Abstract
Free full text

CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function
SUMMARY
CTCF/cohesin play a central role in insulator function and higher-order chromatin organization of mammalian genomes. Recent studies identified a correlation between the orientation of CTCF-binding sites (CBSs) and chromatin loops. To test the functional significance of this observation, we combined CRISPR/Cas9-based genomic-DNA-fragment editing with chromosome-conformation-capture experiments to show that the location and relative orientations of CBSs determine the specificity of long-range chromatin looping in mammalian genomes, using protocadherin (Pcdh) and β-globin as model genes. Inversion of CBS elements within the Pcdh enhancer reconfigures the topology of chromatin loops between the distal enhancer and target promoters, and alters gene-expression patterns. Thus, although enhancers can function in an orientation-independent manner in reporter assays, in the native chromosome context the orientation of at least some enhancers carrying CBSs can determine both the architecture of topological chromatin domains and enhancer/promoter specificity. The findings reveal how 3D chromosome architecture can be encoded by genome sequence.
INTRODUCTION
Interphase chromosomes fold into highly compartmentalized, hierarchical structures, and the topology of chromosome folding is thought to play an important role in critical nuclear processes, including the regulation of gene expression (de Laat and Duboule, 2013; Gibcus and Dekker, 2013; Levine et al., 2014). Individual chromosomes occupy a distinct space in the nucleus, referred to as a “chromosome territory” (Cremer and Cremer, 2001), and within this region are relatively stable chromatin domains containing specific DNA-looping interactions between proximal promoters and distal regulatory DNA elements, such as transcriptional enhancers and silencers, insulators and locus control regions (LCR) (Dixon et al., 2012; Gibcus and Dekker, 2013; Levine et al., 2014; Lieberman-Aiden et al., 2009). Genome-wide studies of mammalian genomes have shown that there are far more enhancers than promoters and that spatiotemporal gene expression is regulated through one or more promoters and multiple enhancers (Bulger and Groudine, 2011; ENCODE Project Consortium, 2012; Zhang et al., 2004). Insulator elements play pivotal roles in orchestrating proper long-range DNA-looping interactions between remote enhancers and their cognate promoters, via mechanisms that are poorly understood (Dowen et al., 2014; Jia et al., 2014; Narendra et al., 2015; Ong and Corces, 2014).
The mammalian CCCTC-binding factor (CTCF), a zinc-finger DNA-binding protein, is the best characterized insulator-binding protein, which also plays a key role in genome looping (Lobanenkov et al., 1990; Ong and Corces, 2014). In addition, the insulator activity of CTCF-binding sites (CBSs) requires the cohesin complex that is recruited by CTCF. Previous studies have implicated CTCF and cohesin complexes in genome-wide chromatin-looping interactions (Handoko et al., 2011; Zuin et al., 2014). Over 100,000 diverse CBSs have been identified in mammalian genomes (Kim et al., 2007; Shen et al., 2012; Xie et al., 2007), and the genome-wide pattern of CTCF occupancy is cell-type specific (Kim et al., 2007; Shen et al., 2012; Wang et al., 2012); however, CBSs are enriched at constitutive boundaries of topologically associated domains (TADs) (Dixon et al., 2012; Gibcus and Dekker, 2013; Zuin et al., 2014). More recently, it was shown that CBSs at the anchors of chromatin loops are arranged in the forward-reverse orientations, suggesting that the relative positions and orientations of CBSs could be important for chromosome architecture (Alt et al., 2013; Guo et al., 2012; Monahan et al., 2012; Rao et al., 2014; Vietri Rudan et al., 2015). However, the underlying molecular mechanisms, through which CTCF-mediated DNA-looping interactions lead to CTCF’s many cellular functions, remain obscure.
