Abstract
Importance
Current prediction models of mortality in idiopathic pulmonary fibrosis (IPF), which are based on clinical and physiological parameters, have modest value in predicting which patients will progress. In addition to the potential for improving prognostic models, identifying genetic and molecular features that are associated with IPF mortality may provide insight into the underlying mechanisms of disease and inform clinical trials.Objective
To determine whether the MUC5B promoter polymorphism (rs35705950), previously reported to be associated with the development of pulmonary fibrosis, is associated with survival in IPF.Design, setting, and participants
Retrospective study of survival in 2 independent cohorts of patients with IPF: the INSPIRE cohort, consisting of patients enrolled in the interferon-γ1b trial (n = 438; December 15, 2003-May 2, 2009; 81 centers in 7 European countries, the United States, and Canada), and the Chicago cohort, consisting of IPF participants recruited from the Interstitial Lung Disease Clinic at the University of Chicago (n = 148; 2007-2010). The INSPIRE cohort was used to model the association of the MUC5B genotype with survival, accounting for the effect of matrix metalloproteinase 7 (MMP-7) blood concentration and other demographic and clinical covariates. The Chicago cohort was used for replication of findings.Main outcomes and measures
The primary end point was all-cause mortality.Results
The numbers of patients in the GG, GT, and TT genotype groups were 148 (34%), 259 (59%), and 31 (7%), respectively, in the INSPIRE cohort and 41 (28%), 98 (66%), and 9 (6%), respectively, in the Chicago cohort. The median follow-up period was 1.6 years for INSPIRE and 2.1 years for Chicago. During follow-up, there were 73 deaths (36 GG, 35 GT, and 2 TT) among INSPIRE patients and 64 deaths (26 GG, 36 GT, and 2 TT) among Chicago patients. The unadjusted 2-year cumulative incidence of death was lower among patients carrying 1 or more copies of the IPF risk allele (T) in both the INSPIRE cohort (0.25 [95% CI, 0.17-0.32] for GG, 0.17 [95% CI, 0.11-0.23] for GT, and 0.03 [95% CI, 0.00-0.09] for TT) and the Chicago cohort (0.50 [95% CI, 0.31-0.63] for GG, 0.22 [95% CI, 0.13-0.31] for GT, and 0.11 [95% CI, 0.00-0.28] for TT). In the INSPIRE cohort, the TT and GT genotypes (risk for IPF) were associated with improved survival compared with GG (hazard ratios, 0.23 [95% CI, 0.10-0.52] and 0.48 [95% CI, 0.31-0.72], respectively; P < .001). This finding was replicated in the Chicago cohort (hazard ratios, 0.15 [95% CI, 0.05-0.49] and 0.39 [95% CI, 0.21-0.70], respectively; P < .002). The observed association of MUC5B with survival was independent of age, sex, forced vital capacity, diffusing capacity of carbon monoxide, MMP-7, and treatment status. The addition of the MUC5B genotype to the survival models significantly improved the predictive accuracy of the model in both the INSPIRE cohort (C = 0.71 [95% CI, 0.64-0.75] vs C = 0.68 [95% CI, 0.61-0.73]; P < .001) and the Chicago cohort (C = 0.73 [95% CI, 0.62-0.78] vs C = 0.69 [95% CI, 0.59-0.75]; P = .01).Conclusions and relevance
Among patients with IPF, a common risk polymorphism in MUC5B was significantly associated with improved survival. Further research is necessary to refine the risk estimates and to determine the clinical implications of these findings.Free full text

Association Between the MUC5B Promoter Polymorphism and Survival in Patients With Idiopathic Pulmonary Fibrosis
Abstract
Importance
Current prediction models of mortality in idiopathic pulmonary fibrosis (IPF), which are based on clinical and physiological parameters, have modest value in predicting which patients will progress. In addition to the potential for improving prognostic models, identifying genetic and molecular features that are associated with IPF mortality may provide insight into the underlying mechanisms of disease and inform clinical trials.
Objective
To determine whether the MUC5B promoter polymorphism (rs35705950), previously reported to be associated with the development of pulmonary fibrosis, is associated with survival in IPF.
Design, Setting, and Participants
Retrospective study of survival in 2 independent cohorts of patients with IPF: the INSPIRE cohort, consisting of patients enrolled in the interferon-γ1b trial (n=438; December 15, 2003–May 2, 2009; 81 centers in 7 European countries, the United States, and Canada), and the Chicago cohort, consisting of IPF participants recruited from the Interstitial Lung Disease Clinic at the University of Chicago (n=148; 2007–2010). The INSPIRE cohort was used to model the association of the MUC5B genotype with survival, accounting for the effect of matrix metalloproteinase 7 (MMP-7) blood concentration and other demographic and clinical covariates. The Chicago cohort was used for replication of findings.
Main Outcomes and Measures
The primary end point was all-cause mortality.
