Abstract
Background
Infection with human immunodeficiency virus type 1 (HIV) is associated with clinical symptoms of accelerated aging, as evidenced by the increased incidence and diversity of age-related illnesses at relatively young ages and supporting findings of organ and cellular pathologic analyses. But it has been difficult to detect an accelerated aging effect at a molecular level.Methods
Here, we used an epigenetic biomarker of aging based on host DNA methylation levels to study accelerated aging effects due to HIV infection. DNA from brain and blood tissue was assayed via the Illumina Infinium Methylation 450 K platform.Results
Using 6 novel DNA methylation data sets, we show that HIV infection leads to an increase in epigenetic age both in brain tissue (7.4 years) and blood (5.2 years). While the observed accelerated aging effects in blood may reflect changes in blood cell composition (notably exhausted cytotoxic T cells), it is less clear what explains the observed accelerated aging effects in brain tissue.Conclusions
Overall, our results demonstrate that the epigenetic clock is a useful biomarker for detecting accelerated aging effects due to HIV infection. This tool can be used to accurately determine the extent of age acceleration in individual tissues and cells.Free full text

HIV-1 Infection Accelerates Age According to the Epigenetic Clock
Associated Data
Abstract
Background. Infection with human immunodeficiency virus type 1 (HIV) is associated with clinical symptoms of accelerated aging, as evidenced by the increased incidence and diversity of age-related illnesses at relatively young ages and supporting findings of organ and cellular pathologic analyses. But it has been difficult to detect an accelerated aging effect at a molecular level.
Infection with human immunodeficiency virus type 1 (HIV) is associated with clinical symptoms of accelerated aging, as evidenced by the increased incidence and diversity of age-related illnesses at relatively young ages and supporting findings of organ and cellular pathologic analyses. But it has been difficult to detect an accelerated aging effect at a molecular level.
Methods. Here, we used an epigenetic biomarker of aging based on host DNA methylation levels to study accelerated aging effects due to HIV infection. DNA from brain and blood tissue was assayed via the Illumina Infinium Methylation 450 K platform.
Here, we used an epigenetic biomarker of aging based on host DNA methylation levels to study accelerated aging effects due to HIV infection. DNA from brain and blood tissue was assayed via the Illumina Infinium Methylation 450 K platform.
Results. Using 6 novel DNA methylation data sets, we show that HIV infection leads to an increase in epigenetic age both in brain tissue (7.4 years) and blood (5.2 years). While the observed accelerated aging effects in blood may reflect changes in blood cell composition (notably exhausted cytotoxic T cells), it is less clear what explains the observed accelerated aging effects in brain tissue.
Using 6 novel DNA methylation data sets, we show that HIV infection leads to an increase in epigenetic age both in brain tissue (7.4 years) and blood (5.2 years). While the observed accelerated aging effects in blood may reflect changes in blood cell composition (notably exhausted cytotoxic T cells), it is less clear what explains the observed accelerated aging effects in brain tissue.
Conclusions. Overall, our results demonstrate that the epigenetic clock is a useful biomarker for detecting accelerated aging effects due to HIV infection. This tool can be used to accurately determine the extent of age acceleration in individual tissues and cells.
Overall, our results demonstrate that the epigenetic clock is a useful biomarker for detecting accelerated aging effects due to HIV infection. This tool can be used to accurately determine the extent of age acceleration in individual tissues and cells.
While antiretroviral therapy for human immunodeficiency virus (HIV) infection is highly effective at preventing AIDS-related complications, treated patients are at a significant risk for a number of diseases typically associated with older age, including cardiovascular disease, osteoporosis, cancer, neurocognitive impairment, and frailty [1–11]. Among the aging HIV-infected population, it has become evident that the incidence of HIV-associated non–AIDS-defining conditions is increasing [12]. In recent years, the Working Group on HIV and Aging published a report for the National Institutes of Health Office of AIDS Research, in which it stated that the cause of this increasing incidence is unclear but proposed that it may be due to an accelerated aging process [13]. To show that HIV infection is associated with accelerated normal aging, one first needs to understand what is meant by normal aging and to find a way of measuring it. Owing to its modest accuracy, telomere length will probably have to be supplemented by additional biomarkers of aging when it comes to understanding and measuring normal aging. DNA methylation levels are particularly promising biomarkers of aging since chronological age (ie, the calendar years that have passed since birth) has a profound effect on DNA methylation levels in most human tissues and cell types [14–23]. Several recent studies support measuring accelerated aging effects by using DNA methylation levels [24, 25]. The noteworthy aspect of our recently developed epigenetic clock (based on 353 dinucleotide markers known as cytosine phosphate guanines [CpGs]) is its broad applicability to most human cell types, tissues, and organs [25]. Predicted age, referred to as DNA methylation age, correlates with chronological age in sorted cell types (CD4+ T cells, monocytes, B cells, glial cells, and neurons) and in tissues and organs, including whole blood, brain, breast, kidney, liver, lung, and saliva [25].
The epigenetic clock is an attractive biomarker of aging because (1) it is more strongly correlated with chronological age than previous biomarkers, including telomere length [26, 27]; (2) it is prognostic of all-cause mortality in later life [28]; and (3) it correlates with measures of physical and mental fitness in older age [29]. The usefulness of the method has been demonstrated in recent case studies, including one showing that obesity accelerates epigenetic aging in liver tissue [27]. Further, trisomy 21 (Down syndrome) accelerates epigenetic aging in blood and brain tissue [30].
