Abstract
Free full text

Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics
Abstract
Antibiotic resistance is a growing problem which threatens modern healthcare globally. Resistance has traditionally been viewed as a clinical problem, but recently non-clinical environments have been highlighted as an important factor in the dissemination of antibiotic resistance genes (ARGs). Horizontal gene transfer (HGT) events are likely to be common in aquatic environments; integrons in particular are well suited for mediating environmental dissemination of ARGs. A growing body of evidence suggests that ARGs are ubiquitous in natural environments. Particularly, elevated levels of ARGs and integrons in aquatic environments are correlated to proximity to anthropogenic activities. The source of this increase is likely to be routine discharge of antibiotics and resistance genes, for example, via wastewater or run-off from livestock facilities and agriculture. While very high levels of antibiotic contamination are likely to select for resistant bacteria directly, the role of sub-inhibitory concentrations of antibiotics in environmental antibiotic resistance dissemination remains unclear. In vitro studies have shown that low levels of antibiotics can select for resistant mutants and also facilitate HGT, indicating the need for caution. Overall, it is becoming increasingly clear that the environment plays an important role in dissemination of antibiotic resistance; further studies are needed to elucidate key aspects of this process. Importantly, the levels of environmental antibiotic contamination at which resistant bacteria are selected for and HGT is facilitated at should be determined. This would enable better risk analyses and facilitate measures for preventing dissemination and development of antibiotic resistance in the environment.
The first antibiotic compound, penicillin, was discovered in 1928 by Alexander Fleming as a product of the fungus Penicillium notatum, and became available for therapeutic use in the 1940s. The therapeutic usage of penicillin was however pre-empted by another class of antibiotics, the sulphonamides, which were introduced in 1937 (1). These new therapeutical agents brought about a paradigm shift in the treatment of bacterial diseases. Not only did deadly infectious diseases become treatable, but the availability of antibiotics also opened up possibilities for new kinds of medical interventions including major surgical interventions and organ transplants (2). For some decades after their introduction, antibiotics seemed to have solved the problem of bacterial infectious diseases forever (3).
Although antibiotic-resistant bacteria started to appear soon after the clinical introduction of antibiotics, the problem was limited and at first dismissed as of little concern. Sulphonamide-resistant Streptococcus pyogenes appeared in hospitals as early as the 1930s and penicillin-resistant Staphylococcus aureus appeared after penicillin had been introduced in the 1940s. In the 1950s, multidrug-resistant enteric bacteria started to cause problems (4). Furthermore, antibiotic resistance capable of being transferred horizontally between bacteria was discovered during this decade (1).
Overuse and misuse of antibiotics are widely regarded as having been major factors in promoting antibiotic resistance (2). In clinical contexts, such misuses include prescription of antibiotics without the infection established to be bacterial (5) and patient non-compliance to the full prescription (6). These problems are further exacerbated in developing countries where socioeconomic factors dictate the handling of antibiotics. Self-medication is prevalent as antibiotics are often sold over-the-counter and a general lack of education and awareness prompts misuse of antibiotics (7, 8). Furthermore, since antibiotics are often sold pill-by-pill in developing countries, poor patients are unlikely to fulfil their prescribed antibiotic regimens once they feel better due to economic reasons (7, 9). Emerging antibiotic resistance in developing countries is not necessarily regionally confined, since today's globalised world allows for resistant bacteria as well as people to travel around the world (2).
The great success of antibiotics at treating infectious diseases prompted the drugs to be used outside of clinical contexts. Antibiotics began to be used in large scale as growth promoters and prophylactics in livestock, usually administered by addition to the feed (10). This new application also meant that the environment began to be massively exposed to antibiotics (5). Other non-clinical large-scale uses of antibiotics include their usage in aquaculture (11) and poultry farming (10). It is widely believed that this excessive use of antibiotics has contributed to the development and dissemination of antibiotic resistance. As a result, a number of antibiotics were banned for usage as growth promoters in the European Union in the 1990s (10, 12).
