Abstract
Free full text

Beclin 1 regulates growth factor receptor signaling in breast cancer
Abstract
Beclin 1 is a haplo-insufficient tumor suppressor that is decreased in many human tumors. The function of Beclin 1 in cancer has been attributed primarily to its role in the degradative process of macroautophagy. However, Beclin 1 is a core component of the Vps34/Class III PI3K (PI3KC3) and Vps15/p150 complex that regulates multiple membrane trafficking events. In the current study, we describe an alternative mechanism of action for Beclin 1 in breast cancer involving its control of growth factor receptor signaling. We identify a specific stage of early endosome maturation that is regulated by Beclin 1, the transition of APPL1-containing phosphatidyIinositol 3-phosphate-negative (PI3P-) endosomes to PI3P+ endosomes. Beclin 1 regulates PI3P production in response to growth factor stimulation to control the residency time of growth factor receptors in the PI3P-/APPL+ signaling competent compartment. As a result, suppression of BECN1 sustains growth factor stimulated AKT and ERK activation resulting in increased breast carcinoma cell invasion. In human breast tumors, Beclin 1 expression is inversely correlated with AKT and ERK phosphorylation. Our data identify a novel role for Beclin 1 in regulating growth factor signaling and reveal a mechanism by which loss of Beclin 1 expression would enhance breast cancer progression.
Introduction
Beclin 1 is the mammalian homolog of the yeast Atg6/Vacuolar protein sorting 30 (Vps30) protein that plays an essential role in macroautophagy (hereafter referred to as autophagy) and vacuolar protein sorting1,2. Beclin 1 is required for normal mammalian development and it has been shown to play a critical role in pathogenesis, including cancer3,4. Mice heterozygous for Beclin 1 expression (BECN1+/-) have an increased incidence of lymphoma and carcinomas of the lung and liver, and these mice also show preneoplastic changes in the mammary gland tissue as they age3,4. Additionally, heterozygous loss of Beclin 1 promotes mammary tumorigenesis in response to parity and WNT1 activation5. These studies support a role for Beclin 1 as a haploinsufficient tumor suppressor gene in a context-dependent manner. In many human tumors Beclin 1 mRNA and protein expression are decreased, the result of both monoallelic loss of the BECN1 gene and aberrant DNA methylation that suppresses gene expression6. Mutation of BECN1 has not been detected in tumors7. Reduced Beclin 1 protein expression is associated with poor prognosis in several cancer types, including breast8,9.
The investigation of Beclin 1 function in cancer has focused primarily on its role in regulating autophagy. BECN1+/- mice show evidence of decreased autophagy and immortalized BECN1+/- mouse mammary epithelial cells exhibit a ROS-mediated DNA damage response that is attributed to the accumulation of damaged mitochondria when autophagy is impaired. This increase in DNA damage has been implicated in transformation10. However, additional membrane trafficking functions of Beclin 1 play significant roles in tissue homeostasis and their dysregulation could contribute to tumor cell biology. Beclin 1 regulates membrane trafficking through its interaction with Vps34/Class III PhosphatidylinositoI-3 Kinase (PI3KC3), a lipid kinase that phosphorylates PI to generate the lipid second messenger PI-3 phosphate (PI3P)11,12. PI3P mediates the recruitment of proteins containing FYVE (Fab1p, YOTB, Vac1p, EEA1) and PX (Phox homology) domains to intracellular membranes to facilitate vesicular fusion and sorting events13-15. Beclin 1 regulates distinct membrane trafficking pathways through mutually exclusive recruitment of additional binding partners. For example, Beclin 1 interacts with Atg14/ATG14L/Barkor (Complex I) to regulate autophagy initiation and Vps38/UVRAG (Complex II) to regulate vacuolar protein sorting, cytokinesis and receptor degradation16,17.
Given the importance of growth factor receptor signaling in normal cell function and the consequences of its dysregulation in cancer, we assessed the role of Beclin 1 in regulating receptor endocytic trafficking and signaling, and sought to determine the mechanism involved. The data we report here reveal a novel role for Beclin 1 in regulating growth factor receptor signaling that involves the control of early endosome maturation. Our findings have important implications for understanding how Beclin 1 loss contributes to cancer.
Results
Intracellular signaling by growth factor receptors is controlled by trafficking through compartments of the endocytic pathway. Factors that regulate receptor movement from the early endosome compartment to either the late endosome/lysosome compartment for signal termination and degradation or the recycling endosome compartment for restoration of surface expression control the strength and duration of signal transduction. To investigate a role for Beclin 1 in the regulation of growth factor receptor signaling, we examined the link between Beclin 1 and two growth factor receptors that have been implicated in breast cancer, the insulin-like growth factor-1 (IGF-1) receptor and the epidermal growth factor (EGF) receptor. The IGF-1R is frequently overexpressed in breast tumors and its activation correlates with poor prognosis18,19, whereas the EGFR is frequently elevated in expression and activation in the triple negative breast cancer (TNBC) subtype20.