Although HIV infection appears to lower the age at which individuals develop age-related illnesses, it is not yet known whether DNA methylation age is a biologically meaningful biomarker of accelerated aging in the context of HIV infection. Here we use both blood and brain tissue from HIV-infected subjects (hereafter, “cases”) and uninfected controls (hereafter, “controls”) to show that HIV infection is significantly associated with increased age acceleration according to the epigenetic clock.
MATERIALS AND METHODS
DNA Methylation Data Sets
An overview of the 11 Illumina DNA methylation data sets used in this article is provided in Table Table1.1. We generated 6 novel DNA methylation data sets and used 5 publicly available data sets. Our 6 novel data sets are available from Gene Expression Omnibus super-series GSE67752. Details on the individual data sets can be found in Table Table11 and Supplementary 1. The 3 brain data sets came from the National NeuroAIDS Tissue Consortium (NNTC) [31]. Informed consent and all study procedures were approved by the institutional review boards (IRBs) at the 4 individual sites composing the NNTC.
Table 1.
Overview of the DNA Methylation Data Sets
| Data Seta | DNA Origin | Description | Illumina Platform | Illumina Arrays, No. | HIV-Infected Subjects, No. | Reference | Public Availabilityb |
|---|---|---|---|---|---|---|---|
| 1 | Brain tissue | Various brain regions from HIV-infected and uninfected subjects | 450 K | 130 | 99 | Novel data | GSE59457 |
| 2 | Brain tissue | Frontal lobe from HIV-infected and uninfected subjects | 450 K | 33 | 8 | Novel data | GSE67749 |
| 3 | Brain tissue | Cerebellum from HIV-infected and uninfected subjects | 450 K | 20 | 8 | Novel data | GSE67748 |
| 4 | Whole blood | HIV-infected and uninfected subjects | 450 K | 92 | 24 | Novel data | GSE53841 |
| 5 | Whole blood | HIV-infected and uninfected subjects | 450 K | 92 | 23 | Novel data | GSE67751 |
| 6 | PBMCs | HIV-infected men | 450 K | 109 | 109 | Novel data | GSE53840 |
| 7 | Whole blood | Healthy controls | 450 K | 335 | 0 | Liu et al [37] | GSE42861 |
| 8 | Blood cell types | 4 healthy subjects | 27 K | 28 | 0 | Accomando et al [39] | GSE39981 |
| 9 | Blood cell types | 6 healthy men | 450 K | 60 | 0 | Reinius et al [40] | GSE35069 |
| 10 | CD4+ T cells and monocytes | 23 healthy subjects | 27 K | 46 | 0 | Rakyan et al [10] | GSE20242 |
| 11 | Blood cell types | 6 healthy subjects | 450 K | 42 | 0 | Zilbauer et al [41] | E−ERAD−179 |
Abbreviations: HIV, human immunodeficiency virus; PBMC, peripheral blood mononuclear cell.
a Illumina data sets used in this article.
b Either Gene Expression Omnibus identifier or ArrayExpress identifier.
Novel Data Set 1: Various Brain Specimens From Cases and Controls
We generated Illumina Infinium 450 K data from 130 fresh frozen brain samples from 84 different subjects (71 cases and 13 controls). Specifically, we considered specimens from the cerebellum (20 cases and 4 controls), frontal lobe (2 cases and 4 controls), hippocampus (4 controls), medial frontal cortex (18 cases), occipital cortex (59 cases and 13 controls), and temporal cortex (4 controls). For the cases, the median year of death was 2005 (range, 1999–2013). Additional details can be found in Table Table2.2. In total, there were 99 samples from cases and 31 samples from controls of similar ages. DNA methylation data from cases and controls were generated at the same time and randomized across plates and chips. HIV load information was available for blood (in the last specimen obtained prior to death) and cerebrospinal fluid (CSF) specimens.
Table 2.
Clinical Characteristics of the Brain Data Set From the National NeuroAIDS Tissue Consortium
| Variable | Subjects, No. | Value |
|---|---|---|
| Age at death, y | 71 | 45.9 ± 9.1 |
| Infection duration, y | 65 | 13 ± 6.9 |
| Nadir CD4+ T-cell count, cells/mm3 | 71 | 27.7 ± 33.2 |
| Plasma viral load at diagnosis, log10 copies/mL | 70 | 3.75 ± 1.35 |
| CSF viral load, log10 copies/mL | 48 | 2.75 ± 1.37 |
| CD4+ T-cell count at death, cells/μL | 70 | 114 ± 141 |
| Global neurocognitive clinical ratinga | 71 | 5.3 ± 2 |
| HIV-related neurocognitive disorderb | 71 | |
| Taking combined antiretroviral therapy | 65 | |
| Neuropathological finding | 31 | |
 Alzheimer type 2 gliosis Alzheimer type 2 gliosis | 7 | |
 Aseptic leptomeningitis Aseptic leptomeningitis | 4 |  |
 Focal infarct Focal infarct | 4 | |
 Hemorrhage Hemorrhage | 2 |  |
 HIV encephalitis HIV encephalitis | 8 | |
 Hypoxic/ischemic damage Hypoxic/ischemic damage | 3 |  |
 Microglial nodule encephalitis Microglial nodule encephalitis | 9 | |
 Lymphoma Lymphoma | 1 | |
 Other noninfectious pathologies Other noninfectious pathologies | 4 |  |
 Other infections Other infections | 2 |  |
 Tuberculosis Tuberculosis | 1 |
The protein abundance levels of the IBA1 marker were assessed as follows: 5-µm-thick, formalin-fixed, paraffin-embedded brain sections were immunostained with mouse monoclonal antibody against Iba-1 (Wako), as previously described [35]. The DAB-stained sections were digitally scanned using a microscopy slide scanner (Aperio ScanScope GL, Leica, Vista, California) equipped with a 20× objective lens (yielding the resolution of 0.5 µm/pixel). Anatomical areas of interest were digitally delineated and the immunoreactivity signals quantified within each of these areas using the Image-Pro Analyzer software (version 6.3; MediaCybernetics, Bethesda, Maryland), as previously described in detail [36].
