Abstract
Free full text

Secretory autophagy
Abstract
Autophagy, once viewed exclusively as a cytoplasmic auto-digestive process, has its less intuitive but biologically distinct non-degradative roles. One manifestation of these functions of the autophagic machinery is the process termed secretory autophagy. Secretory autophagy facilitates unconventional secretion of the cytosolic cargo such as leaderless cytosolic proteins, which unlike proteins endowed with the leader (N-terminal signal) peptides cannot enter the conventional secretory pathway normally operating via the endoplasmic reticulum and the Golgi apparatus. Secretory autophagy may also export more complex cytoplasmic cargo and help excrete particulate substrates. Autophagic machinery and autophagy as a process also affect conventional secretory pathways, including the constitutive and regulated secretion, as well as promote alternative routes for trafficking of integral membrane proteins to the plasma membrane. Thus, autophagy and autophagic factors are intimately intertwined at many levels with secretion and polarized sorting in eukaryotic cells.
Introduction
The majority of eukaryotic secreted proteins are endowed with N-terminal signal peptides, which authorize them to enter the endoplasmic reticulum (ER) and follow a well-defined secretory pathway via the Golgi apparatus for delivery to the extracellular space (Figure 1). However, an exclusive subset of purely cytosolic proteins, lacking signal peptides and thus not capable of entering the ER, are nonetheless actively secreted from the cells to perform their extracellular biological functions. This phenomenon, termed unconventional secretion (Figure 1), includes several distinct processes [1, 2]. One form of unconventional secretion (secretory autophagy [3]) is associated specifically with the autophagy pathway defined by ATG factors that govern the biogenesis of autophagic membranes [4]. The ATG factors directing canonical autophagy include ULKs (mammalian paralogs of yeast Atg1, an upstream protein kinase), Beclins (mammalian paralogs of yeast Atg6, a key component of the lipid kinase VPS34, which generates phosphatidylinositiol 3-phosphate/PI3P), and LC3s and GABARAPs (mammalian paralogs of the yeast Atg8). Classically, the above factors result in the formation of double membrane autophagosomes, which in cooperation with cargo receptors such as p62/SQSTM1 [5], capture and eliminate cytoplasmic components. Conventionally, this occurs through degradation upon fusion of autophagosomes with lysosomes. In contrast to degradative autophagy, the autophagic machinery, through a shared but partially divergent pathway, may lead to secretion/expulsion of cytoplasmic constituents instead of their degradation. Either way, the cell gets rid of the captured cytoplasmic material, but the biological functions and repercussions are different.
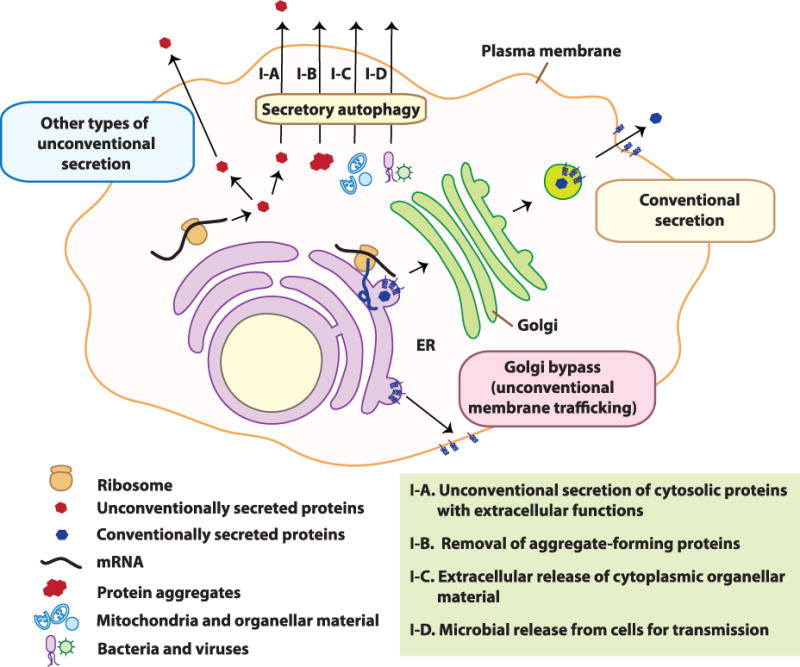
In conventional secretion, proteins possessing a signal peptide enter the lumen of the ER followed by the Golgi apparatus for secretion at the plasma membrane. In unconventional secretion, cytoplasmic proteins that lack a signal sequence and do not get delivered to the lumen of the ER are transported into the extracellular milieu via diverse unconventional secretory pathways, including secretory autophagy. For the latter, depicted re four subtypes (I–A to I–D) differentiated based on the substrates (see also Table 1).
