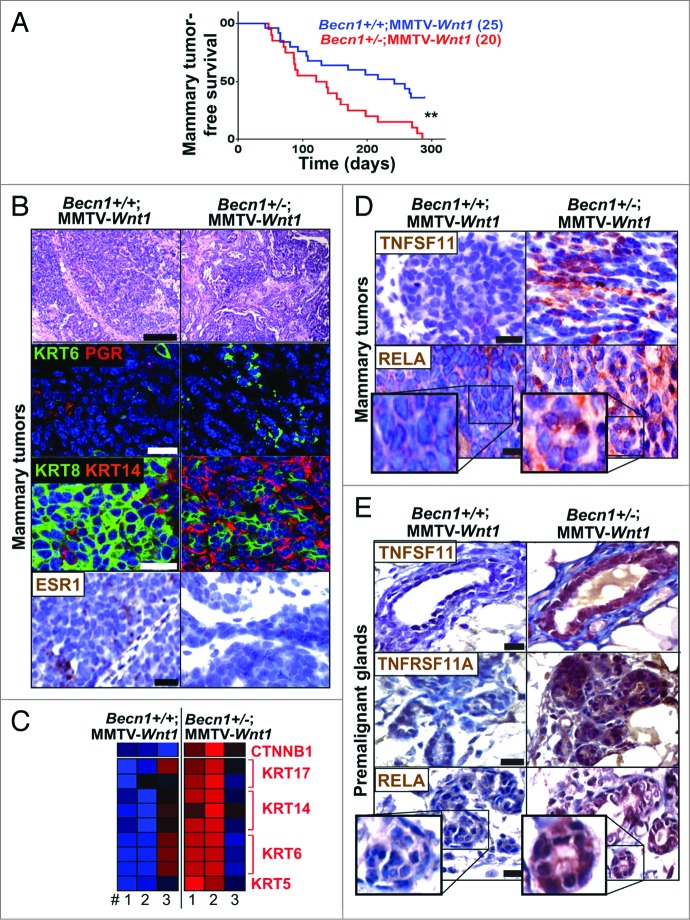
Monoallelic Becn1 loss accelerates WNT1-driven tumorigenesis and gives rise to mammary tumors with TNFRSF11A-NFKB1 pathway activation and basal-like characteristics. Basal-like characteristics and TNFRSF11A-NFKB1 signaling activation are detected in spontaneous mammary tumors that arise faster in Becn1+/−;MMTV-Wnt1 (right) compared with Becn1+/+;MMTV-Wnt1 mice (left). (A) Kaplan-Meier curves for mammary tumor-free survival in Becn1+/+;MMTV-Wnt1 (n = 25) and Becn1+/−;MMTV-Wnt1 (n = 20) mice demonstrates decreased mammary tumor-free survival in Becn1+/−;MMTV-Wnt1 mice. (B–E) Examination of mammary tumors and premalignant MGs from Becn1+/+;MMTV-Wnt1 (left) and Becn1+/−;MMTV-Wnt1 (right) mice. (B) Representative images of H&E staining (top panel), KRT6 (green) and nuclear PGR (red) cell staining (second panel), KRT8 (green) and KRT14 (red) cell staining (third panel), and nuclear ESR1 staining (bottom panel) on tumor sections. (C) Heat map representation of microarray analysis confirms basal-like tumor characteristics along with higher CTNNB1 expression in tumors from Becn1+/−;MMTV-Wnt1 mice. Statistical significance of upregulation of a given signature on a group of samples was determined using a Fischer exact test for enrichment of samples with significant (P < 0.05) signature expression in the group relative to samples outside the group. (D) Representative images of TNFSF11 (top panel) and RELA (bottom panel) expression in mammary tumors, and (E) TNFSF11 (top panel), TNFRSF11A (center panel), and RELA (bottom panel) expression in premalignant MGs from 6- to 10-wk-old cohoused Becn1+/+;MMTV-Wnt1 and Becn1+/−;MMTV-Wnt1 littermates. P < 0.01 determined by a Mantel-Cox test for Kaplan-Meier curves. Scale bar: (B) 200 μm for H&Es; (B, D, and E) 30 μm for all other panels.
Gene expression profiling also showed that mammary tumors from Becn1+/−;MMTV-Wnt1 mice showed higher expression of KRT6 and other basal keratins, including KRT5, KRT14, and KRT17; they also displayed CTNNB1/β-catenin (catenin [cadherin-associated protein], β 1, 88 kDa) upregulation (Fig. 7C), indicating that monoallelic Becn1 loss rendered WNT1-induced mammary tumors basal-like and further enhanced CTNNB1 signaling already present due to WNT1 activation. An earlier publication reported that TP53 expression is regulated by BECN1 and, as such, may contribute to BECN1-associated tumor suppression.41 In our studies, mammary tumors from Becn1+/+;MMTV-Wnt1 and Becn1+/−;MMTV-Wnt1 mice did not exhibit differences in expression of TP53 transcriptional target genes (data not shown), thus suggesting that in the context of WNT1-driven mammary tumorigenesis, differential TP53 activation is not a likely contributor to mammary tumor acceleration in association with monoallelic Becn1 loss.