Novel Data Set 2: Frontal Lobe Specimens From Cases and Controls
Fresh frozen frontal lobe samples from 8 cases and 25 controls were evaluated. The 8 cases had a mean age of 44 years (range, 27–64 years). Years of death ranged from 2000 to 2013. Years of HIV diagnosis ranged from 1989 to 1996. The mean number of years living with HIV (until death) was 12 years (range, 1–19 years).
Novel Data Set 3: Cerebellum Specimens From Cases and Controls
Fresh frozen cerebellar samples from 8 cases and 12 controls were evaluated. The 8 cases correspond to the subjects used in data set 2.
Novel Data Set 4: Blood Specimens From Cases and Controls
Peripheral blood mononuclear cells (PBMCs) isolated from 92 subjects were evaluated. The 24 cases had a mean age of 49 years (range, 29–67 years). The cases were from the National Neurological AIDS Bank study or Multicenter AIDS Cohort Study in Los Angeles. Informed consent and all study procedures were approved by the University of California–Los Angeles Medical IRB. The 68 controls had a mean age of 36 years (range, 18–74 years). The controls were nonmedicated subjects from a previously published study (GSE41169) [19]. DNA methylation data from cases and controls were generated by the same core facility.
Novel Data Set 5: Blood Specimens From Cases and Controls
PBMCs were obtained from the National Neurological AIDS Bank study or Multicenter AIDS Cohort Study in Los Angeles. The 23 cases had a mean age of 45 years (range, 24–68 years). The 69 controls had a mean age of 51 years (range, 35–64 years).
Data Set 6: Adult Male Cases, Novel Data
PBMC samples were obtained from 109 adult male cases to study the relationship between viral load, blood cell counts, and epigenetic age acceleration. This data set did not contain any controls. Cases were from the National Neurological AIDS Bank study or the Multicenter AIDS Cohort Study in Los Angeles. The mean age was 52 years (range, 31–68 years). The following measured blood cell count data were available: mean CD8+ T-cell percentage, 46% (range, 19%–79%); mean CD4+ T cell percentage, 30% (range, 2%–54%); mean granulocyte percentage, 56% (range, 20%–77%); mean CD14+ monocyte percentage, 8.10% (range, 4%–24%).
Data Set 7: Whole-Blood Specimens From Controls in GSE42861
Here, we only used the 335 control samples from the report by Liu et al [37] (ie, from subjects without rheumatoid arthritis). The cell proportions for each control were estimated by Dr Yun Liu [37], using the Houseman method [38].
Data Set 8: Sorted Leukocytes From Individuals in GSE39981
The authors isolated PMBCs by magnetic-activated cell sorting (Miltenyi Biotec) and confirmed purity by fluorescence-activate cell sorting [39]. We focused here on data from 4 individuals for whom complete data on monocytes, neutrophils, B cells, pan T cells, CD4+ T cells, natural killer (NK) cells, and granulocytes were available.
Data Set 9: Blood Cell Type Data From Healthy Males in GSE35069
The authors analyzed sorted blood cells (CD4+ T cells, CD8+ T cells, CD56+ NK cells, CD19+ B cells, CD14+ monocytes, neutrophils, and eosinophils) from 6 healthy male blood donors with a mean age (±SD) of 38 ± 13.6 years [40].
Data Set 10: CD4+ T cells and CD14+ Monocytes From Healthy Subjects in GSE20242
The authors used the Illumina Infinium 27 K array to analyze CD4+ T cells and CD14+ monocytes from the same individuals [16]. We restricted the analysis to the 23 healthy subjects for whom both cell types were available.
Data Set 11: Sorted Blood Cells From 6 Healthy Volunteers in E−ERAD−179
The authors measured DNA methylation levels from whole PBMCs and cell subsets (CD4, CD8, CD14, CD19, and CD16) from 6 healthy volunteers [41].
DNA Methylation Age and the Epigenetic Clock
The epigenetic clock is defined as a prediction method of age, based on the linear combination of the DNA methylation levels of 353 CpG dinucleotides [25]. Predicted age, referred to as DNA methylation age, correlates with chronological age in sorted cell types (CD4+ T cells, monocytes, B cells, glial cells, and neurons) and tissues and organs, including whole blood, brain, breast, kidney, liver, lung, and saliva [25]. By construction, the epigenetic clock (and software) applies to data generated using either the Illumina 450 K or 27 K platform. Mathematical details and software tutorials for the epigenetic clock can be found in the additional files of the article by Horvath [25]. An online age calculator can be found at our website (available at: http://labs.genetics.ucla.edu/horvath/dnamage/).