The degradative canonical autophagy pathway, also referred to as macroautophagy, is classically considered together with microautophagy and chaperone-mediated autophagy as a collection of cytoplasmic self-digestion processes merging with lysosomes, whereby they carry out: (i) nutrient recycling functions at times of starvation by bulk digestion of the cytoplasm [6]; and (ii) cytoplasmic quality control functions [7] by removing a wide spectrum of substrates such as aggregation-prone or aggregated proteins [5, 8], damaged organelles such as irreversibly depolarized mitochondria [9], and invading microbes [10]. These well-studied aspects of degradative autophagy render it an attractive target for disease treatments [11]. In contrast to degradative autophagy, it has become slowly but increasingly apparent that autophagy has other, sometimes biogenesis-associated functions as well as a role in unconventional secretion (secretory autophagy; Figure 2). Secretory autophagy exports a range of cytoplasmic substrates (Table 1) [2, 12–19] [20, 21]. This review primarily examines the developing concept of secretory autophagy including its cargo, biological functions, and the currently limited understanding of its regulation. We will furthermore provide a brief update on the complementary processes of autophagy intersections [15] with the conventional (constitutive and regulated) secretion as well as with vectorial membrane protein trafficking and polarized sorting in mammalian cells. Although these latter processes should not be confused with the sensu stricto secretory autophagy, they complete the picture of the multi-tiered overlaps between autophagy and secretion in eukaryotic cells.
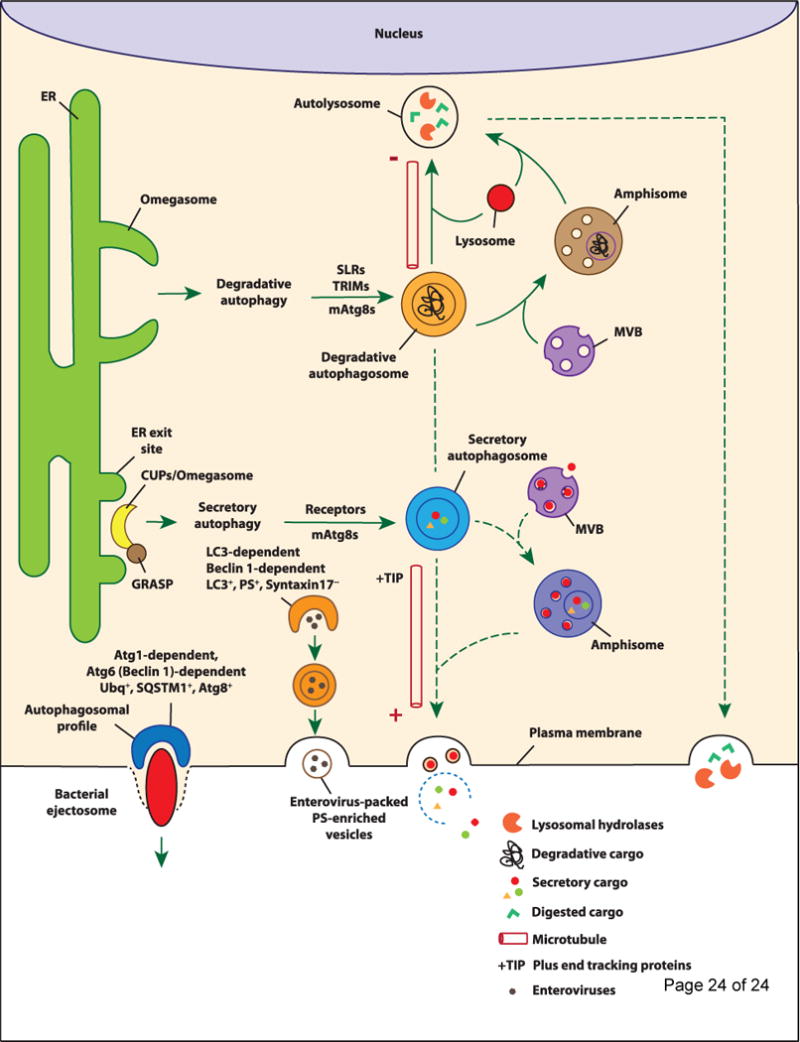
At the ER exit sites, omegasomes (in mammalian cells) facilitate the formation of degradative autophagosomes, of which the elongation and closure require LC3B. The degradative cargo can be captured/delivered into the degradative autophagosomes via autophagy receptors such as SLRs and TRIMs. Upon closure, degradative autophagosomes display motility toward the minus end of the microtubules where they fuse with lysosomes resulting in the degradation of the engulfed contents by lysosomal hydrolases. Conversely in secretory autophagy, CUPS (in yeast) or their putative (see text for discussion) equivalents in mammalian cells that may be omegasomes located near ER exit sites marked by GRASP localization (as shown for CUPS) aid the formation of secretory autophagosomes. The specific members of mammalian Atg8s that facilitate this process and receptors that capture the cargo into secretory autophagosomes have not been defined. Secretory autophagosomes show vectorial motility toward the plus end of the microtubules and eventually fuse with the plasma membrane releasing their contents into the extracellular environment. The vectorial directionality of secretory autophagic organelles may be controlled by +TIP (plus end tracking proteins). Note that both degradative and secretory autophagosomes may fuse with MVBs to generate amphisomes before fusion with lysosome or plasma membrane, respectively. Degradative and secretory autophagosomes may undergo interconversion or redirection in some instances, e.g. when the same substrate can be secreted or degraded. Either way, the destiny of the contents is removal, whether by lysosomal degradation (degradative autophagy) or plasma membrane extrusion (secretory autophagy). At the bottom left, two recently described processes of autophagy-dependent nonlytic extracellular release of cytoplasmic microbes are depicted: Far left, bacterial ejectosome is represented with the associated autophagosomal profile cup shown to be important for providing membrane to prevent host cell lysis during bacterial ejection. The ejectosome-associated autophagosomal cup is positive for ubiquitin (Ubq), sequestosome 1 (SQSTM1), and Atg8 and its formation requires Atg1, 5, 6, and 7. Left, enterovirus release (nonlytic) is depicted within phosphatidylserine-rich vesicles of autophagic origin that are formed within syntaxin 17-negative, LC3 and Beclin 1-dependent compartments. The examples depicted for autophagic secretion represent two extremes of a potential continuum – from delicate cargo of select proteins or their assemblies to large particulates and whole live microbes.