Results
The numbers of patients in the GG, GT, and TT genotype groups were 148 (34%), 259 (59%), and 31 (7%), respectively, in the INSPIRE cohort and 41 (28%), 98 (66%), and 9 (6%), respectively, in the Chicago cohort. The median follow-up period was1.6 years for INSPIRE and 2.1 years for Chicago. During follow-up, there were 73 deaths (36 GG, 35 GT, and 2 TT) among INSPIRE patients and 64 deaths (26 GG, 36 GT, and 2 TT) among Chicago patients. The unadjusted 2-year cumulative incidence of death was lower among patients carrying 1 or more copies of the IPF risk allele (T) in both the INSPIRE cohort (0.25 [95% CI, 0.17–0.32] for GG, 0.17 [95% CI, 0.11–0.23] for GT, and 0.03 [95% CI, 0.00–0.09] for TT) and the Chicago cohort (0.50 [95% CI, 0.31–0.63] for GG, 0.22 [95% CI, 0.13–0.31] for GT, and 0.11 [95% CI, 0.00–0.28] for TT). In the INSPIRE cohort, the TT and GT genotypes (risk for IPF) were associated with improved survival compared with GG (hazard ratios, 0.23 [95% CI, 0.10–0.52] and 0.48 [95%CI, 0.31–0.72], respectively; P<.001). This finding was replicated in the Chicago cohort (hazard ratios, 0.15 [95% CI, 0.05–0.49] and 0.39 [95% CI, 0.21–0.70], respectively; P<.002). The observed association of MUC5B with survival was independent of age, sex, forced vital capacity, diffusing capacity of carbon monoxide, MMP-7, and treatment status. The addition of the MUC5B genotype to the survival models significantly improved the predictive accuracy of the model in both the INSPIRE cohort (C=0.71 [95% CI, 0.64–0.75] vs C=0.68 [95% CI, 0.61–0.73]; P<.001) and the Chicago cohort (C=0.73 [95% CI, 0.62–0.78] vs C=0.69 [95% CI, 0.59–0.75]; P=.01).
Conclusions and Relevance
Among patients with IPF, a common risk polymorphism in MUC5B was significantly associated with improved survival. Further research is necessary to refine the risk estimates and to determine the clinical implications of these findings.
Idiopathic pulmonary fibrosis (IPF) is a chronic progressive disease with a median survival of 3 years.1 The prognosis is variable; patients may remain stable for several years, slowly lose lung function, progress in an intermittent stair-step fashion, or experience precipitous acute exacerbations.2–4 Although clinical and physiological parameters have modest value in predicting which patients will progress,5 serum biomarkers, including chemokine ligand 18, KL6, surfactant protein A (SFTPA), and SFTPD, are independently associated with outcome in IPF.6,7 Recently, elevated plasma concentrations of matrix metalloproteinase 7 (MMP-7), intercellular adhesion molecule 1, and interleukin 8 have been shown to be associated with poor outcomes in IPF, and when combined with clinical features, these plasma proteins predict mortality in IPF.8
Rare mutations in SFTPC, SFTPA2, telomerase reverse transcriptase (TERT), and telomerase RNA component (TERC)9–11 have been associated with development of pulmonary fibrosis. Recently, a common polymorphism in the promoter of a mucin gene (MUC5B) has been found to be associated with an increase in risk of developing both familial and sporadic IPF in an allele dose-dependent manner.12,13 While MUC5B expression in the lung was 14.1 times higher in patients with IPF, the MUC5B promoter polymorphism was associated with up-regulation of this transcript only in unaffected participants.12
The objective of the current study was to determine whether the MUC5B (gene ID: 727897; http://www.ncbi.nlm.nih.gov/gene/727897) promoter polymorphism (rs35705950), previously reported to be associated with the development of pulmonary fibrosis, is associated with survival in IPF.
METHODS
Study Cohorts
The study was approved by the human use committees of National Jewish Health, the University of Colorado Denver, and the University of Chicago. The INSPIRE cohort (derivation cohort) consists of study participants who were enrolled (December 15, 2003–April 12, 2006) in the interferon-γ1b IPF clinical trial and provided written informed consent.14 INSPIRE cohort participants were treated with concomitant agents if they met strict criteria for progression of disease.14 Participants with available genetic and clinical follow-up data from the INSPIRE study were included in this study. Participants in the derivation set were followed up from randomization until death or study termination (a maximum of approximately 3 years after randomization). Vital status was assessed through a combination of patient/family/caregiver contacts and use of death registries (where permitted). Despite early termination of the study, assessable data (including vital status) were obtained for 99% of study patients, including all of the 438 patients in the current analysis.14
The Chicago cohort (validation cohort) consists of IPF participants who were recruited from the Interstitial Lung Disease Clinic at the University of Chicago and provided written informed consent as part of a natural history protocol. Participants in the validation cohort were enrolled in 2007–2010 and followed up from initial evaluation until death, the time of lung transplantation, or censoring on July 9, 2012, and were treated clinically. Vital status was confirmed by the Social Security Death Index as well as by contacting each family/patient at the end of the follow-up period. To minimize any effect of population stratification, only non-Hispanic white patients were included in this study (race/ethnicity data were based on self-report). The majority of participants were observed at 3- to 4-month intervals. All diagnoses met current American Thoracic Society guideline criteria for IPF (ie, definite or probable pattern of usual interstitial pneumonia on computed tomography scan and exclusion of an alternative diagnosis).