Blood Cell Count Estimates
Flow cytometry measures of blood cell counts were assessed by the Multicenter AIDS Cohort Study in Los Angeles as described previously [42]. For data sets involving controls (eg, data set 7), blood cell proportions (CD8+ T cells, CD4+ T cells, NK cells, B cells, and granulocytes) were estimated using Houseman's estimation method [38], which is based on DNA methylation signatures from purified leukocyte samples. The percentage of exhausted CD8+ T cells (defined as CD28−CD45RA− B cells) and the number of naive CD8+ T cells (defined as CD45RA+CCR7+ B cells) were estimated using the advanced analysis option of the epigenetic clock software [25].
RESULTS
Brain Tissue Specimens From Cases and Controls
Using the first brain data set, we observed a strong correlation between DNA methylation age and chronological age in all brain regions (Figure (Figure11A–D). Regression lines through samples from cases only (Figure (Figure11A–C) and controls only (black lines) suggest that cases have a DNA methylation age that is older than that of controls.
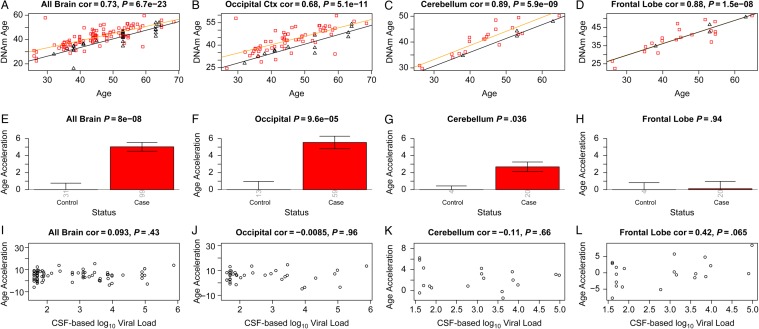
Discovery brain data from human immunodeficiency virus (HIV)–infected subjects (cases) and HIV-uninfected subjects (controls). A–D, DNA methylation (DNAm) age versus chronological age in all brain samples (A) and occipital cortex (B), cerebellum (C), and frontal lobe (D) samples. Red squares and black triangles in the scatterplot denote samples from cases and controls, respectively. The orange line depicts a linear regression line through case samples. Similarly, the black solid line corresponds to a regression line of DNAm age on chronological age in control samples. For each subject (point), age acceleration is defined as the vertical distance to the black regression line (ie, DNAm age minus the expected value based on control samples). E, Mean age acceleration versus control status in all brain samples. Each bar plot depicts 1 standard error around the mean value and reports the Kruskal–Wallis (nonparametric) group comparison test P value. Analogous results can be obtained when restricting the analysis to occipital cortex (B and F) or cerebellar (C and G) samples but not for samples from the frontal lobe (D and G). E–H, The group comparisons in panels E–H involved a total of 130, 72, 24, and 24, samples respectively. The number of samples in each group is reported by the rotated number under each bar. I–L, HIV load (log10 transformed) in cerebrospinal fluid (CSF) of cases versus age acceleration in all brain samples (I) and occipital cortex (J), cerebellum (K), and frontal lobe (L) samples. Abbreviation: cor, correlation.
To perform a formal statistical analysis, we defined a measure of epigenetic age acceleration as the difference between the observed DNA methylation age value and that predicted by a linear model in controls. A positive value indicates that the DNA methylation age is higher than that predicted from the linear model for controls of the same age. On average, brain samples from cases exhibited significant age acceleration effects (P = 8.0 × 10−8; Figure Figure11E), but these effects depended on brain region: significant age acceleration effects were observed in the occipital cortex (P = 9.6 × 10−5; Figure Figure11F) and the cerebellum (P = .036; Figure Figure11G) but not in the frontal lobe (Figure (Figure11H). We caution that differences in age acceleration effects may reflect low sample sizes or technical variability. But the difference between the cerebellum and frontal lobe was reproducible in independent data sets. Using brain data set 3, we observed again a significant age acceleration effect in the cerebellum (P = .031 [Figure [Figure22C]; 7.4 years [Supplementary 1]). But we did not observe an accelerated aging effect in the second frontal lobe data set (data set 2; Figure Figure22D), which is congruent with our findings in data set 1 (Figure (Figure11H). Future research will need to clarify why HIV infection seems to accelerate the age of some brain regions (eg, the occipital lobe and cerebellum) but not that of others (eg, the frontal lobe). To estimate the effect of age acceleration in terms of years, we used a linear regression model that regressed DNA methylation age on chronological age and viral load status (Supplementary 1). The analysis suggested that brain regions of cases were, on average, 7.4 years older than those of controls. The estimated acceleration effect was 9.3 years in the occipital cortex, 5 years in the cerebellum, and 0.1 years in the frontal lobe.