The mammalian protocadherin (Pcdh) α, β and γ gene clusters provide a unique model system to investigate the role of CTCF/cohesin-mediated enhancer-promoter interactions in cell-specific gene expression (Guo et al., 2012; Hirayama et al., 2012; Monahan et al., 2012; Wu and Maniatis, 1999). In the α and γ (but not the β) clusters, the Pcdh “variable regions” contain more than a dozen large and highly-similar “alternately expressed” variable exons followed by two or three “ubiquitously expressed” C-type variable exons in the α and γ clusters, respectively (Figure 1A). By contrast, the downstream “constant regions” of the α and γ clusters are organized into three small exons that encode the intracellular domain of all of the protein isoforms in each cluster (Figure 1A) (Wu and Maniatis, 1999). Previous studies revealed that each “variable” exon (except αc2, β1, γc4 and γc5) is preceded by a promoter containing a highly conserved sequence element (CSE) (Figure 1A) (Tasic et al., 2002; Wu et al., 2001). Subsequently, CTCF was shown to bind to the CSE and to a second CBS within the downstream exon (eCBS) of transcriptionally active α genes, and this binding is required for transcription (Guo et al., 2012; Hirayama et al., 2012; Monahan et al., 2012; Wu et al., 2001).
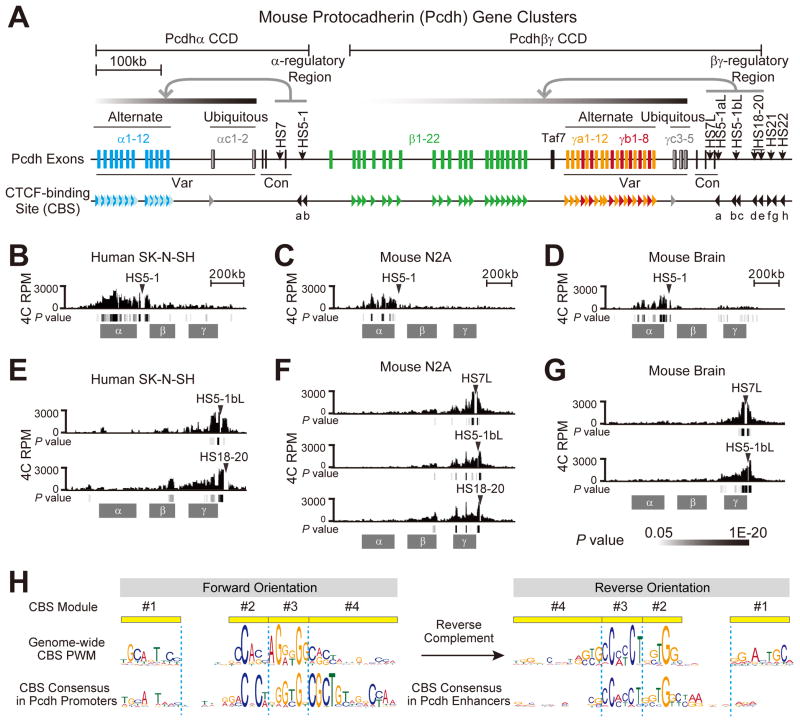
(A) Diagram showing the Pcdh α and βγ CCDs in the three mouse Pcdh gene clusters. The variable (Var) and constant (Con) exons are also indicated. The CBSs and their orientations are indicated as arrowheads. Different types of Pcdh CBSs are represented by differently colored arrowheads. The dark and light blue CBSs represents the CSE and eCBS, respectively, for each of the twelve “alternate promoters” (α1-α12) of the α cluster. The 21 tandem green arrowheads represent the CSE for each member of the β cluster (except β1). The yellow and red arrowheads represent CSEs for γa and γb, respectively. The two gray arrowheads represent the C-type CSEs (αc1 and γc3). The two CBS sites (a, b) downstream of the α cluster and the eight CBS sites (a–h) downstream of the γ cluster are indicated in black arrowheads. The DNaseI hypersensitive sites (HS) in the α and βγ regulatory regions are also shown.
(B–G) Relative distributions of the 4C reads per million (RPM) obtained in human SK-N-SH cells (B), mouse N2A cells (C) and brain tissues (D) using the HS5-1 enhancer as an anchor. 4C interaction profiles in human SK-N-SH cells (E), mouse N2A cells (F) and brain tissues (G) with the regulatory region downstream of the γ cluster as an anchor are also shown. The significance of interactions (P value) is shown under the reads density for each panel.
(H) Showing the forward orientation of CBS sites in Pcdh promoters and reverse orientation of CBS sites in Pcdh enhancers.
See also Figure S1.