Consistent with our finding that monoallelic Becn1 loss results in increased mammary stem and progenitor cell activity in association with TNFRSF11A-NFKB pathway activation (Figs. 1, ,3,3, and 4),4), tumors from Becn1+/−; MMTV-Wnt1 mice exhibited increased TNFSF11 expression and nuclear RELA staining (Fig. 7D). Premalignant MGs from Becn1+/−;MMTV-Wnt1 mice displayed even higher TNFSF11, TNFRSF11A, and nuclear RELA levels (Fig. 7E), indicating that early activation of TNFRSF11A-NFKB signaling may contribute to promotion of WNT1-driven mammary tumorigenesis by monoallelic Becn1 deletion. However, additional studies must be conducted to further investigate whether aberrant TNFRSF11A pathway activation is responsible for the accelerated mammary tumor development in Becn1+/−;MMTV-Wnt1 mice.
Human breast cancers with low BECN1 mRNA expression and an activated WNT pathway gene signature have poor prognosis
To examine the clinical relevance of our mouse tumor studies, we identified human breast cancers exhibiting a gene signature consistent with WNT pathway activation and stratified them for high vs. low (above or below the mean across samples, respectively) expression of BECN1 mRNA. In 2 independent breast cancer cohorts,42,43 tumors with low BECN1 expression and an activated WNT pathway gene signature had significantly worse prognosis than tumors with high BECN1 mRNA and WNT pathway activation (Fig. 8A and B) and also correlated with the triple negative breast cancer (TNBC) subtype (Fig. 8C and D). In a cohort excluding ERBB2-positive cancers,42 tumors with low BECN1 expression and an activated WNT pathway gene signature were almost exclusively TNBCs and showed TNFRSF11A pathway activation (Fig. 8D), similar to mammary tumors from Becn1+/−;MMTV-Wnt1 mice (Fig. 7C).
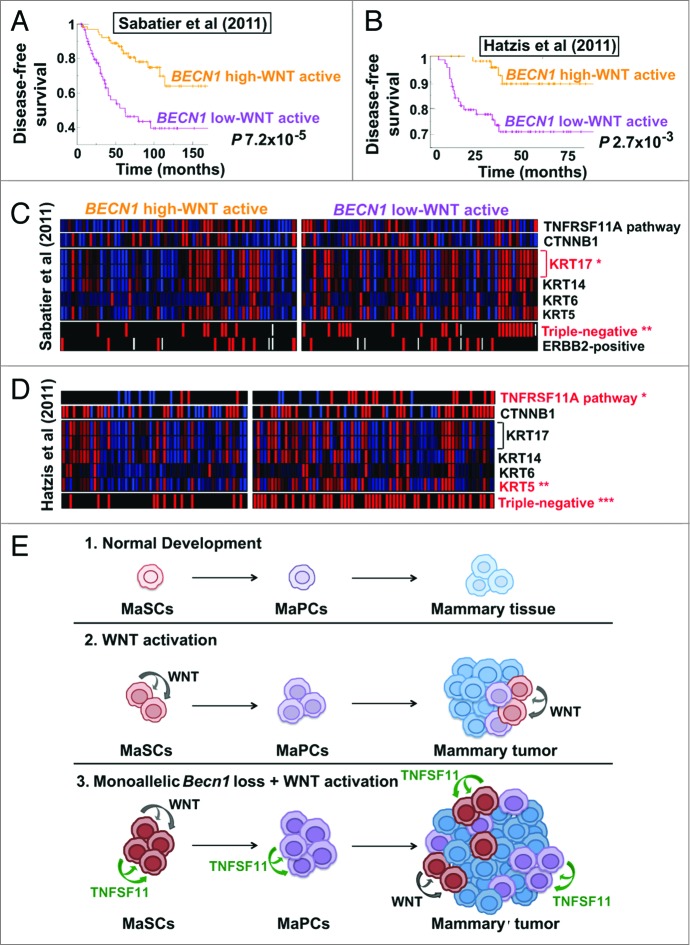
Human breast cancers with low BECN1 expression and an activated WNT pathway gene signature have poor prognosis. (A–D) Kaplan-Meier curves and GSEA for breast tumors displaying an activated WNT pathway gene signature and stratified for BECN1 expression (see Methods). (A) Disease-free survival for patients with breast cancers annotated in the Sabatier cohort (P 7.2 × 10−5). (B) Disease-free survival for patients with ERBB2-negative breast cancers annotated in the Hatzis cohort (P 2.7 × 10−3). Gene expression analysis comparison in (C) the Sabatier cohort and (D) the Hatzis cohort reveals that breast cancers with low BECN1 and an activated WNT pathway gene signature are primarily triple-negative, have basal-like characteristics, and display TNFRSF11A pathway activation and higher CTNNB1. P values were calculated by the Fischer exact test (see Materials and Methods). (E) Model for the cooperation between monoallelic BECN1 loss and WNT pathway activation in mammary tumorigenesis. *P < 0.05, **P < 0.01, ***P < 0.001.
In the Sabatier data set,43 within low BECN1-expressing tumors, an activated WNT pathway gene signature conferred worse disease-free survival (DFS), whereas in high BECN1-expressing tumors, it was associated with improved DFS (Fig. S6A). In tumors not displaying WNT pathway activation, BECN1 expression did not affect DFS (Fig. S6A). In the Hatzis, ERBB2-negative breast tumor database, in low BECN1-expressing and mostly triple-negative tumors, WNT pathway activation did not have an impact on DFS, whereas in high BECN1-expressing tumors, it was again associated with improved DFS (Fig. S6B). In tumors not displaying upregulation of a WNT pathway gene signature, low BECN1 expression conferred worse DFS (Fig. 6B). Examination of additional breast cancer databases and prospective evaluation of clinically annotated breast tumor biopsies will be necessary to further define how the combination of low BECN1 expression and WNT pathway activation affects prognosis within the different breast cancer subtypes.