No patient was dually enrolled in both cohorts; however, all patients in the INSPIRE cohort were included in a recent publication that demonstrated an allele dose-dependent relationship between a common polymorphism in the promoter of a mucin gene (MUC5B) and the risk of developing IPF.12,13 All analyses of both cohorts were performed anonymously. Both cohorts consisted of participants who were considered to be unrelated on the basis of self-report.
Genotyping of MUC5B rs35705950
For the INSPIRE cohort, genotypes of the MUC5B SNP rs35705950 were determined using Taqman genotyping.12,13 For the Chicago cohort, genotyping was performed using the Sequenom iPLEX-Gold assay.
MMP-7 Analysis
The plasma concentration of MMP-7 was determined by enzyme-linked immunosorbent assay in the INSPIRE cohort following manufacturer (R&D Systems) instructions and as previously reported.8
Statistical Analysis
The primary analysis tested for an association of the MUC5B polymorphism with survival using Cox proportional hazards models (Kolmogorovtype supremum tests did not indicate significant departures from the proportional hazards requirement for the MUC5B genotype variable in either the INSPIRE [P=.40] or Chicago [P=.10] cohorts). The model was developed within the INSPIRE cohort by including the MUC5B genotype and then using a stepwise backward elimination process based on the Akaike information criterion to select among potential covariates (sex, age, smoking status, baseline diffusing capacity of carbon monoxide [DLCO], baseline forced vital capacity [FVC], and interferon-γ1b treatment) for inclusion in the final model. Because of power considerations, only main effects were considered. The same model was then fit within the Chicago cohort for validation.
In a secondary analysis, the MMP-7 variable was added to the final survival model for the INSPIRE cohort to determine whether the MUC5B polymorphism was significantly associated with survival in addition to the previously reported effects of MMP-7.8 In preliminary analyses of MMP-7, evidence existed of a threshold effect on survival. Accordingly, MMP-7 was modeled as a binary variable categorized at the first quartile (<5.7 ng/mL vs ≥5.7 ng/mL). An additive genetic model was used for the genotype in all survival models, consistent with previous findings.12,13 Any patient receiving a lung transplant during follow-up was censored at transplant date in the analysis.
A Wald χ2 test was used to test for association of covariates with survival, for which a 2-tailed P<.05 was considered significant. Survival analyses were performed using SAS, version 9.3 (SAS Institute Inc), and the Cox proportional hazards models were applied using the SAS PHREG procedure.
The area under the receiver operating characteristic curve (AUC) was calculated at 100-day intervals for the final, adjusted model in each cohort (with and without the MUC5B genotype covariate). In addition, the integral of time-specific AUC measures (C statistic) was calculated as an overall measure of the predictive value of these nested models.15 We used a bootstrapping procedure with 10 000 samples to calculate 95% confidence intervals for the C statistic within each cohort. We used a permutation test to assess the difference between C statistic values for the nested models in each cohort. For this purpose, we used 10 000 permutations of the genotype values, with respect to the rest of the data set, to calculate empirical P values. All C statistic calculations were performed using R, version 3.0.0, packages “risksetROC” and “boot.”
RESULTS
Study Cohorts
There were 438 patients from the INSPIRE cohort and 148 patients from the Chicago cohort included in the analysis (eFigure). The demographic and pulmonary function characteristics of participants in the cohorts were clinically similar; however, participants in the Chicago cohort were older and had more physiologically advanced disease compared with the INSPIRE cohort (Table 1). The Chicago cohort also had a longer period of follow-up (median, 2.1 years) than the INSPIRE cohort (median, 1.6 years), which contributed to the larger number of deaths among the Chicago participants (Table 1).
Table 1
Clinical and Demographic Characteristics of the Cohorts
| Characteristics | INSPIRE (n = 438) | Chicago (n = 148) | P Value | |
|---|---|---|---|---|
| Period of follow-up, median (IQR), y | 1.6 (1.2–2.1) | 2.1 (1.1–3.9) | ||
| Deaths, No. (%)a | 73 (17) | 64 (43) | ||
| Transplant events, No. (%) | 7 (2) | 10 (7) | .003 | |
| Female, No. (%) | 115 (26) | 35 (24) | .58 | |
| Age, mean (SD), y | 66.6 (7.54) | 69.0 (8.75) | .005 | |
| Ever smokers, No. (%) | 312 (71) | 96 (72) | .93 | |
| FVC, mean (SD), % predicted | 72.2 (12.37) | 64.5 (18.44) | <.001 | |
| FVC, mean (SD), L | 2.9 (0.76) | 2.5 (0.82) | <.001 | |
| DLCO, mean (SD), % predicted | 47.3 (8.89) | 46.2 (17.92) | .51 | |
| DLCO, mean (SD), mL/min/mm Hg | 12.5 (4.40) | 10.7 (4.51) | <.001 | |
| MMP-7, median (IQR), ng/mLb | 7.8 (5.7–11.0) | |||
| MUC5B genotype, No. (%) | ||||
| GG | 148 (34) | 41 (28) |  | |
| GT | 259 (59) | 98 (66) | .31 | |
| TT | 31 (7) | 9 (6) | ||
Abbreviations: DLCO, diffusing capacity of carbon monoxide; FVC, forced vital capacity; IQR, interquartile range; MMP-7, matrix metalloproteinase 7.