A key observation, relevant to the present study, was that the CBSs in the Pcdh HS5-1 enhancers downstream of the α cluster and in each of the promoters and downstream exons are configured in opposite orientations (Guo et al., 2012). Chromosome Conformation Capture (3C) studies revealed that CTCF/cohesin-mediated DNA looping occurs exclusively between paired CBSs within the enhancers and the active promoters (Guo et al., 2012). This organization of CBS sites within the Pcdh clusters was recently shown to reflect the genome-wide organization of CBS pairs at anchors of DNA contact loops (Alt et al., 2013; Rao et al., 2014; Vietri Rudan et al., 2015). This striking organization of oriented CBS sites in Pcdh clusters and the availability of powerful CRISPR genome editing methods provide the opportunity to address the functional significance of the genome-wide CBS organization.
Here, we provide direct functional evidence that the location and relative orientations of CBSs play a critical role in the establishment of chromosome architecture and proper enhancer-promoter interactions. We developed a CRISPR/Cas9-based DNA-fragment in-situ inversion technology (Li et al., 2015), and in conjunction with 3C as well as related 4C (Circularized 3C) and Hi-C methods (Dekker et al., 2002), to study the chromatin organization in the Pcdh clusters. We find that directional CTCF binding to the paired CBSs with a specific combination of forward-reverse orientations determines the formation of specific DNA-looping interactions between enhancers and promoters in mammalian cells. The generality of this observation is demonstrated by showing that the same mechanism operates with CBSs in the β-globin gene cluster and throughout mammalian genomes. This mechanism of CTCF-determined looping directions has important implications regarding chromosomal architecture and the insulator functions of genome-wide CBSs in genome folding and gene regulation.
RESULTS
Two CTCF/cohesin-mediated Chromatin Domains (CCDs) in the Pcdh Locus
We used 3C, 4C and Hi-C to study CTCF/cohesin-mediated DNA looping and chromatin organization in the Pcdh α, β and γ clusters (Figures 1A–1G and S1). Specifically, we performed 4C using the HS5-1 enhancer, the α promoters, or the region immediately upstream of the α cluster as anchors, and showed that promoters within the α cluster interact with HS5-1 in human SK-N-SH cells (Figure 1B) and mouse neuro2A (N2A) cells (Figure 1C) and brain tissues (Figures 1D, S1A and S1B). By contrast, the downstream promoters of the β cluster display virtually no interactions with HS5-1 (Figures 1B–1D).
We also performed 4C using promoters of the γ cluster as anchors, and identified a downstream regulatory region (Figures S1C–S1E). This region contains a cluster of CBSs (CBS sites a–h) located within several DNaseI HS sites (see Figures 1A and S1F) and is enriched with molecular marks typical of enhancers (Figure S1G) (ENCODE Project Consortium, 2012). Similar to the α cluster, this downstream regulatory region interacts with promoters of the γ cluster in human SK-N-SH cells (Figure 1E) as well as in mouse N2A cells (Figure 1F) and brain tissues (Figure 1G). Interestingly, promoters of the β cluster also interacted with this remote enhancer when the β promoters were used as 4C anchors (Figures S1H and S1I), consistent with the previous observation that this region is required for maximum levels of β gene expression, and regulates the γ cluster (Yokota et al., 2011). Finally, we performed Hi-C experiments on SK-N-SH, and analyzed the results along with previously published Hi-C data from H1-hESC and NPC cells (Dixon et al., 2015; Dixon et al., 2012) (Figure S1J). We observed two TAD-like chromatin domains covering α and βγ clusters, respectively, by calculating a Directionality Index with a sliding window of 300 kb (Figure S1J). Taken together, these data show that the HS5-1 enhancer forms a CTCF/cohesin-mediated chromatin domain (CCD) within the α cluster, and the βγ-regulatory region forms a CCD that includes both the β and γ clusters (Figure 1A).