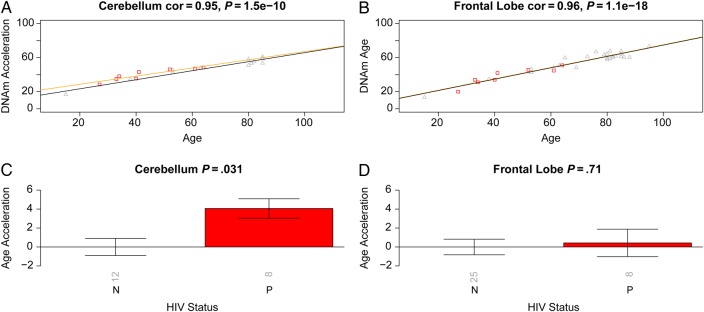
Validation brain data from human immunodeficiency virus (HIV)–infected subjects (cases) and HIV-uninfected subjects (controls). Analogous to Figure Figure1,1, we used independent data sets (data 2 and 3) to relate HIV status to epigenetic age acceleration in the frontal lobe and cerebellum of cases and controls. A and B, DNA methylation (DNAm) age versus chronological age in cerebellum (A) and frontal lobe (B) samples. Points (subjects) are colored by HIV status: cases correspond to red squares. The orange and black lines depict regression lines case and control samples, respectively. The measure of age acceleration is the same as used in Figure Figure1.1. C and D, Age acceleration versus HIV status in cerebellum (C) and frontal lobe (D) specimens. Each bar plot reports the Kruskal–Wallis (nonparametric) group comparison test P value and 1 standard error around the mean. Abbreviation: cor, correlation.
Overall, HIV load in the CSF of cases was not significantly correlated with age acceleration in the brain samples (Figure (Figure11I–K), but there was a marginally significant effect in the frontal lobe (r = 0.42, P = .065; Figure Figure11L). Since viral load was not ascertained in brain tissue, we could not correlate it with epigenetic age acceleration in the respective brain regions.
More-detailed examination is required to determine the cellular and histopathological correlates of age acceleration in HIV-infected brains. Table Table22 provides clinical information about the cases from whom autopsy brain samples were analyzed. Unfortunately, cause of death is not collected by the NNTC, although these data are often unreliable because of the multiple comorbidities of the participants. However, and more relevant, considering that we examined brain DNA, the NNTC conducts neuropathological examination according to a standard protocol. While the brain pathologies from HIV-infected individuals do not necessarily translate to specific diseases or clinical syndromes and are not uncommon in individuals with advanced HIV infection, these data indicated that, of the 31 cases with noted brain pathology, several had >1 type. All cases received a diagnosis ≤1 year prior to death indicating that they were neurocognitively normal or had an HIV-associated neurocognitive disorder (as reported elsewhere [33]). We also included in Table Table22 virologic and clinical variables. Note that in some instances this information was not collected. According to the autopsy results, 8 cases had HIV-associated encephalitis. Greater number of perivascular macrophages are found in the brains of patients with HIV encephalitis [43]. While we cannot rule out that the observed age acceleration effects reflect changes in cellular composition (eg, increased numbers of perivascular macrophages), we could not find a significant relationship between epigenetic age acceleration effects in brain samples from cases and a marker of activated macrophages, namely ionized calcium-binding adapter molecule 1 (IBA1), which is encoded by the gene allograft inflammatory factor 1. The IBA1 marker was assessed using immunohistochemical staining of paraffin embedded tissue, as described in “Methods” section.
Blood Specimens From Cases and Controls
Using 2 independent blood data sets, we observed that cases exhibited a significant age acceleration effect (P = .0048 [Figure [Figure33C] and P = .00036 [Figure [Figure33D]). A linear model analysis revealed that the DNA methylation age of cases was, on average, 5.2 years greater than that of controls (3.7 years in blood data set 4 and 6.7 years in blood data set 5; Supplementary 1). Since we analyzed whole blood (as opposed to sorted blood cells), we cannot rule out that the observed epigenetic age acceleration effects were mediated by changes in blood cell composition.
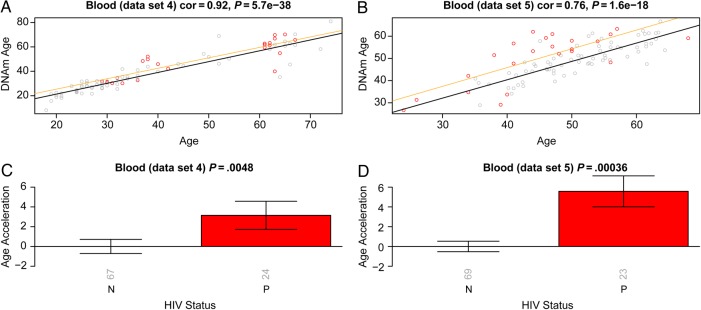
Epigenetic age in blood tissue versus human immunodeficiency virus (HIV) status. The first column (A and C) and second column (B and D) report findings for blood data sets 4 and 5, respectively. A and B, DNA methylation (DNAm) age versus chronological age. Samples from HIV-infected patients (cases; points) are colored in red. The black solid line and the orange line correspond to regression lines through HIV-uninfected control samples and case samples, respectively. For each subject (point), age acceleration is defined as the vertical distance to the black regression line. C and D, Mean age acceleration versus HIV status. The rotated numbers on the x-axis report the group sizes. The bar graphs report 1 standard error and a Kruskal–Wallis test P value. Abbreviation: cor, correlation.