MDA-MB-231 cells (TNBC subtype), MCF-7 cells (luminal breast cancer subtype) and MCF10A cells (immortalized mammary epithelial cells) were stably infected with shRNAs targeting BECN1 (shBECN1) or GFP (shGFP). To monitor signaling, phosphorylation of key downstream signaling effectors of the IGF-1R and EGFR, the serine/threonine kinases AKT and ERK1/2, was assessed in response to a time course stimulation with either IGF-1 (Figure 1A and Figure S1A,B) or EGF (Figure 1B). AKT and ERK1/2 were phosphorylated in a transient manner in response to IGF-1 and EGF stimulation in control shGFP cells. In contrast, phosphorylation levels increased and were sustained at later time points in the shBECN1 cells (quantified in Figures 1C-F and Figure S1C,D). Although similar trends were observed for both AKT and ERK in response to both ligands, the impact on AKT phosphorylation was significant downstream of the IGF-1R and ERK1/2 phosphorylation downstream of the EGFR. These data reflect the dominant signaling pathways that are activated by each of these receptors. In addition, the IGF-1-dependent increase in AKT phosphorylation upon Beclin 1 loss was greater for T308 than for S473. Ectopic expression of Beclin 1 significantly decreased AKT activation in the parental MDA-MB-231 cells (Figure 1G), whereas the elevated signaling observed in the shBECN1 MDA-MB-231 cells decreased upon rescue of Beclin 1 expression (Figure 1H). Taken together, our data reveal that Beclin 1 negatively regulates the intensity and duration of growth factor receptor signaling.

Beclin 1 regulates growth factor-dependent signaling. (A-F) MDA-MB-231 cells expressing shGFP, shBECN1 (#1) or shBECN1 (#2) were stimulated with IGF-1 (100ng/ml) (A) or human EGF (50ng/ml) (B) for the indicated time periods. Phospho-AKT(S473) (C,E), phospho-AKT(T308) (D) and phospho-ERK1/2(T202/Y204) (F) levels were quantified by densitometry. The data shown represent the mean of three (D-F) or four (C) independent experiments. (G) MDA-MB-231 cells expressing Flag-Beclin 1 were stimulated with IGF-1 (100ng/ml) for the indicated time periods. The data shown represent the mean of three independent experiments. (H) MDA-MB-231 cells expressing shGFP, shBECN1 or shBECN1 and Flag-Beclin 1 were stimulated with IGF-1 (100ng/ml) for the indicated time periods. The data shown represent the mean of three independent experiments. *, p < 0.05; **, p < 0.005; ***, p < 0.0005. See also Figure S1.
Growth factor receptor signaling initiates at the cell surface and continues upon receptor internalization into early endosomes21. To investigate how Beclin 1 regulates signaling, we prevented receptor internalization using the clathrin inhibitor chlorpromazine22. IGF-1-dependent activation of AKT was markedly diminished, but equivalent, in both the shGFP and shBECN1 cells in the presence of the clathrin inhibitor (Figure S2). This result suggested that Beclin 1 regulates growth factor receptor signaling post-internalization by controlling the trafficking of receptors from an early endosomal compartment that retains active signaling capacity23. Recent studies have revealed that early endosomes, which are positive for the GTPase Rab5, are comprised of functionally distinct subpopulations of vesicles that differ in binding partners and signaling potential24,25. One such subpopulation is characterized by the presence of the adaptor protein APPL1, which binds both Rab5 and AKT and can regulate both AKT and MAPK activation26-28. APPL1 also interacts with growth factor receptors including the EGFR29 and insulin receptor30. APPL1-positive (APPL1+) endosomes lack the lipid PI3P. When they mature from PI3P negative (PIP3-) endosomes to PI3P+ endosomes, APPL1 is displaced by FYVE-domain containing proteins, such as EEA1 and rabenosyn5, which bind competitively to both Rab5 and PI3P25. In support of a role for Beclin 1 in regulating early endosome trafficking, knockout of Beclin 1 (ATG6) in the Drosophila fat body interferes with the movement of Rab5-positive vesicles from the plasma membrane to the perinuclear compartment31.
To determine if suppression of Beclin 1 inhibits the maturation of PI3P-/APPL+ endosomes, shGFP and shBECN1 cells were stained with APPL1-specific antibodies to evaluate the presence of APPL1+ endosomes. APPL1+ endosomes were more abundant and persisted over the time course of IGF-1 stimulation in the shBECN1 cells, compared with the shGFP cells (Figure 2A,B). We also decreased the expression of ATG5, an essential autophagy gene that is required for the formation and maturation of autophagosomes32. The number of APPL1+ endosomes increased modestly only at the very earliest time point of stimulation in shATG5 cells when compared to control shGFP cells (Figure 2A,B). Suppression of ATG5 significantly impaired autophagy in response to serum deprivation, as monitored by LC3 II levels (Figure 2C), whereas suppression of BECN1 resulted in only a modest reduction in LC3 II conversion. The different results observed with Beclin 1 and ATG5 loss suggest that the regulation of early endosome trafficking involves Beclin 1 functions that are not exclusively related to its role in autophagy. Of note, suppression of APPL1 expression only modestly inhibited IGF-1-dependent signaling to AKT, indicating that it may contribute, but is not required for, activation of this signaling pathway (data not shown).