Today, antibiotic resistance is a well-acknowledged global health problem. While antibiotics are still effective at treating many bacterial infections, some strains are extremely difficult to treat, and therapeutical options are getting fewer (13). This is exacerbated by the fact that the short expected time of usefulness of a new antibiotic compound before resistance arises means that few companies are interested in developing new antibiotics (3). It has been estimated that in the European Union, the United States, and Thailand, antibiotic-resistant bacteria are responsible for more than 25,000, 23,000, and 38,000 deaths every year, respectively (14). In short, antibiotic resistance is getting more prevalent and widely disseminated and few new antibiotics are in development. As the situation stands today, there is a clear risk that mankind will return to clinical conditions resembling those before the therapeutical advent of antibiotics (13).
Since antibiotic resistance causes problems when it complicates the treatment of ill patients, historically it has mainly been studied in the context of clinical settings (15). However, aside from resistance conferred by mutations, the source of antibiotic resistance determinants in pathogenic bacteria must have originated in non-clinical environments (16). Recently, much more attention has been directed at understanding the ecological and environmental processes involved in resistance gene acquisition. While some examples indicating antibiotic resistance gene (ARG) dissemination between environmental and pathogenic bacteria exists (17), the complexity of the processes and relative scarcity of studies done, means knowledge is still lacking in the field. Indeed, it is still unclear what role antibiotics normally play in the social context of the environmental bacterial community (1). In this review, the role of the environment in the dissemination of ARGs is examined in terms of the natural role and prevalence of ARGs, horizontal gene transfer (HGT), dissemination routes, and the correlation between ARGs and anthropogenic contamination with antibiotics.
Origins and roles of antibiotics and resistance genes in the environment
Functions of antibiotics and resistance genes in the environment
Antibiotics were originally discovered as compounds produced by environmental fungi and bacteria capable of killing off other microorganisms. Since these compounds were efficient at eradicating bacteria, it was generally assumed that the purpose of antibiotic production was to stave off competing microorganisms. On the same note, when ARGs were discovered, they were believed to have evolved in bacteria producing the antibiotics and target bacteria in order to protect against the effects of antibiotics (5, 18). While this point of view is not necessarily wrong, recently other aspects of the environmental functions of antibiotics have begun to be explored. The antibiotic concentration levels produced by environmental bacteria are commonly far below the minimum inhibitory concentrations (MICs), which suggests that antibiotics may primarily serve some other function (19, 20). Evidence suggests that sub-inhibitory doses of antibiotics play several roles in the environment as regulatory substances and signalling molecules in inter-bacterial communication (5, 18, 21). Interestingly, sub-inhibitory concentrations of various antibiotics have been shown to induce different states in bacteria, including biofilm formation, the SOS response, and changes in primary metabolism. These states can increase tolerance to antibiotics (22). ARGs have also likely evolved to fulfil other purposes than protecting bacteria from antibiotics. One possibility is that the primary functions of ARGs in the environment are to regulate the responses induced from sub-inhibitory concentrations of antibiotics. Some ARGs may play regulatory roles in the biosynthesis of antibiotics (5). It has been suggested that β-lactamases are enzymes which at one point were involved in peptidoglycan synthesis (18). It is worth pointing out that even though antibiotics and ARGs seem to have functions unrelated to antibiosis in the natural environment, it has been demonstrated that sub-inhibitory concentrations of antibiotics, about 200 times below MIC values, can select for antibiotic-resistant bacteria (23).
Resistance gene recruitment from the environmental gene pool
While ARGs in their environmental context may originally have had other primary functions aside from conferring resistance to antibiotics, these genes have now been recruited as resistance genes in pathogenic bacteria. Furthermore, environmental bacteria harbour an as of yet unexplored pool of genes. Some of these genes may have the potential to be used as ARGs and to be passed on to pathogenic bacteria. ARGs have existed since before humans started to use antibiotics in therapy, and they have likely existed for as long as antibiotics themselves. For example, ARGs encoding resistance to β-lactams, tetracyclines, and glycopeptides have been found in 30,000-year-old Beringian permafrost (24) and multidrug-resistant bacteria have been found in a region of a cave in New Mexico, USA, which had been isolated for more than 4 million years (25). Seeing as how antibiotics and ARGs have existed over such a long period of evolutionary time, it seems likely that the environment is a reservoir of potential ARGs from which pathogenic bacteria may recruit protection against therapeutical agents used against them by humans.