Non-random CBS Orientations in the Two Pcdh CCDs
A CBS motif is located within all of the Pcdh α, β, and γ promoters, except αc2, β1, γc4, γc5 (Figure 1A) (Guo et al., 2012; Kim et al., 2007; Monahan et al., 2012; Nakahashi et al., 2013; Rhee and Pugh, 2011; Schmidt et al., 2012; Wu et al., 2001). We defined the CBS from modules 1 to 4 as being in the forward orientation (Figure 1H). Interestingly, all of the α CSEs and eCBSs are in the forward orientation; by contrast, both HS5-1 CBSs (HS5-1a, b) are in the reverse orientation within the α CCD (Figure 1A). Similarly, all of the βγ CSEs are in the forward orientation, whereas the first 5 CBSs (a–e) in the βγ enhancer complex are in the reverse orientation within the βγ CCD (Figures 1A and S1F). The last three CBSs (f–h) downstream of the βγ-regulatory region are in different orientations, and they do not interact with the βγ promoters (see Figures 1A and S1C–S1F). Thus, the Pcdh chromatin-looping interactions occur between CBS pairs in the forward-reverse orientations in the promoters and enhancers, respectively (Figure 1A). Previously reported weak DNA-looping interactions between two CBSs in the same orientation in the α promoter region may be the consequence of their interactions with common CBSs within the HS5-1 enhancer in the opposite orientation (Guo et al., 2012). Overall, these observations strongly suggest that the relative orientations of CBSs determine the topology of CTCF/cohesin-mediated DNA looping between Pcdh enhancers and promoters (Alt et al., 2013; Guo et al., 2012; Rao et al., 2014; Vietri Rudan et al., 2015).
In-situ Inversion of the Boundary CBS Element Alters DNA Looping and Gene Expression
To directly determine whether CBS orientation is important for enhancer-promoter interactions and DNA looping, we used an efficient in-situ CRISPR inversion of DNA fragment editing method we recently developed (Li et al., 2015) to invert the core HS5-1 element in its endogenous chromosomal location. We screened for CRISPR inversion cell clones derived from HEC-1B cells, which have three alleles at the Pcdh locus (Li et al., 2015) and express a subset of the α (Tasic et al., 2002) and γ clusters (Figure S1F, also see Figure 2D below). Out of 32 clones that were genotyped, we identified a cell clone (V28) in which the orientation of HS5-1 was inverted for two alleles and deleted for one allele (Figure S2A). We then performed 4C using HS5-1 as an anchor. Strikingly, we observed a significant increase in DNA-looping interactions between HS5-1 and promoters in the βγ clusters (from 28% to 79%) and a corresponding decrease in DNA-looping interactions with the promoters driving the expression of the alternate Pcdhα isoforms (from 72% to 21%) (Figure 2A). We confirmed these changes in DNA looping by quantitative 3C assays (Figure 2B). ChIP-qPCR studies showed that CTCF binds to the inverted HS5-1 element; however, a significant decrease in the binding of the cohesin subunit Rad21 to this sequence was observed (Figure 2C). We conclude that inversion of the oriented CBS’s in the HS5-1 enhancer profoundly alters enhancer-promoter interactions in the Pcdh clusters.
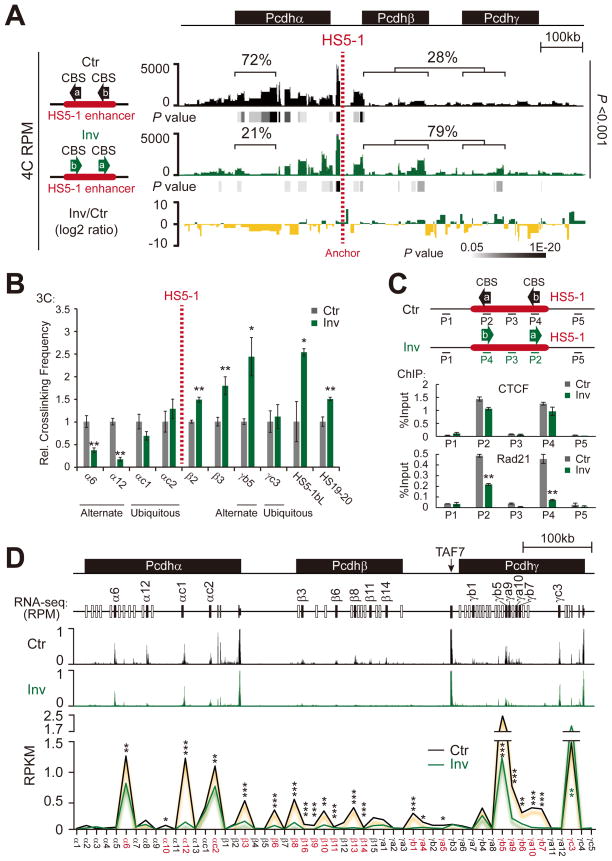
(A) Long-range chromatin-looping interaction profiles of the HS5-1 anchor in wild-type control (Ctr) or in a HS5-1 inversion (Inv) cell line generated from subcloned HEC-1B cells by CRISPR engineering. The log2 ratio between inversion and control is also shown.