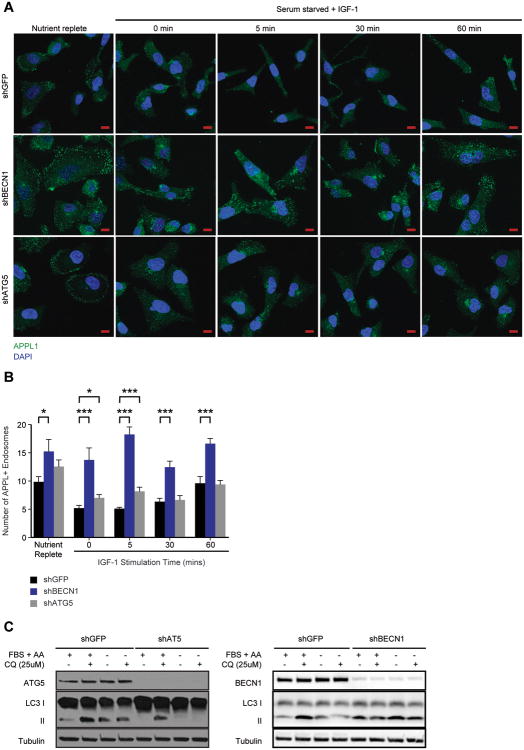
Beclin1 regulates APPL1+ endosomes. (A) MDA-MB-231 cells expressing shGFP, shBECN1 or shATG5 were stimulated with IGF-1 (100ng/ml) for the time periods indicated. Cells were fixed and stained with antibodies that recognize APPL1 (green). Nuclei were stained with DAPI (blue). Scale bar, 10 μm. (B) Quantification of APPL+ endosomes shown in (A). The data shown represent the mean of 23-37 independent cells. (C) Autophagic flux in MDA-MB-231 cells expressing shGFP, shATG5 or shBECN1. Cells were incubated for 4 hours in either complete or serum and amino acid-free media with or without 25 μM Chloroquine diphosphate. FBS, fetal bovine serum; AA, amino acids. *, p < 0.05; ***, p < 0.0005. See also Figure S2.
To investigate the role of Beclin 1 in early endosome receptor trafficking further, we quantified the residency time of growth factor receptors in APPL1+ endosomes using fluorophore-conjugated EGF to follow the EGFR in real-time using Total Internal Reflection Fluorescence (TIRF) microscopy33. EGFR trafficking has been extensively studied using this approach and a corresponding fluorophore-conjugated IGF-1 is not available for these analyses. Control and shBECN1 cells were transfected with TagRFP-T-APPL1 and stimulated with fluorophore-conjugated EGF to track the EGFR (Figure 3A, Movie S1 and Movie S2). Only cells with similar APPL1 transfection efficiency and EGF uptake were used for quantification (Figure 3C). The extent of EGF and APPL1 co-localization was similar between the control and shBECN1 cells initially, but a significant increase in co-localization occurred over time in the shBECN1 cells and this co-localization persisted at an elevated level throughout the analysis (Figure 3B). These results are consistent with Beclin 1 regulating endosome maturation and suggest the possibility that PI3P+ endosomes are required for down-regulation of growth factor signaling.
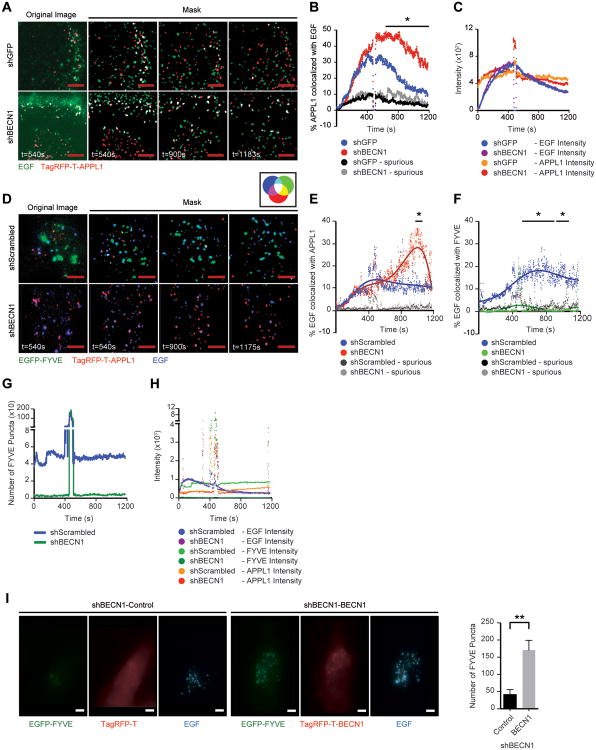
Beclin1 regulates growth factor-dependent early endosome maturation. (A) MDA-MB-231 shGFP or shBECN1 cells expressing TagRFP-T-APPL1 (red) were exposed to Alexa-Fluor-488-EGF (200ng/ml) (green) and imaged for ~20 mins. The far left panels are representative unprocessed images with background removed, while the remaining panels are overlapped masks of images from the individual time points after ligand stimulation. Co-localized signals are represented in white. (B) Percent of APPL1 colocalized with EGF in 100×100 pixels section of each cell (8 sections for shGFP and 7 sections for shBECN1) plotted over the indicated time interval. (C) Average pixel intensity of EGF and APPL1 in cells after ligand stimulation. Spikes represent the wash out of EGF. The data shown represent the mean of two independent experiments performed in duplicate or triplicate. (D) shScrambled and shBECN1 cells expressing EGFP-FYVE (green) and TagRFP-T-APPL1 (red) were exposed to DyLight 650 labelled EGF (200ng/ml) (blue) and imaged for ~20 mins. The far left panels are representative unprocessed images with background removed, while the remaining panels are overlapped masks of images from the individual time points after ligand stimulation. Co-localized signals are represented in colors indicated in the accompanying color wheel. (E,F) Percent of EGF colocalized with APPL1 (E) and FYVE (F) in each cell (n=6). (G) Number of FYVE+ puncta present in the TIRF zone at each time point. (H) Average pixel intensity of EGF, FYVE and APPL1 in cells after ligand stimulation. Spikes represent the wash out of EGF. The data shown represent the mean of three independent experiments performed in triplicate. Spurious, non-specific co-localization. (I) shBECN1 cells expressing either TagRFP-T or TagRFP-T-BECN1 (red) and EGFP-FYVE (green) were stimulated with DyLight 650 labelled EGF (200ng/ml) (blue) for 10 mins before fixation. The number of FYVE positive puncta in each cell was quantified. The data shown represents the mean of two experiments (n=11). Scale bar, 3 μm. *, p < 0.05; **, p < 0.005. See also Movies S1-S4.