Discussion
Role of Becn1 in the homeostasis of the mammary cell hierarchy
Our studies identify monoallelic Becn1 loss as a potential contributor to immature MEC expansion and, thus, deregulation of the mammary cell hierarchy. Our findings come at a time of great interest in the role of autophagy in stem and progenitor cell biology. Recent studies show that autophagy supports stem cell survival in the hematopoietic44 and nervous45 systems, and in the context of an ERα-positive and an ERBB2-positive breast cancer cell lines.10 Here we report that partial Becn1 deficiency leads to increased mammary stem and progenitor cell activities, and we hypothesize that this contributes to increased mammary tumorigenesis in association with monoallelic Becn1 loss under conditions of further mammary stem and progenitor pool expansion due to either parity or activation of particular oncogenes, such as WNT1. Our work identifies 2 specific contexts in which Becn1 heterozygosity promotes mammary tumorigenesis, and provides insight into the seemingly paradoxical roles of BECN1, and autophagy in general, as both tumor-suppressive and tumor-promoting functions. Furthermore, our studies suggest that low BECN1 expression has an impact on tumor histology in the case of WNT1-driven tumorigenesis, giving rise to tumors with more basal and TNBC characteristics.
An important question that arises from this work and the focus of future studies is whether the Becn1 heterozygosity-associated mammary phenotypes described in our study are secondary to nonautophagy-related roles of BECN1 in mammary development and tumorigenesis. Although the impact of conditional Atg7 deficiency on mammary tumor formation following parity or WNT1 activation has not yet been explored, it is intriguing that biallelic Atg7 deletion delays mammary fat pad filling during puberty and does not result in the hyper side-branching phenotype observed in Becn1+/− mice, pointing to a BECN1-specific, and unrelated to autophagy, role in ductal morphogenesis. Alternatively, an autophagy dosage effect on mammary hierarchy may contribute to the observed phenotypes. In that respect, a more severe autophagy defect, such as that conferred by biallelic Atg7 loss, may drastically compromise MaSC and progenitor cell survival, and possibly suppress tumorigenesis, whereas partially deficient autophagy due to Becn1 heterozygosity may result in “injured,” but still surviving, mammary stem and progenitor cells with increased proliferation, which may promote tumor development under conditions that further deregulate the homeostasis of mammary cell hierarchy; an autophagy dosage effect on mammary differentiation and possibly tumorigenesis would be consistent with the results of recent studies on the role of autophagy inhibition in lung cancer.11
Monoallelic Becn1 loss and TNFRSF11A-NFKB signaling activation in mammary tissues
Our studies demonstrate that MGs from pubertal and adult Becn1+/− mice exhibit excessive ductal side-branching in association with abnormal TNFSF11 expression and TNFRSF11A-NFKB axis activation. TNFSF11 is a member of the tumor necrosis factor (TNF) ligand superfamily, primarily known for its role in bone remodeling and metastasis33 and also implicated in MaPC expansion and progestin-induced tumorigenesis.34,46 High TNFRSF11A and TNFSF11 levels were also reported in breast cancers lacking ESR1 and PGR expression and in high-grade and highly proliferative breast tumors;47 consistent with these results, we also found evidence of TNFRSF11A pathway activation in aggressive basal-like tumors exhibiting low BECN1 expression and an activated WNT pathway gene signature. Furthermore, TNFRSF11A overexpression in breast cancer cell lines has been reported to disrupt mammary cell fate, induce epithelial-to-mesenchymal transition (EMT) and promote mammary tumorigenesis and metastasis.47 In our studies, tumors originating in both Becn1+/+;MMTV-Wnt1 and Becn1+/−;MMTV-Wnt1 mice retained epithelial keratin expression, and neither expressed mesenchymal markers, such as VIM/vimentin (data not shown) nor gave rise to visible metastases at the time of necropsy. However, since metastatic lesions are not usually noted in MMTV-Wnt1 mice sacrificed when their primary mammary tumors reach experimental endpoints,40 specifically designed metastasis studies, involving mammary tumor resection survival surgery and/or intracardial injection of tumor-derived mammary tumor cell lines, will be necessary to determine the role of BECN1 in metastasis and EMT in the context of WNT pathway activation.
Our findings, together with the striking similarities in mammary development and tumor phenotypes between Becn1+/− and MMTV-Tnfrsf11a (or MMTV-Tnfsf11) mice, suggest that the TNFRSF11A pathway may mediate the impact of monoallelic Becn1 loss on mammary cell hierarchy, morphogenesis and tumorigenesis. However, whether ectopic TNFRSF11A pathway activation is responsible for, or contributes to, MaSC and progenitor expansion and increased tumor susceptibility following parity or WNT pathway activation upon monoallelic Becn1 loss requires further mechanistic investigation.