Table 1
Autophagy intersection with unconventional and conventional secretion and polarized protein sorting
| Cargo | Methodsa,b | Effects | Functionc | References |
|---|---|---|---|---|
| Pathway I: Secretory autophagy | ||||
| I-A. Unconventional secretion of cytosolic proteins with extracellular functions | ||||
| Acb1 | Analysis of yeasts lacking Atg genes; induction of autophagy by starvation but not with rapamycin | Autophagy induction enhanced and Atg factors required for Acb1 secretion | Necessary for yeast sporulation; in mammals, precursor to neuropeptides and role in metabolism | [12, 13] |
| IL-1β | Pharmacological and starvation modulation of autophagy; knockdowns and knockouts of ATG factors and Rab8 | Autophagy enhanced IL-1β secretion | Key pro-inflammatory cytokine processed by inflammasome | [14, 19, 20, 25, 26] |
| IL-18 | Pharmacological and physiological modulation of autophagy | Autophagy enhanced IL-18 secretion. | Inflammasome-processed cytokine with roles in cell mediated immunity and retinal health | [14] |
| HMGB1 | Atg5 knockout; physiological modulation of autophagy without causing lysis | Autophagy enhanced and Atg5 was necessary for secretion of HMGB1 | Damage-associated molecular pattern – inflammatory mediator | [14, 41] |
| Galectin-3 | Beclin 1 knockdown in β-glucan-stimulated cells via Dectin 1 | Beclin 1 was necessary for secretion | A cytosolic β-galactoside-binding protein with roles in autophagy, cancer, heart disease and stroke | [20] |
| Annexin I, tubulin | As above | As above | Proposed members of Dectin-induced secretome | [20] |
| I-B. Removal of aggregate-forming proteins | ||||
| α-Synuclein | Inhibition of degradative autophagic flux; knockdown of Atg5; analysis of HDAC6 and Rab8 | Inhibition of autophagic maturation enhanced α-synuclein release. | Inter-neuronal transmission of α-synuclein species (modified or aggregated) in Parkinson’s disease | [17] |
| Amyloid beta (Aβ) | Analysis of neuron-specific Atg7-deficient APP transgenic mice and pharmacological stimulation of autophagy | Increased intracellular Aβ accumulation | Alzheimer’s disease pathology and memory impairment | [18] |
| I-C. Extracellular release of cytoplasmic organellar material | ||||
| Mitochondria | Induction of autophagy; autophagic receptor NIX dependent clearance; ultrastructural analyses | Mitochondria removal from developing reticulocytes; release of mitochondrial components | Developmentally regulated clearance of mitochondria | [47] [49] |
| Mixed organellar remnants | Detection of autophagic markers | Release of an LC3+ compartment with organellar remnants (ER, Golgi, plasma membrane) | Final stages of reticulocyte development | [37] |
| Autophagic organelles | Physiological and pharmacological modulation of autophagy | Extracellular export of autophagic vacuoles without cell membrane permeabilization | Linking secretory autophagy with caspase activation | [64] |
| Phagolysosomes | Purinergic stimulation and microtubule perturbation | Release of microbial material captured in autophagolysosomes | Possible immune adjuvant or alternative elimination mechanism | [48] |
| I-D. Microbial release from cells and transmission | ||||
| M. marinum | Genetic and cell biological analyses in the amoeba Dictyostelium as an infected host | Atg1-, Atg6 (Beclin)-, Atg7- dependent, but independent of SQSTM1 (p62 ortholog). | Intercellular spread of infection | [21] |
| B. abortus | Genetic and cell biological analyses | ULK1-, Beclin 1-, ATG14L- and PI3-kinase dependent (but independent of ATG5, ATG16L1, ATG4B, ATG7, and LC3B). | Intercellular spread of infection | [51] |
| Poliovirus Enteroviruses | Genetic and pharmacological manipulation of autophagy and time lapse microscopy. | Virus released nonlytically within phosphatidylserine-rich vesicles of autophagic origin that are formed within lysosomal enzymes-negative syntaxin 17-negative, LC3 and Beclin 1-dependent compartments | Intercellular spread of infection | [16, 52, 57] |
| Morbillivirus | Atg7 knockdown, chloroquine | Virus spread associated with syncytia formation | Spread of infection | [53] |
| Coxsackievirus | Analysis of extracellular microvesicles containing viruses | Released virus detected in LC3II+ extracellular microvesicles. | Intercellular spread of infection | [54] |
| Infleunza A virus | Viral M2 protein association with LC3 | Plasma membrane translocation of LC3 | Supporting alternative (filamentous) mode of budding | [56]. |
| Pathway II: Autophagy and regulated secretion | ||||
| Paneth cell granule contents | ATG16L1- and ATG5-deficient Paneth cells | Autophagy-deficient Paneth cells exhibit abnormalities in the granule exocytosis pathway | Crohn’s disease intestinal abnormalities | [65] |