If suppression of Beclin1 decreases PI3P generation and therefore delays transition of PI3P- endosomes to PI3P+ endosomes, a corresponding decrease in receptor co-localization with the latter should occur. To monitor PI3P+ endosomes, a GFP-tagged FYVE domain from the endosomal protein SARA (EGFP-FYVE) that binds PI3P was co-transfected with TagRFP-T-APPL1 and the cells were stimulated with fluorophore-conjugated EGF (Figure 3D, Movie S3 and Movie S4). Only cells with similar APPL1 and FYVE transfection efficiencies and EGF uptake were used for quantification (Figure 3H). As was observed in the cells expressing TagRFP-APPL1 alone, a significant increase in co-localization of EGF with APPL1 occurred over time in the shBECN1 cells (Figure 3E). In unstimulated cells, PI3P levels are low and EGFP-FYVE is found diffusely in the cytoplasm. Upon stimulation with EGF, EGFP-FYVE was recruited to endosomes in the TIRF zone in the control cells and their co-localization with EGF increased over time (Figures 3F,G). In contrast, EGFP-FYVE remained diffuse in the shBECN1 cells and very few positive endosomes were observed within the TIRF zone (Figures 3F,G). Rescue of Beclin 1 expression in the shBECN1 cells restored the presence of FYVE+ endosomes (Figure 3I).
As an additional measure of PI3P+ endosome maturation, cells transfected with TagRFP-T-APPL1 and stimulated with fluorophore-conjugated EGF were fixed after 25 mins of ligand exposure and stained with antibodies specific for EEA1, a marker of Rab5/PI3P+ early endosomes (Figure 4A). Once again, a significant increase in the amount of APPL1 that co-localized with EGF was observed at this fixed time point in the shBECN1 cells (Figure 4C). A corresponding significant decrease in the co-localization of EGF with EEA1 was also revealed, confirming a delay in the transition of Rab5/PI3P- to Rab5/PI3P+ endosomes (Figure 4D). Total EEA1 levels were equivalent in the shGFP and shBECN1 cells (Figure 4B).
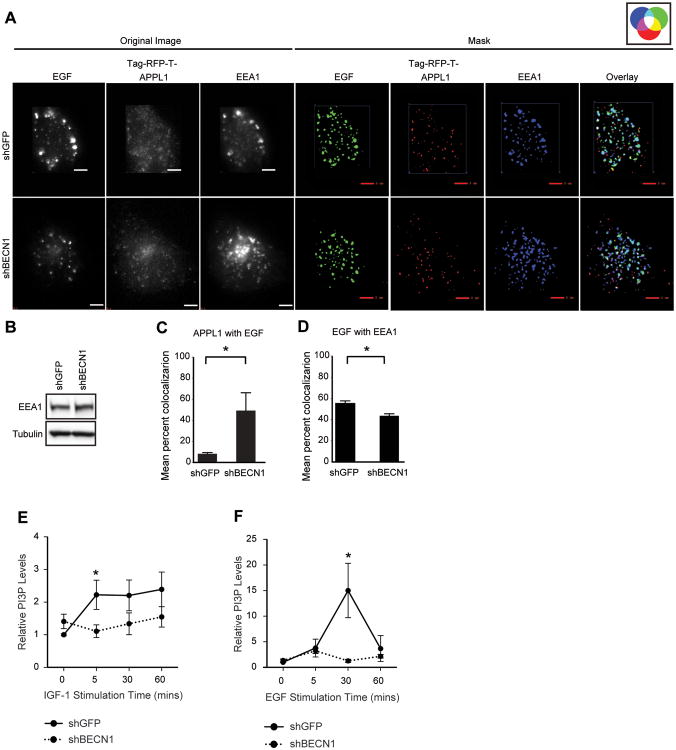
Beclin 1 controls PI3P levels and the maturation of early endosomes. (A) Cells expressing TagRFP-T-APPL1 (red) were stimulated with Alexa-Fluor-488-EGF (200ng/ml) (green) for 25 mins, fixed and stained with antibodies that recognize EEA1 (blue). Original images without background noise (left) were processed to generate masks (right). Co-localized signals are represented by colors indicated in the accompanying color wheel. Scale bar, 3 μm. (B) Immunoblots of cell extracts from (A). (C,D) Quantification of percent co-localization between fluorophores in six independent cells. (E,F) MDA-MB-231 cells expressing either shGFP or shBECN1 were stimulated with IGF-1 (100ng/ml) (E) or EGF (50ng/ml) (F) for the indicated time periods. Total PI3P lipid levels were measured by ELISA assay. The data shown represents the mean of two (EGF; n=4) or three (IGF-1; n=9) independent experiments.