(B) The relative crosslinking frequency measured by quantitative 3C assays in the control or inversion cell lines with HS5-1 as an anchor (HS5-1 is within the same 3C restriction fragment in the genomes of both Ctr and Inv cell lines). Data are means ± SEM (n=4). *P < 0.05 and **P < 0.01.
(C) Control experiments showing functional CTCF/cohesin binding after inversion. **P < 0.01.
(D) RNA-Seq experiments showing expression reduction of the α, β, and γ clusters (except γc3) after inversion. *P < 0.05, **P < 0.01, and ***P < 0.001.
See also Figure S2.
To assess the effects of these alterations we carried out an RNA-Seq analysis on the HEC-1B cells in which the HS5-1 enhancer is inverted. As shown in Figure 2D the decrease of DNA looping between HS5-1 and α promoters resulted in a significant reduction in α transcription. However, a corresponding enhancement in β transcription was not observed in spite of the observed increase of interactions between the inverted HS5-1 enhancer and the β cluster. Similarly, a reduction of γ transcription (except an increase of γc3) was also observed (Figure 2D). Thus, the inappropriate engagement of the HS5-1 enhancer with the downstream β and γ clusters appears to disrupt, rather than enhance transcription.
The function of enhancers tested in mammalian cell transfection experiments with reporter genes is independent of the relative orientations of the enhancer and promoter (Banerji et al., 1981). However, the data of Figures 1 and and22 clearly show that the activity and specificity of enhancers in their normal chromosomal context are highly orientation-specific, likely as a consequence of differences in the altered organization of CCDs caused by the DNA sequence inversion.
Directional CTCF Binding to Pcdh CBS Sequences
A large number of palindromic CBSs have been identified in the human genome (Xie et al., 2007) and yet, intriguingly, CTCF binds to CBSs in a preferred orientation (Nakahashi et al., 2013; Renda et al., 2007; Schmidt et al., 2012). How CTCF binds directionally to large numbers of diverse and seemingly palindromic CBSs therefore remains a mystery. Careful examination of the 17-bp core sequences of the HS5-1b CBS revealed that they are perfectly palindromic (Figure 3A). Considering that the reverse-complement sequences also conform to the CTCF-binding consensus, one would expect that CTCF recognizes HS5-1b in both directions, thus eliminating the apparent asymmetry of CBS pairs in the α promoters and the HS5-1 enhancer. To investigate whether CTCF binding to the HS5-1b CBS is directional, we generated three DNA probes bearing combined 2-bp mutations designed to distinguish between the two putative CTCF-binding directions (Figures 3A and 3B). We also generated a series of 17 CTCF expression constructs encoding two sets of truncated CTCFs in which each zinc finger (ZF) domain was sequentially deleted from either the C- or N-terminus (Figures 3C and S3A).
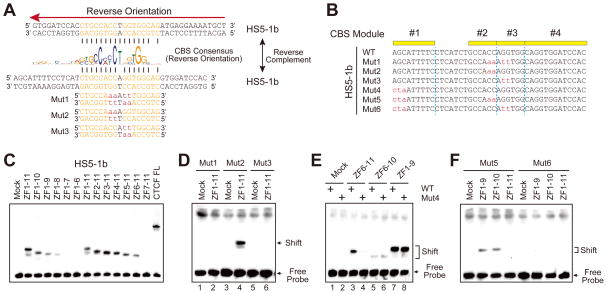
(A) Showing the HS5-1b CBS sequence (double-stranded) of the reverse orientation (indicated above by a red arrow) with the palindromic core highlighted. The double-stranded reverse complement HS5-1b CBS sequences (along with three probes with core sequences mutated) are also shown below the CBS consensus. The nucleotides that match to the CBS consensus are indicated by vertical lines. Note that mut2 and mut3 are exactly the same for the palindrome core sequence.
(B) The wild-type (WT) or mutant (Mut) sequences of HS5-1b probes (shown in the reverse complement).
(C) Gel-shift assays of the wild-type HS5-1b probe using a set of recombinant CTCF proteins with sequentially-deleted zinc-finger domains.