| Cathepsin K | Analysis of Atg5, Atg7, Atg4B, and LC3 in polarized secretion of lysosomal components | Atg defects impaired secretory lysosomal fusion and ruffled border formation | Osteoclast bone resorption and remodeling | [66] |
| Mast cell degranulation | Atg7 defective bone marrow-derived mast cells BMDMC | Impairment in degranulation by Atg7 BMDMC | Allergic disease and anaphylaxis reactions | [67] |
| von Willebrand factor (VWF) | Pharmacological inhibition, knockdowns of Atg5/Atg7, mice with endothelial-specific deletion of Atg5/Atg7 | Atg5-deficient endothelium shows defective secretion of VWF from Weibel-Palade bodies | Platelet adhesion in blood vessel injury | [68] |
| Otoconial core proteins | Atg4b-null mice and Atg5-null neonatal mice | Impaired secretion of otoconial (organic calcium carbonate crystals) core proteins. | Equilibrioception and balance disorders | [69] |
| Mucin | Effects of ATG factors on mucin secretion and effects of nonhematopoietic inflammasome on autophagy | Autophagosomal organelles enriched in NADPH oxidase enhance mucin secretion via ROS, and NLRP6 inflammasome controls autophagy | Goblet cell mucin secretion and control of intestinal microbiota | [70, 71] |
| Pathway III: Autophagy and constitutive secretion | ||||
| IL-6 | Atg5 deletion | Autophagy enabled enhanced secretion during oncogene-induced senescence | Contributing to the senescence-associated secretory phenotype (SASP) | [58] |
| IL-8 | Atg5 deletion, Atg7 depletion, stimulation with tat-BECN1 peptide | Autophagy enhanced senescence associated IL-8 secretion | SASP and potential | [58] [19] |
| LIF, FAM3C, DKK3 | Stimulation with tat-BECN1 peptide and depletion of Atg7. | Autophagy-augmented secretion | Potential biomarkers for autophagy in anticancer therapy | [19] |
| Pathway IV: Autophagy and plasma membrane protein sorting and organelle biogenesis | ||||
| ΔF508-CFTR | Depletion of ATG factors or pharmacological stimulation in AF508 Cftr mice and ΔF508 CFTR cystic fibrosis patients | Autophagy-assisted increase in ΔF508-CFTR cell surface expression | Rescuing CFTR function | [59, 60] |
| Mpl | Pharmacological stimulation and dominant negative expression of Atg5 | Surface expression of immature Mpl linked to mutant Jak2 signaling | Potential role in myeloproliferative cancers | [61] |
| Primary cilium | Genetic and cell biological analyses of multiple ATG, IFT, and centriolar satellite components | Autophagy initiates cilliogenesis on serum starvation but limits ciliary growth under nutrient-rich conditions. | Autophagy and cilliogenesis are regulating each other. Nutrition deprivation is a shared signal. | [62, 63] |
Secretory autophagy as a form of unconventional protein secretion
One of the earliest examples of the unconventional secretion of a cytosolic protein is the non-lytic export from the mammalian cells of the cytosolic protein IL-1β, a proinflammatory cytokine with important biological roles in mammalian systems. IL-1β lacks a leader/signal peptide and resides in the cytosol as an inactive precursor; upon activation of the cytoplasmic protein platform termed the inflammasome [22], IL-1β undergoes proteolytic processing and is exported outside of the cells in membranous carriers first observed by Rubartelli et al., in 1990 [23, 24]. Export is important for IL-1β biological function, as only upon its secretion can IL-1β meet its destination – IL-1 receptors on other cells – thereby evoking pro-inflammatory signaling. The nature of the membranous carriers exporting IL-1β from the cells and the mechanism of the unconventional secretion processes involved have been a matter of debate. Recently, the secretion of IL-1β has been linked to the autophagy machinery [1, 3, 14]. These studies have been extended and expanded by several groups [19, 20, 25, 26]. This was made possible in part by the details of autophagy-assisted unconventional secretion uncovered in yeast through the power of genetics [12, 13]. The yeast studies, although recently re-interpreted by one of the groups [27], have paved the way for reports on autophagic secretion in mammalian cells of a number of cytosolic proteins [3, 14, 19, 20, 25, 26], in keeping with the earlier definition of a large repertoire secretome induced by starvation [28], a classical signal for induction of autophagy.
Mechanisms of secretory autophagy in yeast and mammalian cells
The studies in yeast [12, 13] have identified that secretion of a susbtrate termed Acb1 depends on Atg genes controlling autophagosome formation, Golgi reassembly stacking protein (GRASP in mammals/Grh1 in yeast; known for its conserved role in unconventional secretion [1, 29]), ESCRT proteins involved in endosomal multivesicular bodies (MVB) biogenesis and sorting, and SNAREs important for vesicle fusion at the plasma membrane [12, 13].