Which cell type produces TNFSF11 in MGs from Becn1+/− mice is another topic of interest. TNFSF11 production by MECs is primarily considered the result of progesterone stimulation,28,32 whereas in macrophages, adipocytes, and breast cancer cells, proinflammatory cytokines can also upregulate TNFSF11.36,48,49 Our transplantation studies argue in support of TNFSF11 production by MECs in response to proinflammatory cytokine release by MECs themselves: (1) mammary outgrowths were exposed to same systemic progesterone levels, as CD24+ CD29hi Becn1+/+ and Becn1+/− MECs were transplanted in contralateral fat pads of recipient mice. Increased progesterone signaling in association with monoallelic Becn1 loss must therefore result from a locally-induced expansion of PGR-expressing cells; (2) 2 proinflammatory cytokines known to induce TNFSF11 production, namely IL1B and TNF, were detected in CD24+ CD29hi Becn1+/− MEC-generated outgrowths; (3) CD24+ CD29hi Becn1+/+ and Becn1+/− MEC transplantations were performed in wild-type recipient mice, thus all stromal cells were wild type. The fact that, similar to MGs from Becn1+/− mice, outgrowths from CD24+ CD29hi Becn1+/− MEC transplantation also exhibited increased ductal side-branching and TNFSF11 production indicate that MECs are sufficient to induce these mammary phenotypes. Furthermore, macrophage infiltration was similar in CD24+ CD29hi Becn1+/+ and Becn1+/− MEC-generated outgrowths; (4) increased oxidative stress, a inducer of proinflammatory cytokine production,37 was documented in CD24+ CD29hi Becn1+/− MEC-generated outgrowths; (5) autophagy-deficient status, commonly associated with inflammation and cytokine release,14,37 was maintained in CD24+ CD29hi Becn1+/− MEC-generated outgrowths. This study provides evidence for an essential role of BECN1 in restricting mammary stem and progenitor cell expansion and limiting ectopic TNFRSF11A pathway activation, 2 pathways known to be deregulated in particular contexts of mammary tumorigenesis.18,30,34
Shedding light in the complex role of Becn1 in mammary tumorigenesis
BECN1, and autophagy in general, have complex roles in tumorigenesis: allelic BECN1 deletions have been reported in human cancers,2 and Becn1+/−4,5 and other Atg-deficient50,51 mice are tumor-prone, in support of a tumor suppressor role for autophagy in cancer. However, autophagy also preserves tumor cell survival under stress and likely contributes to tumor progression11 and treatment resistance.52 While apoptosis-deficient Becn1+/− iMMECs, compared with their wild-type counterparts, result in earlier mammary tumor development in nude mice,6 monoallelic Becn1 deletion does not alter ERBB2- or PyMT-driven,8 while it delays Palb2 loss-associated,13 mammary tumorigenesis.
Our studies define specific contexts, either following parity or oncogenic WNT1 activation, in which monoallelic Becn1 deletion promotes mammary tumorigenesis. Spontaneous tumors arising in Becn1+/− mice after parity display mostly squamous cell characteristics, which is similar to the squamous differentiation seen by Wei and colleagues in lung cancers with low BECN1 expression.9 These 2 studies indicate that low BECN1 expression may drive particular lung and breast cancer subsets toward squamous, rather than adenocarcinoma, histologies, thus also raising the possibility of concurrent EGFR activation.
In the case of WNT pathway activation, we propose that WNT1-driven mammary stem and progenitor cell expansion cooperates with Becn1 heterozygosity-induced deregulation of the mammary hierarchy to create more transformation-susceptible immature mammary cells (Fig. 8E). Since monoallelic Becn1 loss results in hormone receptor-negative, basal-like mammary tumors, it is possible that corresponding tumor-initiating cells (TICs) reside within the expanded mammary stem and progenitor cell populations,23 a hypothesis that is worthy of further investigation. If this is the case, other oncogenes deregulating the mammary hierarchy and resulting in expansion of progenitor cell populations may also cooperate with monoallelic Becn1 loss to promote mammary tumorigenesis.
Low BECN1 expression and an activated WNT pathway gene signature define a breast cancer subset with poor prognosis
Despite the early discovery that WNT signaling is highly oncogenic in the mammary gland,18,20 conclusive evidence that it is a tumor-driver in breast cancer lagged behind.53 In contrast to colorectal tumors, which commonly harbor mutations in canonical WNT pathway genes,54 breast cancers lack such genetic alterations.55 However, even in the absence of mutations, canonical WNT pathway activation has been documented in TNBCs by increased CTNNB1 expression and nuclear localization in association with overexpression of the WNT receptor LRP6 (low-density lipoprotein receptor-related protein 6).56 Also, single nucleotide polymorphisms (SNPs) in WNT pathway genes were associated with ESR1-negative breast cancer risk.57
Given (1) that monoallelic Becn1 loss accelerates WNT1-driven mammary tumorigenesis and results in basal-like mammary tumors, and (2) the shift in focus from mutational analysis to collective upregulation of WNT signaling in tumors,58 we interrogated human breast cancer databases for BECN1 expression and WNT pathway activation, using DNA probes for BECN1 and a KEGG pathway-annotated gene signature for WNT pathway (hsa04310; http://www.genome.jp/dbget-bin/www_bget?hsa04310). We found that breast tumors with low BECN1 expression and WNT pathway activation comprise 24–36% of TNBCs and confer poor patient prognosis. Similarities between mammary tumors from Becn1+/−;MMTV-Wnt1 mice and human breast cancers with low BECN1 levels and WNT pathway activation, i.e. basal-like characteristics, TNFRSF11A pathway activation, and tumor aggressiveness, indicate that our mouse tumor model recapitulates molecular features and outcomes of a sizeable TNBC subgroup. Thus, our mouse model is a unique and clinically relevant preclinical model for a particular subset of breast cancers, and provides a valuable tool to develop useful molecular markers and evaluate targeted approaches for the treatment of these aggressive malignancies. Furthermore, our analysis of human breast cancer gene expression profiling indicates that, rather than genomic BECN1 alterations which may not have an impact on breast tumorigenesis,16 BECN1 mRNA expression, which can also be transcriptionally and/or epigenetically regulated, may itself determine breast cancer susceptibility under particular circumstances, such as parity or WNT pathway activation, and thus act as a context-dependent tumor suppressor.