The increase in EGFP-FYVE+ endosomes in response to EGF stimulation and the absence of these endosomes in the shBECN1 cells indicate that Beclin 1 is required for the generation of PI3P in response to growth factor stimulation. To examine directly this role for Beclin 1 in the growth factor-dependent regulation of PI3P generation, we measured total cellular PI3P lipids in response to growth factor stimulation using a competitive ELISA assay. Stimulation of shGFP cells with either IGF-1 or EGF increased PI3P levels significantly (Figures 4E,F). Importantly, PI3P levels did not increase upon stimulation of the shBECN1 cells with either IGF-1 or EGF, which is consistent with the absence of EGFP-FYVE endosomes in the imaging experiments. Combined, our data support the hypothesis that Beclin 1 regulates PI3KC3 activation in response to growth factor stimulation and contributes to the dynamic changes in PI3P levels that accompany endosome maturation.
AKT regulates migration and invasion in many tumor cell types, including breast carcinoma cells34,35. Therefore, we investigated whether loss of Beclin 1, and the resulting sustained activation of AKT, promotes migration or invasion. Suppression of Beclin 1 expression increased the invasion, but not migration, of MDA-MB-231 (Figure 5A) and SUM159 (Figure S3) cells by two-four fold compared to control cells (shGFP) and restoration of Beclin 1 expression in the shBECN1 cells restored invasion to control levels (Figure 5B). Importantly, inhibition of AKT activity using the inhibitor MK2206 suppressed the increased invasion of the MDA-MB-231 shBECN1 cells (Figure 5C). The lack of an increase in migration in the shBECN1 cells suggests that the sustained AKT signaling that occurs upon loss of Beclin 1 selectively regulates pathways important for invasion, such as protease expression, and does not control general cell movement.
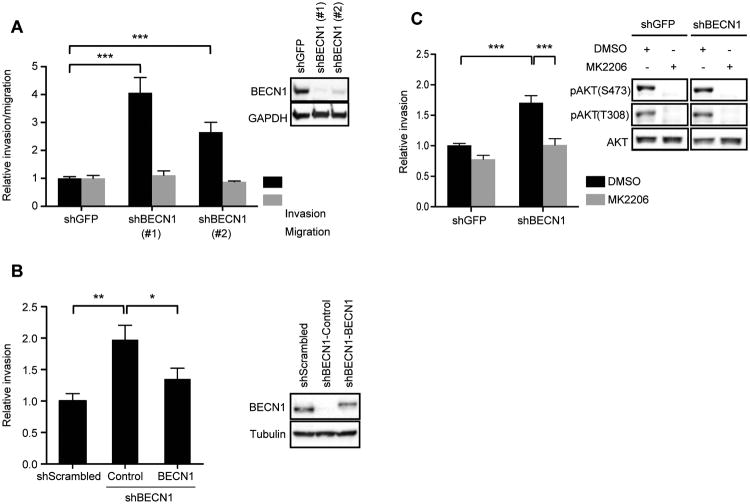
Beclin 1 regulates breast carcinoma cell invasion in an AKT-dependent manner. (A) MDA-MB-231 cells expressing shGFP, shBECN1 (#1) or shBECN1 (#2) were assayed for their ability to migrate (collagen 1) or invade (Matrigel). The data shown represent the mean of three (invasion; n = 7) and two (migration; n = 5) independent experiments performed in duplicate or triplicate. Inset, immunoblots showing Beclin 1 expression in the shBECN1 cells. (B) MDA-MB-231 cells expressing shBECN1 or shBECN1 with restored FLAG-Beclin 1 expression were assayed for their ability to invade Matrigel. The data shown represent the mean of three independent experiments performed in triplicate. Inset, immunoblots showing Beclin 1 expression. (C) MDA-MB-231 cells were assayed for their ability to invade Matrigel after treatment with the AKT inhibitor MK2206 (0.5 μM). The data shown represent the mean of four independent experiments performed in duplicate or triplicate (n = 10). Inset, immunoblots showing phospho-AKT(S473) and (T308) upon treatment with 0.5 μM MK2206 for 1 hour. ***, p < 0.0005. See also Figure S3.
To assess the impact of Beclin 1 on AKT and ERK activation in human breast tumors, we used cBioPortal to analyze the correlation between Beclin 1 expression and AKT and ERK phosphorylation in a cohort of breast cancer patients that had altered levels of Beclin 1 protein expression as measured by reverse phase protein array (RPPA)36,37. A statistically significant negative correlation between Beclin 1 and pAKT (S473; Pearson r = -0.3612) and pERK (T202; Pearson r = -0.3904) was observed (Figure 6).
Discussion
We describe a novel mechanism of action for Beclin 1 in breast cancer involving its control of growth factor receptor signaling. Beclin 1 regulates PI3P lipid levels in response to growth factor stimulation to control the rate at which growth factor receptors transit through a signaling-competent early endosome compartment. In doing so, Beclin 1 controls the intensity and duration of growth factor stimulated AKT and ERK signaling. We demonstrate that suppression of BECN1 sustains growth factor stimulated AKT and ERK activation in breast carcinoma cells in vitro, and that an inverse correlation between Beclin 1 expression and AKT and ERK phosphorylation is also observed in human breast tumors. Functionally, reduction of Beclin 1 expression enhances invasion in an AKT-dependent manner. Taken together, our data reveal a mechanism by which dysregulation of Beclin 1 expression or function contributes to malignant behavior.