The role of HGT in environmental dissemination of ARGs
In order for a pathogenic bacterium to acquire an ARG from the environmental gene pool, it must be transferred via one of the three processes of HGT; conjugation, transformation, and transduction.
Conjugation
Traditionally, conjugation has been regarded as the main facilitator of ARG transfer between bacteria. Antibiotic resistance transferable by bacterial conjugation was discovered in the 1950s (1). Since then, ARGs transferable by conjugation have been recorded several times. The benefits of this mode of transfer include the potential to transfer DNA among a broad host range of species (26). Conjugation has even been demonstrated from bacterial cells to eukaryotic cells (27), and has been seen in many different environments, including soil, marine sediment, seawater, sewage wastewater, and activated sludge (28). The most important genetic elements capable of being transferred by conjugation are the plasmids and the integrative conjugative elements (ICEs) (26).
Transformation
Recently, the importance of transformation in mediating environmental transfer of ARGs has begun to be re-evaluated. Transformation in the environment may intuitively sound like a rare event considering the sensitivity of DNA to degradation by nucleases and the dilution effects in water environments. However, DNA may be stabilised by adhesion to particles from sediment and soil. Dilution effects may also be less important if transformation occurs in biofilms where newly deceased bacteria lyse and allow their neighbouring bacteria to take up their released DNA (28). Natural transformation has been demonstrated in many different environments, including marine water, ground water, rivers, and soil (28), and it has been implicated as responsible for the dissemination of penicillin-resistance genes in Streptococcus spp. (29). In one study, concentrations of extracellular DNA were compared to intracellular DNA in a river basin in China (30). It was found that extracellular DNA (including ARGs) was more abundant than intracellular DNA, implying that extracellular DNA is an important environmental reservoir for genes accessible via transformation.
Transduction
Transduction, transfer of DNA between bacteria via bacteriophages, may also be more important in environmental gene transfer than previously thought (31). Phage particles are well suited for mediating DNA transfer in the environment. Contrary to naked DNA, they are relatively resistant to environmental degradation and their compact size further simplifies their dissemination (28). Furthermore, some bacteriophages are known to have very broad host ranges, some even capable of infecting different bacterial classes (32). These properties make bacteriophages ideal for transferring genes between spatially distant bacterial communities, such as from the environmental communities to human microbiomes (31). Transduction has been shown to be common in marine environments (33). Furthermore, evolutionary studies have demonstrated that considerable parts of the bacterial genomes have prophage origins, implying the importance and magnitude of viral alterations of the bacterial chromosome (34). Through viral metagenome analyses, β-lactamase genes have been detected in activated sludge and urban sewage (35). The gene conferring methicillin resistance in methicillin-resistant S. aureus (MRSA), mecA, has also been found in bacteriophage DNA from a wastewater treatment plant (WWTP) and the receiving water (36).
Integrons
A particularly well-studied genetic transfer element in environmental contexts is the integron. Integrons are genetic assembly platforms capable of capturing and expressing gene cassettes, which can encode antibiotic resistance determinants (Fig. 1). A defining feature of all types of integrons is a gene coding for a site-specific tyrosine recombinase called an integrase which can excise and integrate gene cassettes into the integron. It can also reshuffle the order of gene cassettes which affect the relative rate of expression of the individual cassettes (37, 38). The gene encoding the integrase (intI) is induced by the SOS response. Interestingly, antibiotics such as trimethoprim, quinolones, and β-lactams are known to induce the SOS response. Exposure to any of these antibiotics will thus induce excision, integration, and changes in expression rates of gene cassettes, increasing the likelihood that at least some bacteria in the population carrying the integron will have a high expression rate of a relevant ARG (39). The SOS response has also been shown to be inducible by conjugative DNA transfer, which means that integrons transferred to another host cell on a conjugative plasmid are likely to reshuffle their cassettes and thus increase the phenotype variability in a population (40).