Identifying an aggressive TNBC subtype with low BECN1 expression and activation of both WNT and TNFRSF11A pathways has the potential to greatly advance targeted breast cancer treatment, given that WNT signaling inhibitors are clinically investigated and a TNFRSF11A pathway inhibitor, denosumab, is already FDA-approved for prevention and treatment of osteoporosis and skeletal-related events in patients with bone-metastatic solid malignancies.59 The mechanistic relationship between low BECN1 expression, an activated WNT pathway gene signature and aberrant TNFRSF11A signaling in breast tumors needs to be further investigated to justify pharmacologic targeting of the TNFRSF11A and/or WNT pathways for treatment of such malignancies. Given our finding that monoallelic Becn1 loss results in mammary stem and progenitor cell expansion, which may in turn contribute to acceleration of WNT-driven tumorigenesis, it is conceivable that mammary stem and progenitor tumor cells may arise as promising therapeutic targets in the treatment of aggressive breast cancers exhibiting low BECN1 expression and an activated WNT pathway gene signature.
Altogether our studies identify BECN1 as a mammary tumor suppressor in 2 particular contexts, namely spontaneous mammary tumor formation following parity and WNT1-driven oncogenesis.
Materials and Methods
Mice
For native mammary gland experiments, mice were housed at the Child Health Institute of New Jersey, in pathogen-free facilities, and cared for as per IACUC-approved protocols. For transplantation experiments, recipient mice were housed at Princeton University and procedures were in compliance with IACUC-approved protocols. Becn1+/− mice5 were obtained from Dr S Jin. MMTV-Wnt1 mice were obtained from Dr Y Li. Atg7F/F mice26 were obtained from Dr Komatsu. Gfp-Lc3 mice were obtained from Dr Mizushima. Krt14-Cre mice60 were obtained from Jackson Laboratories.
For CD61 analysis and tumor studies, C57BL/6 Becn1+/− mice were backcrossed to FVB/N for 10 generations. In vitro MaSC and progenitor cell assays were performed in both C57BL/6 and FVB/N backgrounds. For all other studies involving the Becn1+/− genotype, C57BL/6 mice were used. For mammary fat pad filling and ductal side-branching cohoused littermates (for pubertal time points) and cohoused (for at least 3 mo after last pregnancy) multiparous mice were compared. At least 3 animals were compared for each time point. For all immunostaining, littermates or outgrowths from the same recipient mouse were compared. For tumor studies, multiparous mice were cohoused after 2 rounds of pregnancy, nursing, and weaning of their litters. Mice from the multiparous and nulliparous cohorts were palpated weekly for mammary tumor detection. For ovariectomization and stimulation studies procedures were performed as previously described.32
Mammosphere and colony-forming assays
Primary mammosphere assays were performed as previously described.23,25 MaSC viability assays under ultra low attachment were performed as previously described, and mammospheres were counted. 20,000 CD24+ CD29hi MECs per well were plated on ultra-low attachment plates (Costar, 3471) for 10 to 15 d in Dulbecco's modified Eagle's medium (DMEM)/F12, EGF (epidermal growth factor; Sigma, E4127), FGF2 [fibroblast growth factor 2 (basic); Invitrogen, PHG0026], B27 (Gibco, 17504-044), and heparin (Sigma, H6279). Results are presented as means ± standard deviation (SD) from 3 independent experiments. Primary mammospheres (2 or more cells) were counted. For limiting dilution mammosphere assays, 25 to 500 CD24+ CD29hi MECs per well were plated in duplicate.
The 3D colony-forming assays were performed as previously described.23,25 20,000 CD24+ CD29hi MECs per well were plated in reduced growth factor Matrigel (BD, 354230) and supplemented with mammary epithelial growth medium (MEGM) (Lonza, CC3051A), 5% FBS, EGF, and FGF2. Colonies were counted after 9 to 14 d. Results are presented as means ± SDs from 3 independent experiments.
The 2D colony-forming assays were performed as previously described.23,25 1,000 CD24+ CD29lo cells per well were plated on reduced growth factor Matrigel, supplemented with MEGM, 10% FBS, EGF, FGF2. Colonies were counted after 7 to 9 d. Results are presented as means ± SDs from 3 independent experiments.
Transplantation assays
MECs were isolated as previously described.22 FACS was used to collect CD24+ CD29hi MECs, i.e., the MaSC-containing population, and transplantations were performed as previously described into cleared fat pads of 3- to 4-wk-old recipient wild-type C57BL/6 female mice,22 in accordance with IACUC-approved protocols. Primary MEC donors were littermates. CD24+ CD29hi MECs were injected in a 200 μL volume (100 μL reduced growth factor Matrigel + 100 μL 1× phosphate-buffered saline (PBS; Gibco, 14190) at cell numbers specified. Mammary outgrowths were allowed to grow for 6 wk following transplantation and were then dissected, mounted on microscope slides, and processed for histological analysis. Three independent transplantation experiments were performed.