In mammalian cells, secretory autophagy of IL-1β depends upon the autophagy factor Atg5, a GRASP homolog (GRASP55) also affecting general autophagy, and a small GTPase Rab8a, which regulates vectorial sorting to plasma membrane [14]. Rab8a has also been reported to regulate autophagic secretion of other cargo, including α-synuclein [17], a major constituent of Lewy bodies, which are the hallmark protein aggregates associated with Parkinson’s disease. Secretion of α-synuclein required autophagosome formation as demonstrated by the inhibitory effects of 3MA and siRNA-mediated knockdown of Atg5. Secretory autophagy of α-synuclein (a cytosolic cargo) was enhanced by inhibiting fusion of autophagosomes with lysosome using bafilomycin A1 [17]. Likewise, downregulation of HDAC6, with accompanied increased in acetylation of microtubules and decreased autolysosomal fusion, increased α-synuclein secretion [17]. Pharmacological inhibition of degradative autophagic flux, decrease of autophagosomal minus-end motility, and increase of plus end vectorial transport along microtubules may divert autophagosomal organelles (including amphisomes) from degradation into the secretory autophagy pathway. Depending on circumstances, both IL-1β (reviewed in [25]) and α-synuclein [30] can be substrates for autophagic degradation. Thus, degradative and secretory autophagy may in some cases diverge from common precursors, and fate of the cargo (degradation vs. secretion) determined by trafficking directly or via an intermediate towards the lysosomes or plasma membrane. In support of the aforementioned concept, a clear differences between the two pathways identified in yeast is that secretory autophagy requires a SNARE (Sso1) necessary for plasma membrane fusion whereas it is independent of the vacuole fusion SNARE complex (VAM7/VAM3) leading to degradation [12]. Analogous dichotomy is seen in mammalian cells: whereas Rab8a, a regulator of polarized sorting to the plasma membrane, was shown to be crucial for secretory autophagy [14, 17], its closely related isoform Rab8b may be more important for maturation of the degradative autophagosome [31].
The similarity between secretory and degradative autophagosome formation extends to the discovery of a structure that is associated with secretory autophagy in yeast, the compartment for unconventional protein secretion (CUPS) [32], which morphologically resembles the mammalian omegasome, an ER-derived precursor structure that acts as a cradle for the generation of degradative autophagosomes [33]. Yeast GRASP re-localizes to CUPS under starvation conditions [32]. Both CUPS and omegasome can be induced by starvation [32, 33] and their structures are similarly enriched in PI3P and Atg proteins. As CUPS has yet to be demonstrated in mammalian cells and omegasome has not been shown in yeast, whether these two structures are distinct or similar requires further investigation. In addition, whether GRASP, a marker for CUPS, can be found at the mammalian omegasome upon starvation is unknown and this may help resolve the identity of the two structures. If these two structures are the same, this would support the idea that the processes of autophagosome formation for secretory and degradative autophagy share the same set of precursor and Atg proteins and that the two pathways diverge at a later point in the pathway.
In contrast to the above simplistic model, which likens CUPS in yeast to omegasomes (precursors to autophagic organelles in mammals), a recent study in yeast argues that CUPS are distinct from autophagosomes [34]. Further details are now available on CUPS [34]: CUPS are formed by the fusion of at least two classes of membranes. One comes from early Golgi containing Grh1 (yeast equivalent of GRASP), Bug1, the membrane tethering factor Uso1, and the t-SNARE Sed5. The other membrane source are vesicles coming from the late Golgi in a Sec7- and Pik1-dependent manner. The fusion of these two sets of membranes is likely mediated by Sed5 and Uso1. CUPS then mature by addition of membranes from the endosomes by a Vps34-dependent process. More work in both mammalian and yeast systems is needed to determine whether precursors discussed above lead to functionally related (or different) compartments of relevance for secretory processes.
Based on the studies in yeast [12, 13], the cytosolic secretory cargo may be engulfed into an autophagosome but may alternatively include MVB-like intermediates [1] (Figure 2). In mammalian cells, MVBs are affected by autophagic machinery since autophagosomes and MVBs interact to form hybrid structures referred to as amphisomes [35, 36]. Autophagy could export cytosolic substrates directly or deliver them to an MVB intermediate for subsequent release via MVB-plasma membrane fusion. An analogous process has been observed in maturing reticulocytes in which LC3-positive MVB were shown to fuse with the plasma membrane in vitro, and similar vesicles were identified in peripheral blood [37]. Indeed, several overlaps between the MVB-exosome pathway and autophagy exist [38].
Secretory autophagy: cargo selection
Unlike the well-characterized cargo selection process in degradative autophagy [39], nothing is known of how the secretory cargo is selected and engulfed by an autophagosome. The only clue so far comes from the fact that yeast Vps23 (TSG101 in mammals), a component of the endosomal sorting complex required for transport (ESCRT)-I in mammalian cells, is found at the CUPS [32]. As Vps23 is not essential for CUPS formation and as its known mammalian function is to sort ubiquitinated cargo into the MVB, Vps23 may function as a secretory autophagy receptor to capture the unconventional secreted protein substrates into the amphisome. Whether secretory autophagy substrates are ubiquitinated has not been elucidated. In addition, the involvement of other well-known autophagy receptors (e.g. p62, NBR1, NDP52, and optineurin), which facilitate the delivery of ubiquitinated autophagic substrates to degradative autophagosomes, remains unexplored, with the exception of SQSTM1 (the p62 Dictyostelium ortholog), which turned out to be present during but functionally dispensable for secretion of a particulate substrate termed ejectosome [21]. In addition to these well-known autophagy receptors, the tripartite motif (TRIM) family of proteins has been recently reported as a new class of dual autophagic receptor—regulators [40] and it is possible that at least some member(s) of this protein family may act in secretory autophagy.