Beclin 1 is an independent prognostic factor for overall (OS) and distant-metastasis free survival in breast cancer, with low levels of Beclin 1 expression predicting worse OS8. Low Beclin 1 expression together with active WNT signaling also correlates with poor prognosis in breast cancer5. However, the mechanism to explain how loss of Beclin 1 expression would contribute to aggressive tumor behavior remains unresolved. Autophagy, which is thought to support tumor progression by facilitating the survival of tumor cells in stressful microenvironments that may lack sufficient oxygen and nutrients, is inhibited when Beclin 1 expression is decreased. Our study provides evidence for an alternative mechanism of Beclin 1 action to explain how its loss promotes tumor progression. Specifically, we show that Beclin 1 loss enhances oncogenic signaling pathways such as AKT and MAPK that promote survival and invasion. In this direction, an autophagy-independent role for Beclin 1 loss has been suggested to contribute to the enhanced mammary stem and progenitor cell activity and tumorigenesis that is observed in WNT1/BECN1+/- mice5. Taken together with the recent Drosophila study showing that Beclin 1 regulates Notch trafficking, these findings support the possiblitity that multiple receptor trafficking pathways may be impacted by Beclin 1 loss and that this combined dysregulation culminates in enhanced signaling to promote tumor progression38.
We establish an important role for Beclin 1 in the regulation of early endosome maturation. Beclin 1 is required for the growth factor-stimulated generation of PI3P by PI3KC3, which facilitates the recruitment of FYVE domain containing proteins, such as EEA1, that promote endosome fusion and maturation33,39. Another FYVE-domain protein, Fab1/PIKfyve (FYVE-containing phosphatidylinositol 3-phosphate 5-kinase), is recruited to Rab5+/PI3P+ endosomes where it is activated in response to phosphorylation by AKT40. PIKfyve phosphorylates PI3P to generate PI-3,5-P and promote endosome progression to multivesicular bodies where signaling is terminated. Reduced PI3P production in the absence of Beclin 1 would be expected to limit the recruitment of PIKfyve and therefore inhibit maturation. Finally, Beclin 1 recruits UVRAG, which enhances PI3KC3 activation and PI3P production41,42. UVRAG also promotes early endosome maturation by activating C-VPS/HOPS to facilitate Rab7 recruitment and transition to the late endosome, a function that would also be lost in the absence of Beclin 143.
Our studies model the reduction of Beclin 1 expression that occurs in many tumors8,9. Other tumors, however, may exhibit a decrease in Beclin 1 function as a consequence of post-translational modifications, and this disruption would also be expected to impact growth factor receptor trafficking and signaling. For example, activated EGFR binds to Beclin 1 and promotes its phosphorylation on multiple tyrosine residues44. These phosphorylation events disrupt the interaction of Beclin 1 with PI3KC3 and decrease its associated lipid kinase activity. Additionally, Beclin 1 is phosphorylated on S234 and S295 by AKT, which also disrupts PI3KC3 binding 45. These pathways likely function to regulate the rate of receptor trafficking and duration of signaling in normal cells, but when hyperactivated in tumors could further enhance their oncogenic signaling potential. In support of an involvement of this post-translational mechanism of regulation in cancer, expression of a Beclin 1 tyrosine phosphomimetic in NSCLC cells increased proliferation and stimulated enhanced tumor growth44. Similarly, expression of a Beclin 1 mutant that is resistant to AKT phosphorylation inhibited Myr-AKT1-dependent tumorigenesis45. Although these effects of the Beclin 1 phospho-mutants on tumor growth were attributed to the impact of Beclin 1 on autophagy, dysregulated growth factor receptor signaling would also contribute to these phenotypes.
Our study differs from previous work that has focused exclusively on the effect of Beclin 1 loss on autophagy and the contribution of this degradative pathway to cancer. We reveal an alternative mechanism to account for how Beclin 1 loss may impact tumor progression that involves enhancing the magnitude and duration of signals propagated through growth factor receptors, many of which have been identified as oncogenic drivers. Reduction of Beclin 1 expression in tumors would result in dysregulation of both growth factor receptor signaling and autophagy, and these pathways may work in concert to promote tumor progression.
Materials and Methods
Cell lines and antibodies
MDA-MB-231 and MCF-7 human breast carcinoma cell lines were obtained from ATCC. MCF-10A and SUM-159 cell lines were a gift from Dr. A. Mercurio (UMass Medical School). Stable knockdowns were generated using lentiviral vectors containing small hairpin RNAs (shRNA) targeting human BECN1, human ATG5 and GFP, or a scrambled shRNA (Open Biosystems). After infection, cells were selected for stable expression with puromycin (2 μg/ml). For over-expression and Beclin 1 rescue, Beclin 1 was expressed transiently using a lentiviral vector containing Flag-Beclin 1. 48 hours post-infection the cells were stimulated for the indicated time periods.