The observed similar frequencies of the MUC5B polymorphism minor allele (T) among the INSPIRE and Chicago cohorts (37% and 39%, respectively) were similar to what has been observed in IPF.12,13 The MUC5B polymorphism did not meet the expectations of Hardy-Weinberg equilibrium (HWE) (P<.001) for either cohort, indicating its likely role as a true risk allele, as previously reported.12,13
MUC5B rs35705950 Genotype and Survival
The unadjusted 2-year cumulative incidence of death was lower among patients carrying 1 or more copies of the IPF risk allele (T) in both the INSPIRE cohort (0.25 [95% CI, 0.17–0.32] for GG, 0.17 [95% CI, 0.11–0.23] for GT, and 0.03 [95% CI, 0.00–0.09] for TT) and the Chicago cohort (0.50 [95% CI, 0.31–0.63] for GG, 0.22 [95% CI, 0.13–0.31] for GT, and 0.11 [95% CI, 0.00–0.28] for TT). In the single-variable proportional hazards model, INSPIRE participants who were heterozygous and homozygous for the minor allele (GT and TT) were at lower risk of mortality compared with participants with the GG genotype (hazard ratios [HRs], 0.52 [95% CI, 0.34–0.78] and 0.27 [95% CI, 0.12–0.60], respectively; P=.001) (Table 2 and Figure 1). A similar association of the polymorphism with survival was observed in the Chicago cohort, wherein HRs for participants with the GT and TT genotypes were 0.49 (95% CI, 0.30–0.79) and 0.24 (95% CI, 0.09–0.63), respectively (P=.004) (Table 2 and Figure 2). These associations were also significant after independent adjustments for age, sex, smoking history (ever vs never), baseline FVC, and baseline DLCO. Within the INSPIRE cohort, the HR for the MUC5B genotype also remained unchanged (and significant) after independent adjustment for interferon-γ1b treatment (treatment vs placebo); HRs for the GT and TT genotypes were 0.48 (95% CI, 0.31–0.73) and 0.23 (95% CI, 0.10–0.53), respectively (P<.001).
Table 2
Survival Analysis Models of MUC5B
| INSPIRE | Chicago | |||||
|---|---|---|---|---|---|---|
| Model | HR (95% CI) | P Value | HR (95% CI) | P Value | ||
| Univariable | ||||||
| MUC5B genotype | ||||||
| GG | 1 [Reference] |  | 1 [Reference] |  | ||
| GT | 0.52 (0.34–0.78) | .001 | 0.49 (0.30–0.79) | .004 | ||
| TT | 0.27 (0.12–0.60) | 0.24 (0.09–0.63) | ||||
| Adjusted | ||||||
| MUC5B genotype | ||||||
| GG | 1 [Reference] |  | 1 [Reference] |  | ||
| GT | 0.48 (0.31–0.72) | <.001 | 0.39 (0.21–0.70) | .002 | ||
| TT | 0.23 (0.10–0.52) | 0.15 (0.05–0.49) | ||||
| FVC | 0.52 (0.32–0.83) | .006 | 0.53 (0.30–0.93) | .03 | ||
| DLCO | 0.89 (0.85–0.94) | <.001 | 0.90 (0.83–0.98) | .02 | ||
| Sex | ||||||
| Male | 1 [Reference] |  | <.001 | 1 [Reference] |  | .06 |
| Female | 0.29 (0.14–0.59) | 0.47 (0.22–1.02) | ||||
Abbreviations: DLCO, diffusing capacity of carbon monoxide; FVC, forced vital capacity; HR, hazard ratio.
After including all relevant variables (sex, baseline FVC, and baseline DLCO) in a final model, the MUC5B genotype remained a statistically significant predictor of survival. The adjusted HRs for survival associated with the GT and TT genotypes were 0.48 (95% CI, 0.31–0.72) and 0.23 (95% CI, 0.10–0.52) in the INSPIRE cohort and 0.39 (95% CI, 0.21–0.70) and 0.15 (95% CI, 0.05–0.49) in the Chicago cohort (Table 2). In both cohorts, the AUC was higher for the adjusted model that included MUC5B genotype as a predictor than when MUC5B genotype was excluded (Figure 3 and Figure 4). The integral of the time-specific AUC (C statistic) was higher for the adjusted model including the MUC5B genotype compared with the adjusted model that excludes the genotype (C=0.71 [95% CI, 0.64–0.75] and C=0.68 [95% CI, 0.61–0.73], respectively) for the INSPIRE cohort. This result was replicated in the Chicago cohort (C=0.73 [95%CI, 0.62–0.78] and C=0.69 [95% CI, 0.59–0.75], respectively). The increase in the C statistic value was statistically significant in both the INSPIRE (P<.001) and Chicago (P=.01) cohorts, suggesting that the MUC5B genotype improves the predictive accuracy of the model.