Secretory autophagy exports a range of cytosolic proteins in mammalian cells
Autophagic machinery is involved in unconventional secretion of the inflammatory cytokine IL-1β [3, 14, 19, 20, 25, 26]. Moreover, autophagy facilitates active export (without a nonspecific permeabilization of the plasma membrane) of additional unconventionally secreted proteins, which encompass a broader range of cytosolic substrates [14, 19, 20] (Table 1; section IA). Among the secretory autophagy cargo are IL-18 [14] and HMGB1 [14, 41]. Similarly to IL-1β, IL-18 and HMGB1 do not contain a signal peptide and their secretion can be triggered by inflammasome activation [28]. IL-18 is a conventional inflammasome substrate, whereas HMGB1 is most likely indirectly associated [28]. However, secretory autophagy is not restricted to inflammasome substrates. Additional leaderless cytosolic proteins have been reported in the autophagy-dependent secretome, including galectins (galectin-3), cytoskeletal proteins (tubulin), and others such as annexin-I [20].
Secretory autophagy of aggregation-prone proteins
Among the conventional cargo for degradative autophagosomes are protein aggregates [5]. Mirroring this, secretory autophagy may play a role in extracellular export of protein aggregates or aggregation-prone protein species (Table 1, I-B). Induction of autophagy in conjunction with inhibition of degradative autophagic flux induces secretory autophagy of α-synuclein species or aggregates associated with Parkinson disease [17]. Secretion of α-synuclein may be associated with inter-neuronal transmission mechanisms of α-synuclein species and the spreading pattern of α-synuclein inclusion body disease throughout the nervous tissues.
In addition to α-synuclein aggregate expulsion, secretory autophagy has also been implicated in extrusion of amyloid beta (Aβ) peptide aggregates associated with Alzheimer’s disease [18]. By crossing amyloid precursor protein transgenic mice with neuron-specific Atg7-deficient mice, Nilsson et al. observed a paradoxical increase in intracellular Aβ aggregate accumulation in the perinuclear regions of neurons and a concurrent decrease in Aβ secretion and extracellular Aβ plaque formation [18]. This neurotoxic phenotype, which could be further exacerbated through inhibition of autophagy by eventual extracellular amyloidosis due to the overall Aβ accumulation in neural tissues, resulted in impaired memory. Transduction of neurons isolated from these mice with Atg7 rescued their ability to release Aβ [18]. Moreover, rapamycin (an autophagy inducer) increased whereas spautin-1 (an autophagy inhibitor) decreased secretion of Aβ from wild-type primary neurons [18]. These data indicate a role of secretory autophagy in toxic protein aggregate extrusion and protection against their intracellular accumulation.
Secretory autophagy and extracellular release of intracellular organellar contents
One distinct feature of autophagy is that large cytoplasmic objects such as organelles including surplus peroxisomes [42], excess mitochondria [43], depolarized mitochondria [9, 44], damaged endosomes and lysosomes [45, 46], or invading microbes [10] can be engulfed by autophagosomes. Some of the above autophagy targets, typically associated with degradative autophagy, can also become secretory autophagy cargo [43, 47–49] (Table 1, I-C). The massive developmentally-regulated mitochondrial clearance during reticulocyte maturation depends on autophagy and can terminate either in degradation or in a readily observable exocytosis of the captured mitochondria [47]. This programmed process of mitochondrial release relies on the mitochondrial autophagic receptor Nix [47] and seems to differ from sporadic mitochondrial depolarization and Parkin-dependent mitophagy as an ongoing quality control process in most cells [44]. Nevertheless, mitochondrial material can be released from other cells in association with autophagy. Proteomic analysis of extracellular membranous material released upon starvation of endothelial cells shows enrichment of autophagy proteins (LC3 and Atg16L1) along with mitochondrial components (VDAC and COX4) [49]. In other instances, extracellular release of mitochondrial material [50] has been noted, although mechanisms have not been determined.
Autophagolysosomes and their content instead of being fully processed by degradation can be extruded from cells [48]. This is an active process dependent on stimulation of the purinergic receptor P2X7R with its agonist ATP and the associated microtubule rearrangement into radial extension from the centrosome toward the cell periphery [48]. The control of the divergence between continuation of degradation vs. extrusion of the autophagocytosed material from cells remains to be fully analyzed and the specific biological functions of these phenomena remain to be understood.