Epigenetic Age Acceleration Versus Blood Cell Counts in Adult Male Cases
We used PBMCs from 109 adult male cases (data set 6) to study the relationship between epigenetic age, HIV load, and blood cell counts. DNA methylation age was closely related with chronological age in these cases (r = 0.80; median error, 3.9 years; Figure Figure44A). We defined a measure of epigenetic age acceleration as the residual from a linear regression line (Figure (Figure44A). A multivariate regression analysis suggested that cases with detectable viral load (ie, >35 copies/mL) had a DNA methylation age that was on average 3.6 years greater than that of adult male cases with a nondetectable viral load, but the association was only marginally significant (P = .033) and requires further validation studies. About 92% of cases were receiving antiretroviral therapy, but we were unable to assess who was adherent to their medications. Thus, detectable viral load could be due to a lack of adherence or to treatment failure. Epigenetic age acceleration correlated with several cell count measures in these cases, including NK cells (r = 0.33, P = 4 × 10−4; Figure Figure44B), monocytes (r = 0.2, P = .05; Figure Figure44C), granulocytes (r = −0.37, P = .00019; Figure Figure44D), CD4+ T cells (r = −0.23, P = .023; Figure Figure44E), CD8+ T cells (r = 0.24, P = .017; Figure Figure44F), exhausted (CD28−CD45RA−) CD8+ T cells (r = 0.36, P = 1.0 × 10−4; Figure Figure44H), and the number of naive (CD45RA+CCR7+) CD8+ T cells (r = −0.26, P = .0059; Figure Figure44G).
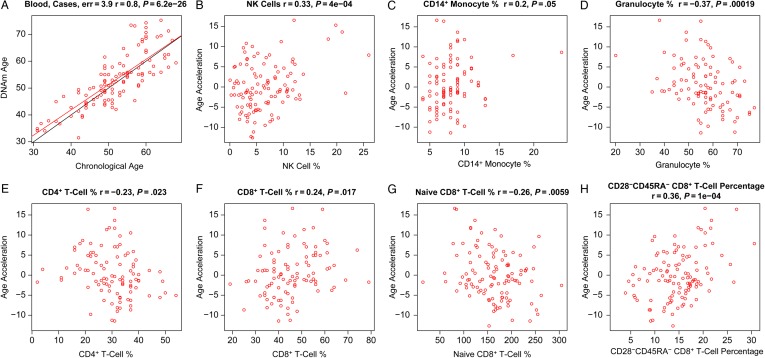
Age acceleration versus blood cell counts in human immunodeficiency virus (HIV)–infected subjects (cases). Here we used DNA methylation (DNAm) data from peripheral blood mononuclear cells from cases (data set 6). A, DNAm age versus age. The red line indicates the regression line. The black line corresponds to y = x. Epigenetic age acceleration (defined as the residual) versus the percentage of natural killer (NK) cells (B), CD14+ monocytes (C), granulocytes (D), CD4+ T cells (E), CD8+ T cells (F), naive CD8+ T cells (G), and exhausted (CD28−CD45RA−) CD8+ T cells (H).
Epigenetic Age Acceleration Versus Blood Cell Counts in Controls
In contrast to our findings for cases, epigenetic age acceleration exhibited weaker correlations with blood cell count measures in controls (Figure (Figure5).5). The highest correlation was observed for the percentage of exhausted CD8+ T cells in the controls from blood data set 5 (r = 0.29, P = .016; Figure Figure55H), but this finding was not validated in an independent data set (data set 7; Figure Figure55P). These findings are congruent with the fact that sorted blood cells appear to have similar DNA methylation ages (Supplementary 2). The relatively small sample sizes provided by data sets 8–11 did not provide sufficient statistical power for detecting significant differences in DNA methylation age between blood cells (B cells, CD4+ T cells, CD8+ T cells, eosinophils, granulocytes, monocytes, NK cells, and neutrophils) isolated from the same controls (Supplementary 2).
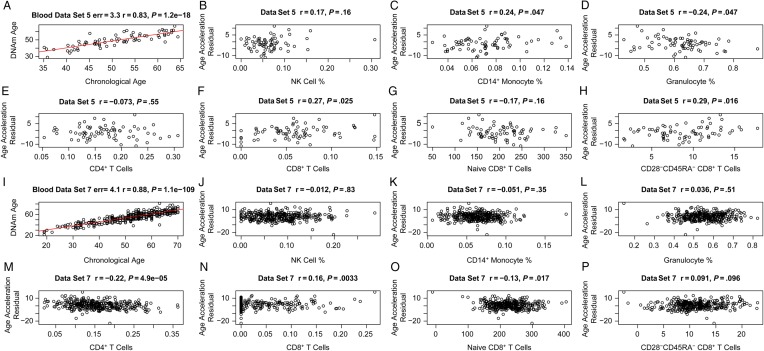
Age acceleration versus blood cell counts in HIV-uninfected control subjects (controls). The first 2 rows (A–H) and the last 2 rows (I–P) present results for controls from blood data sets 5 and 7, respectively. A and I, DNA methylation (DNAm) age versus chronological age. Age acceleration was defined as the residual resulting from the regression line (red line). Age acceleration versus natural killer (NK) cells (B and J), CD14+ monocytes (C and K), granulocytes (D and L), CD4+ T cells (E and M), CD8+ T cells (F and N), naive CD8+ T cells (G and O), and exhausted (CD28−CD45RA−) CD8+ T cells (H and P). The blood cell abundance measures were estimated on the basis of DNAm data (“Methods” section).