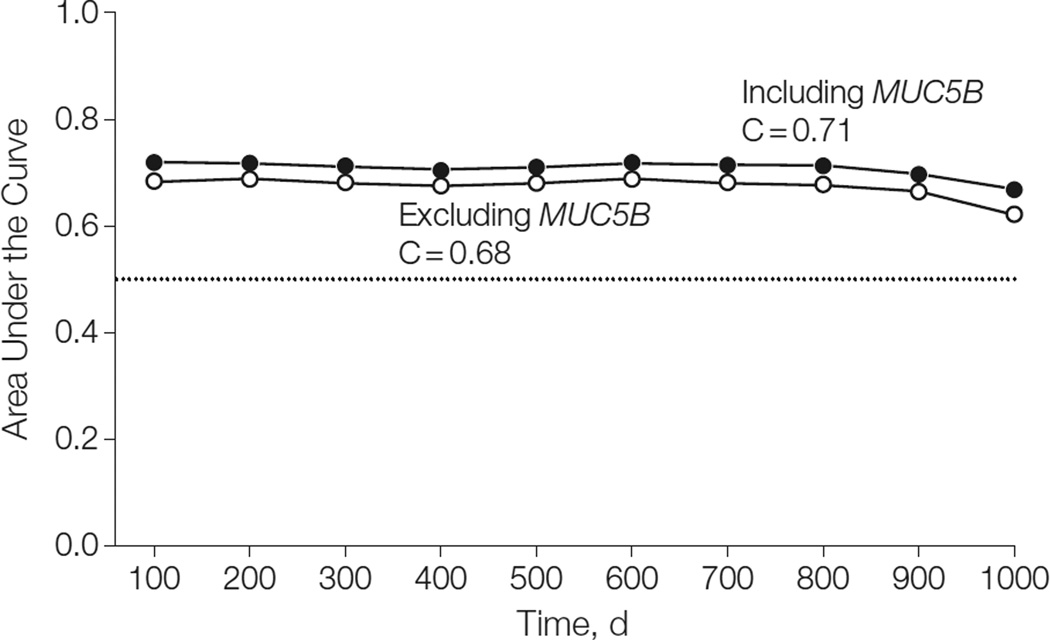
Time-Dependent Area Under the Curve for Adjusted Models Including and Excluding the MUC5B Genotype Predictor, INSPIRE Cohort

Time-Dependent Area Under the Curve for Adjusted Models Including and Excluding the MUC5B Genotype Predictor, Chicago Cohort
In the secondary analysis, the observed association between the MUC5B promoter polymorphism and survival was explored after accounting for the effect of MMP-7, which has recently been reported as prognostic in IPF.8 Matrix metalloproteinase 7 was significantly associated with survival when added to the final model for the INSPIRE cohort. The hazard was 2-fold higher for patients with MMP-7 levels of 5.7 ng/mL or higher than for patients with MMP-7 levels of less than 5.7 ng/mL (HR, 2.06; 95% CI, 1.05–4.07; P=.04) (Table 3). However, the HR for the MUC5B genotype remained constant and statistically significant, suggesting that the MUC5B promoter polymorphism explains a portion of the variation in survival among IPF participants beyond that explained by MMP-7 levels.
Table 3
Survival Analysis Model in INSPIRE Cohort, Adjusted for MMP-7
| Variables | Hazard Ratio (95% CI) | P Value | |
|---|---|---|---|
| MMP-7, ng/mL | |||
| <5.7 | 1 [Reference] |  | .04 |
| ≥5.7 | 2.06 (1.05–4.07) | ||
| MUC5B genotype | |||
| GG | 1 [Reference] |  | |
| GT | 0.46 (0.30–0.70) | <.001 | |
| TT | 0.21 (0.09–0.49) | ||
| FVC | 0.55 (0.34–0.89) | .01 | |
| DLCO | 0.89 (0.85–0.95) | <.001 | |
| Sex | |||
| Male | 1 [Reference] |  | <.001 |
| Female | 0.25 (0.12–0.53) | ||
Abbreviations: DLCO, diffusing capacity of carbon monoxide; FVC, forced vital capacity; MMP-7, matrix metalloproteinase 7.
No significant association between treatment and survival was observed in the INSPIRE cohort (eTable 1), and none of the genotype groups was associated with improved survival in the treatment stratum compared with placebo. However, exploratory analyses showed evidence of a possible interaction between interferon-γ1b treatment and MUC5B genotype. When stratified by treatment group, the effect of the genotype appeared to be stronger in the treatment group than in the placebo group. For the GT and TT groups, the unadjusted HRs were 0.75 (95% CI, 0.35–1.63) and 0.56 (95% CI, 0.12–2.65), respectively, among the placebo group and the unadjusted HRs were 0.44 (95% CI, 0.27–0.71) and 0.19 (95% CI, 0.07–0.50) among the treatment group. In post hoc analyses, the interaction was not statistically significant (P=.07) when added to the final survival model (eTable 2).