Secretory autophagy in microbial egress and dissemination
Intracellular pathogens such as bacteria [21, 51] and viruses [16, 52–54] can be released from infected cells in a variety of process assisted by the autophagic machinery (Table 1, I-D). A recent study with Mycobacterium marinum [21] showed that autophagosomal organelles chaperone a structure termed the ejectosome [55], an actin-tail based apparatus enabling cytosolic bacteria to exit the host cell with the purpose of potentially contributing to microbial cell-to-cell spread (Fig. 2). A functional (non-lytic to the host cell) ejectosome in M. marinum-infected amoeba Dictyostelium is seen [21] as escorted by a distinct polar autophagic profile, which is Atg8 positive, ubiquitin-positive, and SQSTSM1 (an autophagic receptor)-positive. Although the host-sparing function of the ejectosome was SQSTM1-independent (thus indicating possible involvement of other unknown receptors) it was Atg5, Atg7 and Atg6 (Beclin)-dependent. Atg18 (a PI3P-recognizing autophagic regulator) and PI3P fluorescent probes colocalized with these profiles. Whereas the bacteria could still form an ejectosome and leave the host cell in Atg1-negative amoeba, the autophagic machinery was important to seal the plasma membrane and protect the host cell from leaking cytoplasm and undergoing death [21]. Other forms of autophagy-assisted bacterial (Brucella) egress from infected cells without causing cell death have been reported [51]. Thus, autophagy enables non-lytic egress of bacteria from host cells. This may support spread of infection while avoiding excessive inflammatory signaling and immune clearance elicited by host’s own alarmins released from necrotic cells.
Viral budding or release from cells can be assisted by ATG factors. The M2 protein of influenza contains a motif termed LIR (LC3 interacting region), which enables it to associate with LC3 at the plasma membrane to assist filamentous mode of viral budding [56]. Autophagy assists other viruses, such as poliovirus and coxsackievirus B, in their egress from cells. Although classically poliovirus spreads by lysing host cells, autophagy has been proposed to support an alternative, non-lytic exit of the poliovirus [16, 52]. Using quantitative time-lapse microscopy, Bird et al., demonstrated that poliovirus can be released from cells in tissue culture without cell lysis, a process augmented by autophagy induction and diminished by knockdown of the key autophagic membrane protein LC3 [16]. This process was validated in vivo in a mouse model of polio infection [16]. A significant new study [57] has shown that enteroviruses (note: poliovirus is a prototypical enterovirus) are non-lytically secreted from cells in membrane-bound phosphatidylserine (PS)-positive vesicles originating from autophagosome-like organelles (Fig. 2). The precursor compartments for these virus-laden secreted vesicles is enriched for PS and is LC3-positive, whereas their formation depends on LC3 and Beclin1 and can be stimulated by tat-Beclin 1 peptide [57]. Cells can also shed nonpolio viruses such as coxsackievirus B in LC3-positive vesicles [54], a phenomenon that may contribute to infection spread and avoidance of neutralizing antibodies.
Intersections between autophagy and conventional secretion
In addition to the secretory autophagy pathway discussed above that involves the secretion of leaderless proteins and possibly whole organelles and microbes via an autophagosome intermediate to the extracellular space (Table 1, pathway I), autophagy also affects three other general secretory and plasma membrane protein trafficking pathways in the cell (Table 1, pathways II–IV). In pathway II, autophagy affects (probably via indirect mechanisms) the regulated secretion of cargo within pre-formed secretory granules. In pathway III, autophagy affects (selectively) the output for subsets of constitutively secreted proteins (Table 1, pathway III) [58]. The above pathways, recently covered in a review [15], are updated in Table 1. The pathway IV is addressed in the following section. Importantly, these pathways should not be confused with unconventional secretion but consist of (a) conventional (ER-to-Golgi-to plasma membrane) secretion of lumenal cargo (both regulated or constitutive) or (b) alternative pathways of trafficking of integral membrane proteins to the plasma membrane and organelle biogenesis at the plasma membrane.
Autophagic machinery in polarized membrane protein sorting and ciliogenesis
Autophagy has also been shown to be associated with the unconventionally trafficking of proteins to the plasma membrane, first shown in the case of CFTR (Table 1, pathway IV) [59, 60]. Cell surface expression of ΔF508-CFTR, the most prevalent CFTR mutant associated with cystic fibrosis, can be restored in a GRASP-dependent manner [59]. In addition, expression of the autophagy protein Beclin 1 or treatment with the autophagy-inducer cystamine can increase ΔF508-CFTR cell surface expression in vitro and in vivo and reduce lung inflammation and mortality in the CftrF508del homozygous mice [60].
Mpl, the receptor for thrombopoietin and a major player in megakaryocytes differentiation and maturation, is another example of transmembrane protein being exported to the plasma membrane via unconventional trafficking pathways (Table 1, pathway IV) [61]. While the fully glycosylated, mature, receptor follows the canonical ER-to-Golgi-to-membrane pathway, the unconventional, autophagy-related secretory pathway allows the core-glycosylated, immature, protein to reach the cell surface through a GRASP-dependent pathway. Aberrant Mpl intracellular trafficking has been linked to mutant Jak2 signaling associated with myeloproliferative neoplasms (progressive blood cancers). Mpl was observed to be loaded onto autophagic vesicles directly from the ER, as shown by high-resolution correlative light/electron microscopy technique [61]. Pharmacological inhibition of autophagy or expression of mutant Atg5K130R led to a decrease cell surface expression of the ER form of Mpl. Complementing these observations, autophagic route for delivery of immature (core-glycosylated) Mpl was significantly activated by pharmacological induction of autophagy with rapamycin and by overexpression of GRASP55 [61]. Thus, autophagy may provide a target in certain hematologic malignancies treatments.