The basic structure of a class 1 integron. The gene intI1 encodes a site-specific integrase which can excise and integrate gene cassettes at the site-specific integration site attI. In this example, the integron contains three gene cassettes denoted as GC1, GC2, and GC3. Expression of the gene cassettes is induced by the promoter PC. Class 1 integrons also consist of two conserved genes at the 3′-end, quarternary ammonium compound resistance gene qacEΔ1 and sulphonamide resistance gene sulI.
Integrons are typically divided into mobile integrons and chromosomal integrons. While chromosomal integrons are usually stationary in the bacteria, mobile integrons are readily disseminated between bacteria. While mobile integrons cannot mobilise and transfer themselves per se, they are often associated with genetic elements which can, such as plasmids (37, 38, 41). Recent studies have also indicated that natural transformation may be important in the dissemination of integrons (42). Mobile integrons are often capable of changing genetic locations within the host cell as well, since they are commonly associated with transposable genetic elements such as insertion sequences (ISs) and transposons (41). Class 1 integrons, a class of mobile integrons commonly found among clinical isolates, are associated with transposons derived from Tn402, which in turn can be carried by larger transposons, such as Tn21. Although mobile integrons usually only have a few gene cassettes in their cassette arrays, they often encode antibiotic resistance functionality and other phenotypes which give the host bacteria an adaptive advantage (37, 38).
Integrons are ubiquitous in the environment, and the ability of integrons to excise and acquire new gene cassettes has led to the notion that the sum of all gene cassettes in a given environment constitutes a metagenome which resident integrons can access. The number of different gene cassettes in 50 m2 soil has been estimated to be more than 2,000 (43). One study found about 1,000 different integron gene cassettes in marine sediments (44). While the large majority of these environmental cassette genes encode unknown functions, their variety suggests that bacteria carrying integrons have a vast pool of functions to tap into in order to adapt to changing conditions. Several studies have correlated high class 1 integron concentrations to anthropogenic activity. In one study, concentrations were found to be higher in aquatic environments contaminated with metal and antibiotics compared to unexposed environments (45). Another study showed that class 1 integrons were more abundant in detergent and antibiotic-contaminated sewage sludge and pig slurry compared to unexposed agricultural soils (46). Class 1 integron concentrations have also been shown to increase in the river Ravi as it flows through the city of Lahore, Pakistan (47). The capacities of mobile integrons to disseminate among bacteria, to confer adaptive advantages in changing conditions, and to utilise the environmental metagenome of gene cassettes, make them likely facilitators of environmental antibiotic resistance dissemination.
Genetic context and environmental dissemination of ARGs
The environmental presence of ARGs is well documented for several different types of environments. The extent of ARG dissemination is likely dependent upon the genetic context of the particular gene.
Sulphonamide resistance
Sulphonamide resistance genes sulI and sulII are examples of ARGs which are widespread in the environment. As sulI is often found as a conserved part of class 1 integrons (29), it can be expected to be found wherever these widespread mobile genetic elements are ubiquitous. sulII is most commonly found on plasmids of the incQ group (48, 49). sulI and sulII have been found in river water from Colorado, USA (50), Danish pigs (51), Australian and German surface waters (52), and in freshwater and marine water in the Philippines (53). sulI has also been found in wastewater (54, 55).
Trimethoprim resistance
Trimethoprim resistance genes of the dfr family generally appear to be found on integrons as cassettes (48, 56). For instance, dfrA1 has been found as a gene cassette on both class 1 and class 2 integrons (49, 56). The dfr genes’ propensity for being carried on integrons is likely to have facilitated their widespread dissemination in the environment (48, 56). dfrA1, dfrA5, dfrA6, dfrA12, and dfrA17 have been detected as cassettes in class 1 integrons in a river in India (57). In Portugal, dfrA1, dfrA7, dfrA12, and dfrA17 were found as integron cassettes in a polluted lagoon (58), and dfrA1 and dfrA12 were found in a WWTP connected to a slaughterhouse (59). drfA1 has also been detected in surface waters from Germany and Australia (52).