DISCUSSION
These findings suggest that the common polymorphism in the promoter of MUC5B (rs35705950), previously reported to be strongly associated with the development of familial interstitial pneumonia and idiopathic pulmonary fibrosis,12,13 is significantly associated with improved survival in IPF. These findings are consistent with a previous report of an association between the MUC5B variant and less severe pathological changes in familial interstitial pneumonia,16 as well as another report of slower decline in FVC for patients with IPF.17 This study is, to our knowledge, the first to demonstrate that a genetic variant is associated with survival in IPF.
While the minor (T) allele is the risk allele for development of IPF, it is also the minor allele that confers a survival advantage among patients with IPF. These results suggest that the IPF phenotype is heterogeneous and consists of at least 2 clinical subtypes that are, in part, separable by the MUC5B genotype. This would imply that the IPF phenotypes associated with other genetic and/or environmental risk factors (separate from the MUC5B promoter polymorphism) have poorer prognosis and possibly different pathogenic mechanisms. If there are several pathogenic mechanisms that lead to lung fibrosis, it is conceivable that the genotype-phenotype relationships are also unique and, consequently, give rise to specific, gene variant–directed clinical outcomes.
However, it is important to consider how and under what circumstances the promoter polymorphism of MUC5B might create a survival advantage in patients with IPF. Although individuals with the MUC5B promoter polymorphism could present earlier in the course of disease (lead time bias) or have preferential loss of sicker participants (survivor bias), these explanations are unlikely. In this study, patients with IPF with or without the MUC5B promoter polymorphism presented at similar ages and with similar degrees of lung impairment (Table 4). Moreover, the 2 cohorts in this manuscript are representative of physiologically mild (INSPIRE) and moderate (Chicago) stages of IPF, suggesting that the MUC5B polymorphism may be associated with outcome at different stages of disease. Previous findings indicate that patients with IPF have enhanced mucus production regardless of the MUC5B promoter genotype.12
Table 4
Patient Characteristics by MUC5B Genotype
| INSPIRE | Chicago | |||||
|---|---|---|---|---|---|---|
| Characteristics | GG (n = 148) | GT (n = 259) | TT (n = 31) | GG (n = 41) | GT (n = 98) | TT (n = 9) |
| Deaths, No. (%) | 36 (24) | 35 (14) | 2 (6) | 26 (63) | 36 (37) | 2 (22) |
| Transplant events, No. (%) | 2 (1) | 5 (2) | 0 (0) | 3 (7) | 7 (7) | 0 (0) |
| Female, No. (%) | 39 (26) | 68 (26) | 8 (26) | 6 (15) | 27 (28) | 2 (22) |
| Age, mean (SD), y | 65.7 (8.22) | 67.0 (7.34) | 68.5 (4.80) | 68.7 (10.22) | 69.1 (8.25) | 68.4 (7.70) |
| Ever smokers, No. (%) | 114 (77) | 174 (67) | 24 (77) | 25 (66) | 66 (76) | 5 (56) |
| FVC, mean (SD), % predicted | 71.9 (12.15) | 71.7 (12.07) | 77.7 (14.74) | 64.0 (19.39) | 64.1 (18.62) | 72.3 (8.48) |
| FVC, mean (SD), L | 2.9 (0.72) | 2.8 (0.77) | 3.0 (0.82) | 2.5 (0.75) | 2.5 (0.85) | 2.8 (0.73) |
| DLCO, mean (SD), % predicted | 47.1 (9.05) | 47.0 (8.36) | 50.7 (11.65) | 45.3 (18.70) | 45.5 (17.67) | 60.0 (12.52) |
| DLCO, mean (SD), mL/min/mm Hg | 12.7 (4.22) | 12.4 (4.49) | 12.6 (4.59) | 11.0 (5.27) | 10.4 (4.21) | 13.6 (3.18) |
| MMP-7, median (IQR), ng/mLa | 7.5 (5.1–10.6) | 7.9 (5.8–11.6) | 8.1 (5.2–10.2) | |||
| Interferon-γ1b treatment, No. (%)a | 98 (66) | 176 (68) | 21 (68) | |||
Abbreviations: DLCO, diffusing capacity of carbon monoxide; FVC, forced vital capacity; IQR, interquartile range; MMP-7, matrix metalloproteinase 7.
Although the specific mechanisms that convey the observed survival advantage to patients with the MUC5B promoter polymorphism are not yet apparent, enhanced mucosal host defense, reduction in infectious complications, a beneficial drug response, and a potential dual role for MUC5B in wound repair should all be considered. Regardless of the mechanism, the results suggest that patients with IPF with the MUC5B promoter polymorphism may represent a pathogenically distinct disease entity that incorporates both a significantly higher predisposition to disease and a significantly longer survival. It is conceivable that specific pharmacologic agents may work well in a subset of patients with IPF who are separable by genomic and biochemical markers. The findings of this study suggest that there may be value in measuring the MUC5B polymorphism in future clinical therapeutic trials.