Autophagy has other polarized sorting and plasma membrane organelle biogenesis roles. Starvation is a common signal for autophagy and biogenesis of the primary cilium, a sensory organelle continuous with but functionally delimited from the plasma membrane. Autophagy plays both a positive and a negative role in the formation and length of the primary cilium [62, 63], whereas cilium can serve as a staging ground for autophagic machinery components [63]. Autophagy helps initiate cilium formation under starvation conditions by reducing levels of OFD1, a protein that accumulates at the centriolar satellites where it inhibits ciliogenesis [63]. Basal autophagy limits uncontrolled cilium growth under nutrient-rich conditions by diminishing intraflagellar transport protein IFT20 levels [62]. Finally, signaling from the cilia regulates autophagy [62]. Thus, not only does autophagy provide alternative trafficking routes for integral membrane proteins to reach the plasma membrane where they carry out their functions, but it also controls biogenesis of complex domains and organelles at the plasma membrane.
Concluding remarks
The concept of secretory autophagy, discovered less than half a decade ago, is still in its infancy. Although some progress has been made, new questions emerge whereas previously identified ones remain to be answered. What differentiates a secretory autophagosome from a degradative autophagosome? For instance, what aspects of the molecular machinery of secretory autophagy overlap with degradative autophagy, and how and where do the two mechanisms diverge? What are the molecular tags/signals that identify a secretory substrate and what are the receptors that bring it into the autophagosomes? Can cells switch between secretory and degradative autophagy modes for the same cargo? Is conversion into amphisome a mandatory sorting station before secretory autophagy can occur? What is the composition and biological function spectrum of the autosecretome, how does its composition change with physiological needs, and how does it influence cellular, tissue, and organismal functions? Many questions remain before we can define secretory autophagy as a specific pathway, but it is an attractive concept deserving further testing.
Acknowledgments
MP was supported by National Science and Technology Development Agency, Development and Promotion of Science and Technology Talents Project research grant 023/2557, Thailand Research Fund, Office of the Higher Education Commission, and Mahidol University. TK was supported in part by Manpei Suzuki Diabetes Foundation and Uehara Memorial Foundation. This work was supported by NIH grants AI042999 and AI111935.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest have been highlighted as:
*of special interest
**of outstanding interest
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.ceb.2015.04.016
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4529791?pdf=render
Citations & impact
Impact metrics
Article citations
Plekhg5 controls the unconventional secretion of Sod1 by presynaptic secretory autophagy.
Nat Commun, 15(1):8622, 04 Oct 2024
Cited by: 0 articles | PMID: 39366938 | PMCID: PMC11452647
Gasdermin D and Gasdermin E Are Dispensable for Silica-Mediated IL-1β Secretion from Mouse Macrophages.
Immunohorizons, 8(9):679-687, 01 Sep 2024
Cited by: 0 articles | PMID: 39264735 | PMCID: PMC11447662
Nanoscale ZnO doping in prosthetic polymers mitigate wear particle-induced inflammation and osteolysis through inhibiting macrophage secretory autophagy.
Mater Today Bio, 28:101225, 06 Sep 2024
Cited by: 0 articles | PMID: 39309162 | PMCID: PMC11415586
Autophagy-related proteins measured in umbilical blood cord samples from human newborns: what can we learn from?
Pediatr Res, 96(5):1120-1122, 16 Jul 2024
Cited by: 0 articles | PMID: 39014242 | PMCID: PMC11521990
Discovery of Cell Number-Interstitial Fluid Volume (CIF) Ratio Reveals Secretory Autophagy Pathway to Supply eHsp90α for Wound Healing.
Cells, 13(15):1280, 30 Jul 2024
Cited by: 0 articles | PMID: 39120311 | PMCID: PMC11312289
Go to all (262) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation.
Trends Cell Biol, 22(8):397-406, 05 Jun 2012
Cited by: 135 articles | PMID: 22677446 | PMCID: PMC3408825
Review Free full text in Europe PMC
Autophagy for secretory protein: Therapeutic targets in cancer.
Adv Protein Chem Struct Biol, 133:159-180, 24 Nov 2022
Cited by: 2 articles | PMID: 36707200
Secretory Autophagy and Its Relevance in Metabolic and Degenerative Disease.
Front Endocrinol (Lausanne), 11:266, 05 May 2020
Cited by: 41 articles | PMID: 32477265 | PMCID: PMC7232537
Review Free full text in Europe PMC
Autophagy and Protein Secretion.
J Mol Biol, 432(8):2525-2545, 21 Jan 2020
Cited by: 31 articles | PMID: 31972172
Review
Funding
Funders who supported this work.
NIAID NIH HHS (5)
Grant ID: R37 AI042999
Grant ID: R01 AI111935
Grant ID: AI042999
Grant ID: R01 AI042999
Grant ID: AI111935
NIGMS NIH HHS (2)
Grant ID: GM085273
Grant ID: P50 GM085273
NIH (2)
Grant ID: AI111935
Grant ID: AI042999
National Science and Technology Development Agency (1)
Grant ID: 023/2557