Mammary gland whole mounting
Native mammary glands from pubertal mice and mammary outgrowths generated by CD24+ CD29hi MEC transplantation were mounted onto microscope slides and fixed in 10% formalin overnight. Mammary glands from aged mice were mounted on microscope slides and fixed in Carnoy fixative for de-fatting. Mounts were then subjected to graded ethanol to water washes, and stained with carmine solution (Sigma, C1022 prepared as directed, with Aluminum Potassium Sulfate, Sigma, A7167) overnight with shaking. Stained slides were then subjected to graded water to ethanol washes, destained and stored in Histo-Clear II (National Diagnostics, HS-202). Imaging was done on Olympus SZ61 dissecting microscope (Rutgers Cancer Institute of New Jersey) using the ProRes Capture Pro2.5 software.
Stable cell lines and culture conditions
Cells were maintained in F12 (Gibco, 11765), 10% FBS, 1% penicillin/streptomycin, 5 ng/mL EGF, 1 μg/mL hydrocortisone (Sigma, H0888), 5 μg/mL insulin (Sigma, I9278). Cells were plated on glass coverslips, and allowed to attach overnight. For immunofluorescence (IF), cells were fixed with 1:1 methanol:acetone at −20 °C for 10 min and rinsed with 1× PBS. Slides were incubated with 50 μL KRT6 primary antibody 1:200 in 3% bovine serum albumin (BSA; Benchmark, 100-106), 0.01% Tween 20 (BIO-RAD, 170-6531), 1× PBS at 37 °C for 90 min with shaking. After washing with 0.01% Tween 20, 1× PBS, slides were incubated with 50 μL FITC-conjugated anti-rabbit antibody (Jackson ImmunoResearch, 711-095-152) 1:100 in 3% BSA, 0.01% Tween 20, 1× PBS at 37 °C for 30 min with shaking. Cells were stained with DAPI (Invitrogen, P36931) and mounted with ProLong Gold Antifade Mounting Media (Life Technologies, P363930). Slides were imaged using the Olympus 1×51 scope and Metamorph Image program.
Primary MEC isolation and FACS for MEC subpopulations
Primary MECs were isolated as previously described.22,23,25 MECs were stained as previously described22,23 and sorted using FACSVantage SE with DiVa (BD) at Princeton University. LIN− refers to cluster differentiation 31 (CD31)−, cluster differentiation 45 (CD45)−, and erythroid-specific LY76 (lymphocyte antigen 76)− negative cells, i.e. MECs. CD24+ CD29hi MECs refer to a MaSC-containing population, CD24+ CD29lo to a mammary progenitor cell-containing, and CD24− to differentiated MECs, as previously described.22,23 FACS profiles were analyzed using FlowJo software (TreeStar, Inc.). Five independent experiments were performed.
Primary and stable cell lines
Immortalized mouse mammary epithelial cells were generated as previously described.6 MaSC and MaPC colony- and mammosphere-forming assays were performed as previously described.23,25 MaSC transplantations were performed as previously described.22
Histology of tissues and tumors
Mammary tissues were processed and imaged as previously described.15 Following Carmine staining, mammary glands and outgrowths from CD24+ CD29hi MEC transplantation were transferred to 70% ethanol overnight, and processed as above. Hematoxylin and eosin (H&E) staining and imaging were performed by the Histopathological and Imaging Core Facility at Rutgers Cancer Institute of New Jersey.
Antibodies
For flow cytommetry, CD31 (BD558737), CD45 (B553078), LY76/TER119 (BD553672), and CD24-PE (BD553262) antibodies were from BD Biosciences, CD61-APC (MCD6105) from Invitrogen, and CD29-FITC (MCA2298FB) from Serotech were used. For immunohistology, KRT6 (PRB-169P) from Covance, KRT17 (ab53707), TNF/TNFα (ab8871), F4/80 (ab6640), and 8-O-dG (ab48508) from Abcam, KRT14 (gift from Dr Segre), PGR (gift from Dr Clarke), CASP3 (9661) from Cell Signaling Technology, MKI67 (NCL-L-Ki67-MM1) from Leica, ESR1/ERα (sc-542) and RELA/p65 (sc-8008) from Santa Cruz Biotechnology, TNFRSF11A/TNR11 (AF692) and TNFSF11/TNF11 (AF462) from R&D Systems, SQSTM1 (BML-PW9860) from Enzo, LC3 (NB100-2331) and IL1B/IL-1β (NBP1-19775) from Novus Biologics, and KRT8 (TROMA-1) and BrdU (G3G4) from the Developmental Studies Hybridoma Bank were used.