It may also be useful to assess whether future prognostic models that incorporate MUC5B and other molecular features have the potential to affect clinical care of patients. The clinical-radiographic-physiologic scoring system5 as well as a simpler index18 including pulmonary function and, most recently, a model that incorporates sex, age, FVC, and DLCO19 have all been found to be predictive of mortality to some extent. Additional studies have evaluated change in pulmonary function indexes over 6- to 12-month periods in IPF and have concluded that the decline in FVC is predictive of survival.20–23 Although these clinical assessments of IPF are modestly successful in correlating measures of disease severity with eventual mortality and potentially allow better staging of patients with IPF, they do not approach the predictive accuracy required to guide personalized management. In particular, these approaches cannot predict which patients, despite similar clinical presentations, will have a better or worse clinical outcome, a task that is necessary because of the variable and unpredictable course of the disease.24
Moreover, these approaches have limited utility because they focus on measures of disease severity (ie, FVC and DLCO) to predict future disease progression. Given that the addition of MUC5B resulted in relatively small gains in predictive accuracy in patients with established disease, it is premature to consider routine clinical genotyping of patients with IPF. However, there may be value in assessing scoring systems that use the potential predictive value of MUC5B combined with other genetic and molecular factors to predict prognosis in patients with subclinical and early-stage disease, before a notable decline in lung function is observed. This is especially important because there are currently no IPF pharmacological therapies approved for use in the United States,25 and opportunities for early genetic counseling or lung transplantation may be a patient’s only recourse. Recently, several other genetic loci have been identified as risk factors for fibrotic lung disease.26 Future studies are needed to determine whether these loci account for the apparent phenotypic heterogeneity and have added predictive value for survival in IPF that could be used to develop improved scoring systems.
Although the MUC5B polymorphism did not meet the expectations of HWE, this was not an unexpected finding and is not likely to bias the results of this analysis. Previous studies have reported similar observations among patients with IPF.12,13 In the study by Seibold et al,12 the minor allele frequency among patients with IPF was very similar to those observed in this study (38% compared with 37% for INSPIRE and 39% for Chicago), while the frequency among controls was reported to be much lower (9%) and consistent with HWE. In addition, it has been shown that these departures from HWE in the cases but not controls are consistent with the disease model wherein the minor (T) allele is associated with disease.12,27 Regardless, the findings presented here are robust to departures from HWE because a genotype-based test of MUC5B was used as opposed to an allelic test, which does assume HWE.
This study had several limitations. First, although analyses in both cohorts were well powered to detect the association of genotype with survival, the relatively low numbers of events in each cohort led to wide confidence intervals for the estimated HRs. Larger samples would be needed to obtain a more precise estimate of the HRs for the MUC5B genotype.
Second, while there was not statistically significant evidence of an interaction effect between the MUC5B genotype and interferon-γ1b treatment, it is possible that a true interaction exists that this study was not powered to detect. This would imply that the strength of the association between genotype and survival is dependent on interferon-γ1b treatment. However, if there was a true interaction effect, replication of the strength of the association between genotype and survival within the Chicago cohort would not have been expected, since none of these patients were treated with interferon-γ1b. Further studies would be needed to determine whether an interaction effect exists between the MUC5B genotype and other clinical treatments. Nevertheless, this emphasizes the need to account for genotype in clinical trials and prediction models of IPF.
Third, this study used all-cause mortality as the primary end point, as opposed to IPF-related mortality, to maintain generalizability and comparability across cohorts since cause of death information was not complete for both cohorts. Among the INSPIRE patients, 66 of the 73 deaths were confirmed to be IPF related and 7 (2 GG and 5 GT) were possibly or definitely unrelated to IPF. This should have little effect on the results of this study because the vast majority of patients with IPF die within a relatively short period owing to the nature of the disease.
Fourth, data for additional genetic markers and serum biomarkers that have been reported to be associated with outcome in IPF were not available for these cohorts. More comprehensive predictive models may be developed by including these biomarkers and other genetic risk factors.
Idiopathic pulmonary fibrosis is etiologically heterogeneous and biologically dynamic, and sequence changes in MUC5B, transcriptional signatures in the fibrotic lung,2,28 and serum biomarkers, including chemokine ligand 18, KL6, SFTPA, SFTPD, MMP-7, intercellular adhesion molecule 1, and interleukin 8, are potential predictors of disease activity and outcome in patients with IPF.6–8,29 Further study is necessary to refine the risk estimates and to determine the research and clinical implications of these findings.
Acknowledgments
Drs Seibold and D. A. Schwartz have filed a patent for MUC5B genetic variants, methods, and compositions for risk prediction, diagnosis, prognosis, and treatment of pulmonary disorders. Drs Kossen and Seiwert are employee shareholders of InterMune Inc, which is pursuing a license to the aforementioned patent, and have filed patents on use of peripheral blood biomarkers in IPF. Dr Noth has contracts for clinical trials with Boehringer Ingelheim, Sanofi, and Stromedix; has served on the speakers’ bureau of GlaxoSmithKline; is a consultant to Boehringer Ingelheim and Immuneworks; and has patents related to gene expression profiling and plasma proteins. Dr Kaminski is a consultant to Sanofi, InterMune, Stromedix, Celgene, Promedior, and Vertex; is a recipient of investigator-initiated research grants from Gilead, Celgene, and Centocor; and has filed a patent on use of peripheral blood biomarkers in IPF.