DISCUSSION
In the following, we discuss several models that could explain the relationship between HIV status and epigenetic age acceleration.
Model 1 assumes that changes in telomere length mediate the effect of HIV infection on epigenetic age acceleration (ie, HIV infection → telomere length → epigenetic age acceleration). This model is probably incorrect because (1) there seems to be at best a very weak correlation between leukocyte telomere length and epigenetic age acceleration [44], and (2) it is difficult to adapt this model to explain age acceleration effects in brain tissue.
Model 2 posits that the effect of HIV load on age acceleration is mediated by increases in the amount of senescent or exhausted T cells, such as CD28−CD45RA− CD8+ T cells (ie, HIV infection → exhausted/senescent T cells → age acceleration). Our blood data support this model to some extent: we found that the amount of exhausted CD8+ T cells correlated with epigenetic age acceleration in cases (Figure (Figure44H) and, to a lesser extent, in controls (Figure (Figure55H and and55P). But it is difficult to use this model for explaining accelerated aging effects in brain tissue, owing to the blood-brain barrier.
Model 3 is the independent model, in which HIV infection causes increases in exhausted T-cell counts and age acceleration independently (ie, exhausted/senescent T cells ← HIV infection → age acceleration). In other words, HIV infection confounds the relationship between the exhausted T-cell count and age acceleration. This is a plausible model, but it leaves us with the question of how HIV infection leads to epigenetic age acceleration. At this point, it is difficult to address this question since it is unknown what is being measured by DNA methylation age. Several lines of indirect evidence suggest that DNA methylation age might measure the cumulative work done by an epigenomic maintenance system [25]. In support of this hypothesis, HIV-1 infection is known to induce double-strand breaks of chromosomal DNA [45] and to induce chromosomal DNA damage responses by activating Rad3-related or ataxia-telangiectasia mutated proteins and by promoting phosphorylation of their downstream substrates [46, 47]. If one assumes that viral integration into the host genome leads to epigenomic instability (related to genetic instability), then the observed accelerated aging effects could reflect the protective actions of the epigenomic maintenance system.
Our study has several limitations, including the following. First, we analyzed relatively few controls in our brain data sets (n = 13 in data set 1) because it is difficult to obtain brain samples from relatively young, deceased subjects. Second, our observational study may be biased by hidden confounders that distinguish controls from cases. Third, we were not able to assess whether detectable viral load in cases was due to treatment failure or lack of adherence to the treatment. The weak association between age acceleration and viral load may reflect the fact that viral load does not capture the history of viral load burden. Controlled in vitro studies would be helpful for understanding the relationship between viral load and age acceleration.
In conclusion, we demonstrate that HIV infection is associated with a significant increase in DNA methylation age in brain and blood tissue. Our results are consistent with the reported clinical manifestations of accelerated aging effects among HIV-infected adults despite apparent viral control. Future studies will need to explore whether the epigenetic clock can serve as a useful tool for the study of and/or inform therapeutic strategies aimed at preventing HIV-associated non–AIDS-defining conditions such as cardiovascular disease and HIV-associated neurocognitive disorder [6, 48].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. S. H. conceived of the study and performed the statistical analysis. S. H. and A. J. L. wrote the article. A. J. L. provided the blood and brain data and assisted with the clinical interpretation.
S. H. conceived of the study and performed the statistical analysis. S. H. and A. J. L. wrote the article. A. J. L. provided the blood and brain data and assisted with the clinical interpretation.
Financial support. This work was supported by the National Institute on Aging, National Institutes of Health (NIH; grant 5R01AG042511-02 to S. H.); the University of California, Los Angeles (UCLA) AIDS Institute; the UCLA Center for AIDS Research (AI28697); the UCLA Clinical and Translational Science Institute; the National Center for Research Resources; the National Center for Advancing Translational Sciences (UL1TR000124); the National Institute for Drug Abuse, NIH (grant R01DA030913 to A. J. L. and S. H.); the National Institute of Allergy and Infectious Diseases, NIH (grant U01-AI-35040 to the Los Angeles site of the Multicenter AIDS Cohort Study [Roger Detels, principal investigator, UCLA]); and the National NeuroAIDS Tissue Consortium, which consists of the National Neurological AIDS Bank (U01-MH08021 and R24-NS38841; Singer), the Texas NeuroAIDS Research Center (U01-MH083507 and R24-NS45491; Benjamin Gelman, principal investigator, University of Texas Medical Branch), the Manhattan HIV Brain Bank (U01-MH083501 and R24-MH59724; Susan Morgello, principal investigator, Mt. Sinai Medical Center), and the California NeuroAIDS Tissue Network (U01-MH083506 and R24-MH59745; David Moore, principal investigator, UCSD).