Immunostaining
For immunohistochemistry (IHC), slides were deparrafinized with xylenes, rehydrated with ethanol to water washes, and boiled at 97 °C for 16 min in 1× Antigen Unmasking Solution (Vector, H-3300). For PGR staining, retrieval was done by autoclaving at 121 °C, 16 to 17psi, for 30 min in Vector 1× unmasking solution. For ESR1 staining, retrieval was done by microwaving slides in 1× Vector Unmasking Solution for 20 min. Slides were washed in water, followed by endogenous peroxidase blocking with 3% H2O2. Washes in 0.05% Tween 20 in 1× PBS or 0.05% Triton X-100 (Sigma, X100) (for PGR) were performed before blocking for 1 h at room temperature in 5% goat serum albumin for rabbit antibody, 5% BSA for goat antibody, or MOM Blocking Reagent (Vector, MKB-2213) for mouse or rat antibodies. For ESR1 staining, retrieval was done by microwaving slides in 1× Vector Unmasking Solution for 20 min. For rabbit and goat antibodies, primary antibodies were added in blocking reagent overnight at 4 °C. Biotin-conjugated anti-rabbit 1:1000 (Novus, NB 730-B) in 5% goat serum albumin (Jackson ImmunoResearch, 005-000-001)or anti-goat 1:500 (Jackson ImmunoResearch, 711-065-152) in 5% BSA were added to slides for 1 h at room temperature. For rat and mouse antibodies, primary antibodies were added in MOM diluent (prepared as per MKB-2202) for 30 min at room temperature as directed. For rat antibody, biotinylated anti-rat (Dako, E0468) 1:500 was added for 1 h at room temperature. For mouse antibody, biotinylated anti-mouse antibody (MKB-2225) 1:250 in MOM diluent was added at room temperature for 10 min (15 min for PGR). For all antibodies, sections were washed with 0.05% Tween 20 in 1× PBS, then incubated in Vector Elite ABC reagent (PK-7100) for 30 min at room temperature. For ESR1, an additional Streptavidin-HRP (Vector, SA-5704) incubation step was performed for 30 min as room temperature. All sections were incubated in DAB + chromogen substrate (Dako, K3468) for 10 min (15 min for PGR and ESR1), and counterstained with Harris Modified Hematoxylin 1:5 (Fischer Scientific, SH26-500D). Slides were mounted with Permount medium (Fisher Scientific, SP15-100). Imaging was done with an Olympus 1×51 microscope, Jenoptik Laser Optik camera system (Rutgers Cancer Institute of New Jersey), and ProgRes C10Plus software.
For immunofluorescence (IF), slides were deparrafinized with xylene, rehydrated with ethanol to 1× PBS washes, and boiled at 97 °C for 16 min in 1 M Urea (Bio-Rad, 161-0730) or 1× Vector Unmasking Solution (TNFSF11 only). For PGR staining, retrieval was done by autoclaving slides at 121 °C, 16 to 17 psi, for 30 min in 1× Vector Unmasking Solution. Slides were washed 4 times in water and once in 1× PBS, followed by blocking for 1 h at room temperature in 5% BSA (0.5% Tween-20, 1× PBS) or MOM Blocking Reagent (1× PBS for MKI67 and 0.05% Triton X-100, 1× PBS for PGR). Nonmouse primary antibodies were added in 5% BSA at room temperature for 2 h, except for the TNFSF11 antibody, which was added in 1% BSA, 0.5% Tween 20 overnight at 4 °C. PGR antibody was used in MOM diluent in 0.05% Triton X-100, 1× PBS for 2 h at room temperature. Anti-rat, anti-chicken, and anti-rabbit fluorescent antibodies 1:100 (Jackson ImmunoResearch) were used in 5% BSA for 1 h at room temperature. For TNFSF11, biotinylated anti-goat 1:500 (Jackson ImmunoResearch, 711-065-152) was used for 1 h at room temperature, followed by streptavidin-Texas Red 1:100 (Invitrogen, SA1017) for 30 min. For PGR, biotinylated anti-mouse 1:250 in MOM diluent in 0.05% Triton X-100, 1× PBS was used at room temperature for 15 min, followed by streptavidin-Texas Red 1:100. For KRT6, CD29 and CD24 costaining, slides were first stained with KRT6 and CD29 antibodies, anti-rabbit Alexa-647 1:100 (Invitrogen, A20991) and anti-rat TRITC, then with the CD24-PE. After all antibody incubations, sections were washed with 1× PBS 3 times and mounted with ProLong Gold antifade with DAPI. Confocal imaging was done using the LSM510 Meta Confocal Laser Scanning Microscope at the Keck Collaborative Neuroscience Center at Rutgers University.
Data quantification and statistical analysis
For all in vitro studies statistics, 2-tailed Student t tests were performed using http://graphpad.com/quickcalcs/ttest1/. Immunostaining quantification was performed on at least 3 individual mammary gland specimens, including all TEBs and ducts per slide. For mammary side-branching quantification, results are presented as means with standard deviation (± SD) from 3 mammary gland specimens per genotype from C57BL/6 littermates. Same results were obtained in FVB/N background. For statistics on Kaplan Meier curves, Graph Prism software and Mantel-Cox analysis were utilized.
Mouse mammary tumor gene expression analysis
A minimum of 1 million cells were harvested and lysed directly from culture plates using 2 mL Trizol (Invitrogen, 15596-026). Cell lysates were incubated on ice for 5 min, then vortexed for 1 min. Tumors were dissected from mice and snap frozen in liquid nitrogen. Cell lysates and tumor specimens were given to the Functional Genomics Core Facility at Rutgers CINJ for RNA extraction and gene expression profiling on Affymetrix Mouse Genome 430 2.0 microarray. Using GeneSpring GX 11.5 (Agilent Technologies, Inc., Santa Clara, CA, USA), raw gene expression signals were summarized with MAS5. The data was further normalized using baseline transformation by the median of all samples and log scaling. Further, the normalized expression signals were averaged between biological replicates where applicable. Gene expression data was first filtered by flags (labeled as “present,” “marginal,” or “absent”). Probes were only kept if at least one sample was labeled as “present.” Gene lists were generated by pair-wise comparison. Differentially expressed genes were designated by looking for a significant fold change (> 2.0) between the normalized expression values of 2 experiments. Genes were considered significantly stagnant if the fold change was low (< 1.25).