Quinolone resistance
The first quinolone resistance gene discovered, qnrA, was found to be carried on the plasmid pMG252. Since then, many qnr genes have been found associated with plasmids and related mobile genetic elements. qnrA and qnrB are often found on class 1 integrons, making quinolone resistance a trait often associated with other resistance determinants co-carried on the integron (60). Several studies have reported the isolation of qnr genes from environmental sources. qnrS has been isolated from several different sources, including the activated sludge of a WWTP in Germany (61), from the river Seine in France (62), a lake in Switzerland (63) and river water in Turkey (64). qnrB has been found in wastewater effluent from a WWTP in Italy (65), qnrB and qnrS have been found in Mexican soils irrigated with wastewater (66), and qnrA, qnrB, and qnrS have been found in an urban coastal wetland close to the US–Mexico border (67).
Tetracycline resistance
While many tetracycline resistance determinants are chromosomally encoded, the majority of tet genes are found on plasmids, transposons, and ICEs. Many of the mobile genetic elements which carry tet genes are conjugative and carry genes encoding resistance to other antibiotic compounds. For instance, tetM can be found on the ICE Tn2009 which also carries the macrolide–lincosamide–streptogramin B resistance gene ermB, and macrolide efflux genes mefA and mfrD. It is likely that great diversity of tet genes and the diversity and mobilisability of the genetic elements in which they reside have contributed significantly to their dissemination among many different bacterial genera (68). tetA, tetB, tetC, tetD, tetE, tetG, tetM, tetO, tetS, and tetQ have been found in wastewater from two WWTPs in Wisconsin, USA (69), tetO and tetW in river water from Colorado, USA (50), and tetB, tetL, tetM, tetO, tetQ, and tetW in archived soil collected in the Netherlands (70). tetA, tetC, tetG, tetM, tetS, and tetX have been detected in activated sludge from 15 different sewage treatment plants in China (71), and tetA and tetB in surface water from Germany and Australia (52).
Vancomycin resistance
The vanA operon, which encodes vancomycin resistance, is typically found to be carried on Tn1546 or Tn1546-like elements. While the former is non-conjugative, the latter is often found on conjugative plasmids. Dissemination of the vanB operon is believed to be mainly due to the spread of Tn916-like ICEs and related elements carrying the gene cluster (72). Both vanA and vanB have been found in influent wastewater in England (73) and Sweden (74). vanB has also been reported in wastewater effluent and a receiving river in Sweden (55). Additionally, vanA and vanB have been found in meat from swine and bovine sources (75), and in marine water in the United States (76). vanA has also been found in wastewater in Portugal (77), and wastewater and drinking water in Germany (78). Poultry have been particularly well studied with regard to vancomycin resistance prevalence, and vanA has been found in poultry from Norway (79) and Sweden (80). Interestingly, a variant of the vanA operon has been found in 30,000-year-old Beringian permafrost (24), which suggests that vancomycin resistance is both ancient and widespread in the environment.
Macrolide resistance
ermB is the most widespread of the macrolide resistance genes, and it is linked with a variety of different mobile genetic elements including ICEs located on both chromosomes and plasmids as well as non-conjugative transposons (81). The ICEs among which ermB have been found to be carried include Tn1545, Tn2010, and others of the Tn916 family, indicating that ermB is often linked with other antibiotic resistance determinants on a conjugative platform (81, 82). erm genes are prevalent in the environment, and they have been found in a variety of different environments. ermA and ermB have been found in milk from cows in Brazil (83) and in poultry production environments along the eastern seaboard of the United States (84). Additionally, ermB have been found in poultry samples (85), wastewater in Portugal (77), and German and Australian surface waters (52). ermA, ermB, ermC, ermF, ermT, and ermX have also been found in bovine and swine manure as well as a swine waste lagoon (86).