This work was supported by the National Institute on Aging, National Institutes of Health (NIH; grant 5R01AG042511-02 to S. H.); the University of California, Los Angeles (UCLA) AIDS Institute; the UCLA Center for AIDS Research (AI28697); the UCLA Clinical and Translational Science Institute; the National Center for Research Resources; the National Center for Advancing Translational Sciences (UL1TR000124); the National Institute for Drug Abuse, NIH (grant R01DA030913 to A. J. L. and S. H.); the National Institute of Allergy and Infectious Diseases, NIH (grant U01-AI-35040 to the Los Angeles site of the Multicenter AIDS Cohort Study [Roger Detels, principal investigator, UCLA]); and the National NeuroAIDS Tissue Consortium, which consists of the National Neurological AIDS Bank (U01-MH08021 and R24-NS38841; Singer), the Texas NeuroAIDS Research Center (U01-MH083507 and R24-NS45491; Benjamin Gelman, principal investigator, University of Texas Medical Branch), the Manhattan HIV Brain Bank (U01-MH083501 and R24-MH59724; Susan Morgello, principal investigator, Mt. Sinai Medical Center), and the California NeuroAIDS Tissue Network (U01-MH083506 and R24-MH59745; David Moore, principal investigator, UCSD).
Potential conflict of interest. Both authors: No reported conflicts.
Both authors: No reported conflicts.
Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
Articles from The Journal of Infectious Diseases are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/infdis/jiv277
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/jid/article-pdf/212/10/1563/16866722/jiv277.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/infdis/jiv277
Article citations
Muscle Quality and Physical Function in Men With and Without HIV.
J Gerontol A Biol Sci Med Sci, 79(11):glae229, 01 Nov 2024
Cited by: 0 articles | PMID: 39288937
Epigenetic aging differentially impacts breast cancer risk by self-reported race.
PLoS One, 19(10):e0308174, 24 Oct 2024
Cited by: 0 articles | PMID: 39446903 | PMCID: PMC11500918
Immunoaging at Early Ages Could Drive a Higher Comorbidity Burden in People with HIV on Antiretroviral Therapy Compared with the Uninfected Population.
Int J Mol Sci, 25(20):10930, 11 Oct 2024
Cited by: 0 articles | PMID: 39456715 | PMCID: PMC11507496
Microglia and macrophages alterations in the CNS during acute SIV infection: A single-cell analysis in rhesus macaques.
PLoS Pathog, 20(9):e1012168, 16 Sep 2024
Cited by: 2 articles | PMID: 39283947 | PMCID: PMC11426456
A systematic review of phenotypic and epigenetic clocks used for aging and mortality quantification in humans.
Aging (Albany NY), 16(17):12414-12427, 30 Aug 2024
Cited by: 0 articles | PMID: 39215995 | PMCID: PMC11424583
Review Free full text in Europe PMC
Go to all (331) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders.
J Neurovirol, 22(3):366-375, 21 Dec 2015
Cited by: 74 articles | PMID: 26689571 | PMCID: PMC4900944
Perinatally acquired HIV infection accelerates epigenetic aging in South African adolescents.
AIDS, 32(11):1465-1474, 01 Jul 2018
Cited by: 53 articles | PMID: 29746298 | PMCID: PMC6026068
Accelerated epigenetic aging in Werner syndrome.
Aging (Albany NY), 9(4):1143-1152, 01 Apr 2017
Cited by: 100 articles | PMID: 28377537 | PMCID: PMC5425119
DNA Methylation as a Biomarker of Aging in Epidemiologic Studies.
Methods Mol Biol, 1856:219-231, 01 Jan 2018
Cited by: 14 articles | PMID: 30178254
Review
Funding
Funders who supported this work.
California NeuroAIDS Tissue Network (1)
Grant ID: U01-MH083506 and R24-MH59745
Manhattan HIV Brain Bank (1)
Grant ID: U01-MH083501 and R24-MH59724
NCATS NIH HHS (2)
Grant ID: UL1TR000124
Grant ID: UL1 TR000124
NIA NIH HHS (3)
Grant ID: R01 AG042511
Grant ID: 5R01AG042511-02
Grant ID: R21 AG046954
NIAID NIH HHS (5)
Grant ID: U01 AI035040
Grant ID: UM1 AI035043
Grant ID: P30 AI028697
Grant ID: U01-AI-35040
Grant ID: AI28697
NIDA NIH HHS (2)
Grant ID: R01DA030913
Grant ID: R01 DA030913
NIH (3)
Grant ID: R01DA030913
Grant ID: U01-AI-35040
Grant ID: 5R01AG042511-02
NIMH NIH HHS (14)
Grant ID: R24 MH059724
Grant ID: U01 MH083506
Grant ID: U24 MH100928
Grant ID: U01 MH083500
Grant ID: U01 MH083501
Grant ID: U01-MH083501
Grant ID: U01-MH083506
Grant ID: R24-MH59724
Grant ID: R24-MH59745
Grant ID: U24 MH100929
Grant ID: R24 MH059745
Grant ID: U01 MH083507
Grant ID: U01-MH08021
Grant ID: U01-MH083507
NINDS NIH HHS (4)
Grant ID: R24 NS045491
Grant ID: R24-NS38841
Grant ID: R24-NS45491
Grant ID: R24 NS038841
National Center for Advancing Translational Sciences (1)
Grant ID: UL1TR000124
National Institute on Aging
National Institutes of Health
National Neurological AIDS Bank (1)
Grant ID: U01-MH08021 and R24-NS38841;
Texas NeuroAIDS Research Center (1)
Grant ID: U01-MH083507 and R24-NS45491;
UCLA Center for AIDS Research (1)
Grant ID: AI28697