The generated gene list data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). The Functional Analysis identified the biological functions and/or diseases that were most significant to the data set. Molecules from the data set that met the fold change cutoff of 2-fold and were associated with biological functions and/or diseases in Ingenuity's Knowledge Base were considered for the analysis. The right-tailed Fisher exact test was used to calculate a P value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
Human breast cancer cohorts
The Sabatier43 cohort included 266 breast tumor samples analyzed with Affymetrix U133 Plus 2.0 Arrays [PMID: 20490655]. The data was downloaded from Gene Expression Omnibus, series GSE21653. The Hatzis42 cohort included 508 ERBB2-negative breast tumor samples analyzed with Affymetrix U133A Arrays [PMID: 21558518]. The data was downloaded from Gene Expression Omnibus, series GSE25066. Raw CEL files were processed using the justRMA function in R Bioconductor. The samples were classified into BECN1 high or BECN1 low depending on whether BECN1 mRNA expression (probe 208945_s_at) was above or below the average across samples. WNT pathway gene signature was based on KEGG pathway: hsa04310 for WNT signaling pathway-Homo sapiens (http://www.genome.jp/dbget-bin/www_bget? hsa04310). An activated WNT pathway gene signature was attributed to tumors with significant (P < 0.05) upregulation of WNT pathway gene signature expression relative to the rest of tumors within each breast cancer cohort; a Fischer exact test was used for this calculation.
Pathway gene signatures
Pathway gene expression signatures (according to KEGG pathway annotations) were analyzed using Gene Set Enrichment Analysis (GSEA) [PMID: 16199517]. For each sample and signature, GSEA reports a signature expression score between 0 and 1 and the statistical significance (P value) for signature overexpression. The statistical significance of upregulation of a given signature on a group of samples was determined using a Fischer exact test for enrichment of samples with significant (P < 0.05) signature expression in the group relative to samples outside the group.
Acknowledgments
We thank Z Yue and S Jin (Mount Sinai School of Medicine and Rutgers University), Dr Komatsu (Tokyo Metropolitan Institute of Medical Science), Dr Mizushima (University of Tokyo), and Y Li (Baylor College of Medicine) for providing Becn1+/−, Atg7F/F, GFP-LC3B, and MMTV-Wnt1 mice, respectively. We thank Dr Segre (National Institute of Health) for the KRT14 antibody and Dr Clarke (Westmead Millennium Institute) for the PGR antibody. We also thank N Goldsmith-Kane for assistance with confocal microscopy, the Histopathological, Imaging, and Functional Genomics Core Facilities at Rutgers Cancer Institute of New Jersey; also, the Flow Cytometry Facility at Princeton University. We are indebted to Ed and Sue Goldstein for their unconditional support and encouragement. This work is dedicated to the memory of Stacy Goldstein.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
We are grateful for financial support from a New Jersey Commission on Cancer Research Predoctoral Fellowship (MC), NIH grant R00-CA133181 (VK) and a Damon Runyon Clinical Investigator Award (VK).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
Articles from Autophagy are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.4161/auto.34398
Read article for free, from open access legal sources, via Unpaywall:
https://www.tandfonline.com/doi/pdf/10.4161/auto.34398?needAccess=true
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.4161/auto.34398
Article citations
Autophagy is required for mammary tumor recurrence by promoting dormant tumor cell survival following therapy.
Breast Cancer Res, 26(1):143, 18 Oct 2024
Cited by: 0 articles | PMID: 39425240 | PMCID: PMC11488247
Autophagy Deficiency Induced by SAT1 Potentiates Tumor Progression in Triple-Negative Breast Cancer.
Adv Sci (Weinh), 11(36):e2309903, 29 Jul 2024
Cited by: 1 article | PMID: 39073262 | PMCID: PMC11423137
Amentoflavone regulates the miR-124-3p/CAPN2 axis to promote mitochondrial autophagy in HCC cells.
Toxicol Res (Camb), 13(4):tfae110, 23 Jul 2024
Cited by: 1 article | PMID: 39050595
BECN1 mRNA expression in breast cancer tissue; significant correlation to tumor grade.
Mol Genet Genomics, 299(1):56, 24 May 2024
Cited by: 0 articles | PMID: 38787424 | PMCID: PMC11126480
Elucidating the crosstalk between endothelial-to-mesenchymal transition (EndoMT) and endothelial autophagy in the pathogenesis of atherosclerosis.
Vascul Pharmacol, 155:107368, 26 Mar 2024
Cited by: 1 article | PMID: 38548093 | PMCID: PMC11303600
Review Free full text in Europe PMC
Go to all (100) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus (2)
- (1 citation) GEO - GSE21653
- (1 citation) GEO - GSE25066
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
ERBB2 overexpression suppresses stress-induced autophagy and renders ERBB2-induced mammary tumorigenesis independent of monoallelic Becn1 loss.
Autophagy, 10(4):662-676, 30 Jan 2014
Cited by: 27 articles | PMID: 24492513 | PMCID: PMC4091153
Keratinocyte autophagy enables the activation of keratinocytes and fibroblastsand facilitates wound healing.
Autophagy, 17(9):2128-2143, 18 Sep 2020
Cited by: 71 articles | PMID: 32866426 | PMCID: PMC8496719
RARα1 control of mammary gland ductal morphogenesis and wnt1-tumorigenesis.
Breast Cancer Res, 12(5):R79, 05 Oct 2010
Cited by: 15 articles | PMID: 20923554 | PMCID: PMC3096972
Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Prog Neurobiol, 106-107:33-54, 01 Jul 2013
Cited by: 171 articles | PMID: 23827971
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P30 CA072720
Grant ID: R00-CA133181