Anthropogenic contamination with antibiotics and ARGs
Dissemination routes of antibiotics and ARGs
Antibiotics of human origin can enter the environment through a number of different routes (Fig. 2). Antibiotics and their metabolites are released from hospitals with urine and faeces from patients as hospital wastewater effluent. Similarly, antibiotics are released into the wastewater treatment system via people taking antibiotics from home. From the WWTPs, the antibiotics can end up in sludge dispersed on fields as fertilizer, or released as run-off directly into the receiving surface waters (8, 87–89). Wastewater can also be treated by releasing it into wetlands (90). In such cases, the wetlands will be exposed to antibiotic contaminants in the wastewater. Antibiotics are also used therapeutically or as growth promoters in livestock and poultry. Antibiotics and their metabolites will spread through animal excrements and end up in fields and ground water, or in the case of antibiotic use in fish farms, directly into the aquatic environment (8, 87). It is also worth noting, that wherever antibiotics are spread, it is also likely that resistant bacteria follow the same routes of dispersal (91). This results in environments where antibiotics, ARGs, resistant bacteria, and the environmental bacterial flora (which may also harbour ARGs and potential ARGs) are mixed. These types of environments are likely resistance hotspots where ARGs proliferate and new resistant strains are created by HGT. The routes by which humans may come into contact with these bacteria are numerous. They include consumption of crops grown by contaminated sludge used as fertilizer, drinking of water drawn from contaminated ground or surface water and frolicking in marine water linked to contaminated surface water. When these resistant bacteria enter humans, they have the opportunity to spread their ARGs to the human microbiome (8).
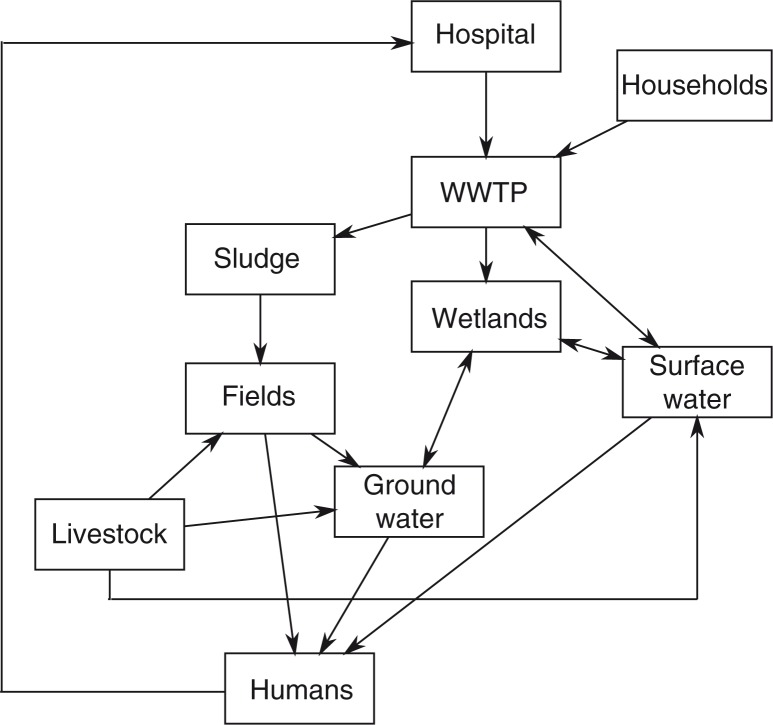
Antibiotics and antibiotic resistance genes (ARGs) can enter and re-emerge in humans via the environment by a number of different routes. For example, antibiotics and ARGs from patients taking antibiotics can end up in various environments (e.g. surface water) via excrements which pass through the wastewater treatment system. The commixture of antibiotics, ARGs and resident environmental bacteria at these locations provide an ideal opportunity for ARGs to develop and disseminate in the bacterial community.
Environmental antibiotic contamination and correlation to antibiotic resistance
Antibiotic concentrations in aquatic environments generally have been found to range from ng L−1 to low µg L−1 levels (88, 89, 92). In a summary of studies reporting antibiotic concentrations in aquatic environments (88), median concentrations in surface and ground water were reported as 30 and 71 ng L−1, respectively. Antibiotic concentrations in wastewater were largely dependent on the source of the waste. For hospital wastewaters, a median concentration of 2,100 ng L−1 was reported. For urban wastewaters, the median concentration was found to be 300 ng L−1. It is unknown what effect such levels of antibiotics have on bacteria in the environment. It has been demonstrated in vitro that resistant bacteria can be selected for at antibiotic concentrations lower than the MIC (93). For the strains used in that study, the minimum selective concentrations were 15,000 ng L−1 for tetracycline and from 2,500 ng L−1 to as low as 100 ng L−1 for ciprofloxacin depending on what mutation the particular strain was carrying. It is likely that the concentrations necessary for selection in a complex environmental community are different than compared to in vitro experiments. Regardless, it suggests that adverse effects of antibiotic contamination may be observed even at concentrations ubiquitous in wastewater. Furthermore, several studies have demonstrated that some antibiotics can induce conjugation and recombination in bacteria, even at sub-lethal concentrations (94–96), suggesting that transfer of ARGs by HGT may be common in environments contaminated with antibiotics.