Funding/Support: This research was supported by the National Heart, Lung, and Blood Institute (grants R01-HL095393, R01-HL097163, R01-HL095397, P50-HL0894932, P01-HL092870, RC1-HL09961, and RC2-HL101715) and the Dorothy P. and Richard P. Simmons Endowed Chair for Pulmonary Research (University of Pittsburgh School of Medicine).
Role of the Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Additional Contributions: We gratefully appreciate all participating IPF patients for their contributions to IPF research.
Footnotes
Author Contributions: Dr Peljto had full access to the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Fingerlin, Garcia, Lindell, Loyd, Noth, Schwarz, Kaminski, Schwartz.Acquisition of data: Zhang, Ma, Lindell, Steele, Gibson, Brown, Talbert, Markin, Kossen, Murphy, Noth, Schwarz, Kaminski, Schwartz.
Analysis and interpretation of data: Peljto, Fingerlin, Ma, Richards, Silveira, Loyd, Seibold, Brown, Talbert, Kossen, Seiwert, Noth, Schwarz, Kaminski, Schwartz.
Drafting of the manuscript: Peljto, Fingerlin, Richards, Silveira, Gibson, Markin, Noth, Schwarz, Kaminski, Schwartz.
Critical revision of the manuscript for important intellectual content: Peljto, Zhang, Fingerlin, Ma, Garcia, Richards, Lindell, Steele, Loyd, Seibold, Brown, Talbert, Kossen, Seiwert, Murphy, Kaminski, Schwartz.
Statistical analysis: Peljto, Fingerlin, Ma, Richards, Silveira, Noth, Schwartz.
Obtained funding: Seiwert, Noth, Schwarz, Kaminski, Schwartz.
Administrative, technical, or material support: Zhang, Lindell, Steele, Loyd, Gibson, Brown, Talbert, Markin, Seiwert, Murphy, Schwartz.
Study supervision: Fingerlin, Loyd, Schwarz, Kaminski, Schwartz.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. No other disclosures were reported.
Online-Only Material: The eFigure and 2 eTables are available at http://www.jama.com.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1001/jama.2013.5827
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4545271?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Serum soluble isoform of receptor for advanced glycation end product is a predictive biomarker for acute exacerbation of idiopathic pulmonary fibrosis: a German and Japanese cohort study.
Respir Res, 25(1):405, 11 Nov 2024
Cited by: 0 articles | PMID: 39529063 | PMCID: PMC11552171
Associations of circulating matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases with clinically relevant outcomes in idiopathic pulmonary fibrosis: Data from the IPF-PRO Registry.
PLoS One, 19(10):e0312044, 17 Oct 2024
Cited by: 0 articles | PMID: 39418259 | PMCID: PMC11486396
Bleomycin-Induced Pulmonary Fibrosis in Transgenic Mice Carrying the Human MUC5B rs35705950 Variant.
Cells, 13(18):1523, 11 Sep 2024
Cited by: 0 articles | PMID: 39329706 | PMCID: PMC11430646
Highlights on Future Treatments of IPF: Clues and Pitfalls.
Int J Mol Sci, 25(15):8392, 01 Aug 2024
Cited by: 1 article | PMID: 39125962 | PMCID: PMC11313529
Review Free full text in Europe PMC
Genetics and Genomics of Pulmonary Fibrosis: Charting the Molecular Landscape and Shaping Precision Medicine.
Am J Respir Crit Care Med, 210(4):401-423, 01 Aug 2024
Cited by: 1 article | PMID: 38573068
Review
Go to all (271) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs
- (3 citations) dbSNP - rs35705950
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The minor T allele of the MUC5B promoter rs35705950 associated with susceptibility to idiopathic pulmonary fibrosis: a meta-analysis.
Sci Rep, 11(1):24007, 14 Dec 2021
Cited by: 5 articles | PMID: 34907291 | PMCID: PMC8671516
Review Free full text in Europe PMC
Prognostic role of MUC5B rs35705950 genotype in patients with idiopathic pulmonary fibrosis (IPF) on antifibrotic treatment.
Respir Res, 22(1):98, 01 Apr 2021
Cited by: 17 articles | PMID: 33794872 | PMCID: PMC8017848
MUC5B rs35705950 minor allele associates with older age and better survival in idiopathic pulmonary fibrosis.
Respirology, 28(5):455-464, 26 Dec 2022
Cited by: 7 articles | PMID: 36571111
Idiopathic Pulmonary Fibrosis Is a Genetic Disease Involving Mucus and the Peripheral Airways.
Ann Am Thorac Soc, 15(suppl 3):S192-S197, 01 Nov 2018
Cited by: 32 articles | PMID: 30431344 | PMCID: PMC6322034
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NHLBI NIH HHS (15)
Grant ID: P01 HL092870
Grant ID: P01-HL092870
Grant ID: P50 HL084932
Grant ID: RC2 HL101715
Grant ID: F32 HL009961
Grant ID: P50-HL0894932
Grant ID: R01 HL097163
Grant ID: RC2-HL101715
Grant ID: R01-HL095393
Grant ID: R01-HL095397
Grant ID: R01-HL097163
Grant ID: RC1-HL09961
Grant ID: RC1 HL099619
Grant ID: R01 HL095393
Grant ID: R01 HL095397