Antibodies used for immunoblotting or immunofluorescence staining, with the exception of tubulin (Sigma), were purchased from Cell Signaling Technology, Inc: pIGF-1Rβ (Y1135/1136) (Catalog # 3024), IGF-1Rβ (Catalog # 3027), pEGFR (Y1086) (Catalog # 2220), EGFR (Catalog # 4267), pAKT (S473) (Catalog # 9271), pAKT (T308) (Catalog # 2965), AKT (Catalog # 9272), pERK1/2 (T202/Y204) (Catalog # 9106), MAPK (Catalog # 9102), Beclin 1 (Catalog # 3738), ATG5 (Catalog # 2630), LC3B (Catalog # 3868) and APPL1 (Catalog # 3858). Antibodies recognizing EEA1 were generated as described previously46.
Plasmids
Lentiviral Flag-Beclin 1 was constructed by cloning Flag-Beclin 1 (Dr. Stephen Doxsey, UMass Medical School) into the EcoRI cloning site of the lentiviral expression vector pCDH-CMV-MCS-EF1-copGFP (System Biosciences). TagRFP-T-BECN1 was generated by cloning Flag-Beclin 1 in frame with TagRFP-T at the N terminus of the protein using the EcoRI cloning site46. For rescue of Beclin 1 expression, silent mutations were introduced into the region of BECN1 targeted by the shRNA using the QuikChange site-directed mutagenesis kit (Stratagene). EGFP-FYVE containing the FYVE domain of human SARA (589-656) and TagRFP-T-APPL1 have been described previously47.
Growth factor stimulation assays
Cells were serum-starved for either 2 hrs or overnight in media containing 0.1% BSA and then stimulated with IGF-1 (100 ng/ml) or hEGF (50ng/ml) for the time periods indicated in the Figures. To inhibit internalization, cells were incubated with 10ug/ml chlorpromazine for 1 hour prior to and during stimulation. Cells were infected with lentiviruses containing shBECN1-resistant Flag-BECN1 for over-expression and rescue experiments and stimulated 48hrs post-infection. Cells were extracted at 4°C for 10 minutes in a 20 mM Tris buffer, pH 7.4, containing 0.15 M NaCl, 1% NP-40, 10% Glycerol, 1 mM sodium orthovanadate, 10 mM NaF and protease inhibitors (Complete Mini Tab; Roche). Cell extracts were immunoblotted as described previously48. Bands were detected by chemiluminescence using a ChemiDoc XRS+ system (BioRad Laboratories) and band intensities were quantified by densitometry using Image Lab (Beta 1) (Bio-Rad Laboratories). PI3P lipid levels were measured using the PI3P Mass ELISA Kit (Catalog # K-3300) according to manufacturer's instructions.
For autophagic flux assay, MDA-MB-231 cells expressing shGFP or shATG5 were incubated in either RPMI 1640 supplemented with 10% fetal bovine serum or Hank's Balanced Salt Solution (GIBCO®), with and without 25 μM Chloroquine diphosphate (Catalog # C6628; SIGMA) for 4 hours.
Invasion and migration assays
Invasion and migration assays were performed as described previously48. Cells were infected with lentiviruses containing shBECN1-resistant Flag-BECN1 48hrs prior to the assays. Cells were pre-incubated with 0.5 μM MK2206 for 1 hour prior to the assays. Inhibitors were also present in the upper and lower wells of the Transwell chamber during the assays.
Image Analysis
Cells were plated on glass coverslips, serum-starved overnight and stimulated with IGF- 1 (100 ng/ml) before fixing in 4% formaldehyde for 30 mins. Fixed cells were blocked for an hour in 1× DPBS containing 3% BSA and 0.5% Triton X-100 and then incubated in the same buffer containing primary antibody for 1 hour. Following three washes with 1× DPBS, cells were incubated in secondary antibody (Alexa Fluor 488 or Alexa Fluor 660 from Molecular Probes; Life Technologies Corporation) diluted in blocking buffer for 1 hour. Stained cells were imaged using a Zeiss LSM700 confocal microscope with a 63×, 1.4 NA Plan Apochromat oil objective equipped with a AxioVision Imager.Z2 camera. Zen 2012 software was used to acquire the images.
The Total Internal Reflection Fluorescence (TIRF)/Epi-fluorescence Structure-illumination Microscopy (TESM) is a custom-built microscope system that simultaneously combines TIRF and wide-field epi-fluorescence modes46. The latter incorporates structured illumination for fast optical sectioning and enhanced spatial resolution. Two diode-pumped solid-state 100mW lasers (Cobalt) produced 491 nm and 561 nm light while a 120mW diode laser (Blue Sky Laser) produced 660 nm light. The system uses a modified Olympus IX71 inverted microscope. The TIRF illumination is adjusted such that it passes through the edge of an Olympus TIRF 60× objective with an NA of 1.49 at an angle so as to visualize 100-200nm from the coverslip. Light form the fluorophores was collected and relayed onto a 1004×1002 Andor iXon 885 EMCCD camera, binned by 2. A Physik Instruments PIFOC was used for fine focus control. Focus stabilization was performed by pgFocus consisting of software and electronics developed by the Biomedical Imaging Group at UMass Medical School (http://big.umassmed.edu/wiki/index.php/PgFocus).