Extremely high concentrations of antibiotics have been reported in wastewater connected to antibiotic-manufacturing facilities. Oxytetracycline concentrations in wastewater effluent from a WWTP treating waste from an oxytetracycline-manufacturing facility in China was found to be as high as 20,000,000 ng L−1. Oxytetracycline concentrations in the receiving river were as high as 640,000 ng L−1 (97). Comparisons between bacterial strains isolated downstream and upstream of the oxytetracycline waste discharge point indicated that strains isolated downstream frequently had significantly higher MICs to seven different antibiotic classes. They were also more frequently multidrug resistant and many different tet genes were detected in downstream isolates (98). High concentrations of antibiotics have also been found in effluent from a WWTP treating wastewater from a large antibiotic-manufacturing site in India. Ciprofloxacin was found in concentrations as high as 31,000,000 ng L−1, more than 1,000 times the lethal concentration for some bacterial strains and higher than therapeutic levels in human plasma (99). ARGs were found to be more abundant in the water downstream of the production site than upstream (100). In a study in Pakistan, elevated concentrations of antibiotics in a river downstream a pharmaceutical formulation facility were measured (39). At this site, the concentration of sulfamethoxazole was found as high as 49,000 ng L−1. The trimethoprim concentration was found to be 28,000 ng L−1. The corresponding ARGs sulI and dfrA1 were also measured at high concentrations, 0.80 and 0.43 genes/16S rRNA genes, respectively.
In the aforementioned studies, antibiotic resistance indicators were found to be elevated in sites exposed to very high levels of antibiotic contamination. However, there have also been indications that ARG abundances correlate to antibiotic contamination in environments with comparatively lower antibiotic exposure. Examples include a river in Colorado, USA, where higher concentrations of ARGs were reported at anthropogenically impacted sites compared to pristine sites located upstream (50) and a Swedish river in which higher ARG concentrations were determined downstream a WWTP discharging treated wastewater from the adjacent city (55). In a study in Pakistan, it was found that ARG concentrations increased in the river Ravi as it passed through the city of Lahore (39). The observed increase in ARG concentrations in these studies may be due to accumulation from anthropogenic sources rather than proliferation. In one study, a series of controlled experimental wetlands were exposed to environmental levels of antibiotics and no increases in ARG concentrations could be observed (101). In another study, ARG concentration increases were not observable in microcosms composed of lake water exposed to antibiotic concentrations up to 1,000 times those commonly measured in wastewater (102). While neither of these studies evaluated phenotypic resistance, ARGs were present in similar concentrations from the start to end of the experiments, indicating that selection did not take place. Clearly, more studies are needed to elucidate the complex interactions involved in antibiotic resistance proliferation and dissemination in environmental microbial communities.
Conclusions
It is becoming increasingly clear that the environment plays an important role in the dissemination of ARGs. A growing body of evidence suggests that ARGs are ubiquitous in the environment. HGT events are likely to be common in aquatic environments; integrons in particular are well suited for mediating environmental dissemination of ARGs. Many studies have managed to correlate higher environmental concentrations of ARGs and integrons to locations which are affected by human activities. These elevated levels are likely to arise from routine discharge of antibiotics, ARGs, and bacteria into aquatic environments, e.g. via wastewater and run-off from livestock facilities. These anthropogenic contaminants create a veritable hotspot for antibiotic resistance dissemination as they co-mingle with each other and the environmental pool of ARGs and bacteria. Contamination with high levels of antibiotics is of obvious concern as such levels are likely to directly select for resistant bacteria. While it is unclear how low levels of antibiotics affect the microbial community in complex environments, caution is warranted as studies have demonstrated the in vitro-capacity of sub-lethal concentrations of antibiotics to both induce resistance and facilitate HGT. More studies are needed to investigate at which concentrations antibiotic resistance is developed and disseminated, and also how these concentrations differ in different environments with different bacterial communities. Knowledge of these levels may prevent an environmental proliferation of ARGs which may eventually exacerbate resistance prevalence in pathogenic bacteria. Finally, understanding the role of the environment in the dissemination of ARGs is essential to effectively combat the rising threat of antibiotic resistance.