Cells were transfected using Cell Line Nucleofector® Kit V (Lonza Cologne AG, Germany). For imaging, cells were plated on glass coverslips and incubated at 35°C in KRH buffer (125 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 1.2 mM MgSO4, 25 mM HEPES pH 7.4, 2 mM sodium pyruvate and 0.5% BSA). Cells transfected with TagRFP-T-APPL1 alone were stimulated with Alexa-Fluor-488 EGF complex (Invitrogen) and cells transfected with EGFP-FYVE47 and TagRFP-T or TagRFP-T-BECN1 were stimulated with biotinylated EGF (Catalog # E3477; Life Technologies) complexed with streptavidin DyLight 650 conjugate (Catalog # 84547; Thermo Scientific). EGF was washed out after 8 mins of stimulation and replaced with KRH buffer. For Figure 3A the time series protocol used lasers emitting 491 nm (EGF) and 561 nm (TagRFP-T-APPL1) while for Figure 3D the time series protocol used lasers emitting 491 nm (EGFP-FYVE), 561 nm (TagRFP-T-APPL1) and 660 nm (EGF).
Quantitative TIRF image analysis was performed as described previously33,46. Time series raw images were first corrected by subtracting the background fluorescence followed by performing a temporal running average of three time points to reduce noise with negligible effect on the data. In order to quantitatively analyze structured fluorescence (i.e. vesicles) without interference from diffuse fluorescence, images were convolved with a small, 2D Gaussian spot (σ = 150 nm) that preserved the mean intensity and a larger, 2D Gaussian spot (σ = 300 nm) that estimated and subtracted the local background. A binary masking image was generated by visually thresholding the image so as to only select pixels belonging to the fluorescent structures (which were given a value of one) and eliminate noise (pixel value was set to zero). Masked images were generated by multiplying the binary mask with the original image to display structures with intensity greater than the average local background. Co-localization was determined by two-way overlap of the masked images of each label and co-localized pixels are shown as white (Fig. 3A) or as indicated by the accompanying color wheel (Fig. 3D, ,4A).4A). Co-localization values have been reported as percent of pixels of a label that co-localized with another label. Co-localization seen when pixel-rich regions were rotated 180 degrees relative to each other have been labeled spurious.
Epifluorescence images were generated from structured illumination data sets according to methods described previously46. Raw images were processed as described above and binary masked images were used for quantifying the number of EGFP-FYVE puncta (between 10-10000 pixel size) in each cell (Fig. 3I) and for quantitative analysis of co-localization between labels (Fig. 4A).
Statistical Analysis
Statistical analyses between two groups were performed using the two-tailed unpaired Student's t-test. Data are represented as mean ± standard error unless mentioned otherwise. A p value of 0.05 was considered statistically significant.
Supplementary Material
1
2
3
4
supp fig legends
Acknowledgments
This work was supported by National Institute of Health (NIH) grants CA142782 and CA177167 (LMS), DK60564 (SC) and CA159314 (EHB). We thank Lawrence M. Lifshitz and Clive Standley of the UMass Biomedical Imaging Group for their contributions to the programs used to analyze the TESM data. We thank Sha Zhu for assistance with cloning, Usha Acharya for assistance with the PI3P lipid measurements and Chung-Cheng Hsieh for statistical advice.
Footnotes
Conflict of Interest: The authors declare no competing financial interests.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/onc.2014.454
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/onc2014454.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/onc.2014.454
Article citations
To eat or not to eat: a critical review on the role of autophagy in prostate carcinogenesis and prostate cancer therapeutics.
Front Pharmacol, 15:1419806, 07 Jun 2024
Cited by: 0 articles | PMID: 38910881 | PMCID: PMC11190189
Review Free full text in Europe PMC
Beyond self‑eating: Emerging autophagy‑independent functions for the autophagy molecules in cancer (Review).
Int J Oncol, 64(6):57, 12 Apr 2024
Cited by: 1 article | PMID: 38606507 | PMCID: PMC11087037
Review Free full text in Europe PMC
Advances in the study of autophagy in breast cancer.
Breast Cancer, 31(2):195-204, 05 Feb 2024
Cited by: 1 article | PMID: 38315272 | PMCID: PMC10901946
Review Free full text in Europe PMC
The nonautophagic functions of autophagy-related proteins.
Autophagy, 20(4):720-734, 08 Sep 2023
Cited by: 1 article | PMID: 37682088 | PMCID: PMC11062363
Review Free full text in Europe PMC
The Role of Autophagy in the Development of Pathological Conditions of the Body.
Acta Naturae, 15(3):37-49, 01 Jul 2023
Cited by: 0 articles | PMID: 37908768 | PMCID: PMC10615188
Go to all (41) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic.
Exp Cell Res, 316(20):3368-3378, 17 Jul 2010
Cited by: 119 articles | PMID: 20643123
Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity.
Autophagy, 10(11):2036-2052, 30 Oct 2014
Cited by: 100 articles | PMID: 25483966 | PMCID: PMC4502817
The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function.
Autophagy, 1(1):46-52, 17 Apr 2005
Cited by: 269 articles | PMID: 16874027
Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Prog Neurobiol, 106-107:33-54, 01 Jul 2013
Cited by: 171 articles | PMID: 23827971
Review
Funding
Funders who supported this work.
NCI NIH HHS (6)
Grant ID: CA177167
Grant ID: R01 CA159314
Grant ID: R01 CA177167
Grant ID: CA159314
Grant ID: CA142782
Grant ID: R01 CA142782
NIDDK NIH HHS (2)
Grant ID: DK60564
Grant ID: P01 DK060564