Acknowledgements
This review is partly based on the PhD thesis ‘Deliberations on the impact of antibiotic contamination on dissemination of antibiotic resistance genes in aquatic environments’ by Björn Berglund, Linköping University, 2014, ISBN: 978-91-7519-361-8. The author thanks Per-Eric Lindgren and Jerker Fick for their help and supervision during his PhD project.
Conflict of interest and funding
The author has no conflict of interest. Funding of the studies included in the author's PhD thesis was partly provided by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas) and the Swedish Foundation for Strategic Environmental Research (MISTRA).
References
Articles from Infection Ecology & Epidemiology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.3402/iee.v5.28564
Read article for free, from open access legal sources, via Unpaywall:
https://www.tandfonline.com/doi/pdf/10.3402/iee.v5.28564?needAccess=true
Citations & impact
Impact metrics
Article citations
Whole genome analysis of multidrug-resistant <i>Escherichia coli</i> isolate collected from drinking water in Armenia revealed the plasmid-borne <i>mcr-1.1-</i>mediated colistin resistance.
Microbiol Spectr, 12(10):e0075124, 21 Aug 2024
Cited by: 0 articles | PMID: 39166856 | PMCID: PMC11448411
Antibiotic-resistant <i>Escherichia coli</i> from treated municipal wastewaters and Black-headed Gull nestlings on the recipient river.
One Health, 19:100901, 22 Sep 2024
Cited by: 0 articles | PMID: 39399230 | PMCID: PMC11470789
Colistin Resistance Mediated by Mcr-3-Related Phosphoethanolamine Transferase Genes in Aeromonas Species Isolated from Aquatic Environments in Avaga and Pakro Communities in the Eastern Region of Ghana.
Infect Drug Resist, 17:3011-3023, 15 Jul 2024
Cited by: 0 articles | PMID: 39050833 | PMCID: PMC11268572
An overview of the occurrence, impact of process parameters, and the fate of antibiotic resistance genes during anaerobic digestion processes.
Environ Sci Pollut Res Int, 31(29):41745-41774, 10 Jun 2024
Cited by: 0 articles | PMID: 38853230 | PMCID: PMC11219439
Review Free full text in Europe PMC
Evaluation of Antibiotic Biodegradation by a Versatile and Highly Active Recombinant Laccase from the Thermoalkaliphilic Bacterium Bacillus sp. FNT.
Biomolecules, 14(3):369, 19 Mar 2024
Cited by: 1 article | PMID: 38540787 | PMCID: PMC10968384
Go to all (83) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Research progress on distribution, migration, transformation of antibiotics and antibiotic resistance genes (ARGs) in aquatic environment.
Crit Rev Biotechnol, 38(8):1195-1208, 28 May 2018
Cited by: 32 articles | PMID: 29807455
Review
Antibiotic resistant genes in the environment-exploring surveillance methods and sustainable remediation strategies of antibiotics and ARGs.
Environ Res, 215(pt 1):114212, 26 Aug 2022
Cited by: 3 articles | PMID: 36037921
Review
Dissemination of antibiotic resistance genes in representative broiler feedlots environments: identification of indicator ARGs and correlations with environmental variables.
Environ Sci Technol, 48(22):13120-13129, 03 Nov 2014
Cited by: 57 articles | PMID: 25338275
Dissemination of antibiotic resistance genes (ARGs) via integrons in Escherichia coli: A risk to human health.
Environ Pollut, 266(pt 2):115260, 15 Jul 2020
Cited by: 40 articles | PMID: 32717638
Review