(D–F) Gel-shift assays using recombinant CTCF proteins with probes of Mut1-3 (D), Mut4 (E), Mut5 and Mut6 (F).
See also Figure S3.
Remarkably, electrophoretic mobility shift assay (EMSA) experiments revealed that CTCF recognizes palindromic HS5-1b in only one direction relative to its sequences, because mutation of “GG” to “tt” (mut1 and mut3, Figures 3A and 3B) abolished CTCF binding (lanes 2 and 6, Figure 3D) whereas mutation of “CC” to “aa” (mut2, Figures 3A and 3B) did not abolish CTCF binding (lane 4, Figure 3D). To further investigate the directional CTCF recognition, we generated combinations of these mutations with 3-bp mutations in the HS5-1b module1 (mut5 and mut6 with mut4 as the control, Figure 3B). We found that the first three nucleotides of module1 are recognized by the C-terminal ZF11, and this recognition determines the direction of CTCF binding to the CBS with palindromic core sequences (Figures 3E and 3F). In particular, introduction of mutations into the first tri-nucleotide from “AGC” to “cta” did not alter the binding to ZF6-10 (compare lanes 6 and 5 in Figure 3E), but did reduce the binding of ZF6-11 to levels similar to those of ZF6-10 (compare lanes 4 and 3 with lanes 5 and 6 in Figure 3E). Thus, the C-terminal ZF11 of CTCF determines its directional binding to the HS5-1b CBS with palindromic core sequences, suggesting that module1 is the key directional element in CBSs with palindromic core sequences.
To further determine the directionality of CTCF binding at the Pcdh CBS repertoire, and the recognition profile of the 11 ZF domains of CTCF, we mutated distinct sets of 3-bp sequences in modules 1, 2, or 4 of a large set of Pcdh CBSs (Figure S3B). We found that the C-terminal ZF domains of CTCF recognize module1 of the CBS, and that the N-terminal ZF domains recognize module4 (Figures S3C–S3W). For example, CTCF ZF3 and ZF2 recognize the CGC and TGT tri-nucleotides of the α8 CBS, respectively, because mutations of these tri-nucleotides reduced CTCF binding only when ZF3 and ZF2 were present (Figures S3B–S3F). In addition, module2 of the β3 CBS appears to be bound by CTCF ZF6/7 (Figures S3B, S3G–S3I). Moreover, CTCF ZF2-11 and ZF4-11 are essential for binding the CSE of γa10 (Figures S3B, S3J, S3K and S3N) and γb7 (Figures S3B, S3L and S3N), respectively. In particular, ZF11 of CTCF is absolutely required for CTCF binding of the CSE of γa10 and γb7, as deletion of ZF11 abolished CTCF binding to these two CBSs (Figure S3J and S3L). Furthermore, CTCF ZF11 recognizes the first tri-nucleotide TGC in module1 of the βγ-b CBS (Figures S3B, S3M and S3N). Finally, we show that different types of Pcdh CBSs are recognized by distinct combinations of the CTCF ZF domains (Figures S3O–S3W).
Taken together, these observations clearly show that CTCF recognizes CBSs in only one direction relative to its target sequences, and that distinct combinations of CTCF ZF domains recognize different types of Pcdh CBSs. Thus, the configuration of directional CTCF binding determines the topology of CTCF/cohesin-mediated DNA looping in the Pcdh clusters. Although the nature of the interactions between CTCF/cohesin complexes on the active Pcdh alternate promoters and the HS5-1 enhancer is not known, these observations suggest that functional interactions require directional binding of CTCF/cohesin in the forward-reverse orientations to the Pcdh CBS pairs.
Directional CTCF Binding in Genome-wide DNA Looping Specificity
Recent whole genome Hi-C experiments revealed that the vast majority of DNA loops correlate with the presence of CBS pairs arranged in a convergent orientation (Rao et al., 2014; Vietri Rudan et al., 2015). However, because chromatin contacts detected by Hi-C are unbiased, and do not specifically relate to CTCF/cohesin binding, these loops may or may not be established by CTCF and the associated cohesin complex. Based on our observations that directional CTCF binding to forward-reverse CBS pairs determines topological looping domains in the Pcdh clusters, we investigated genome-wide CBS orientation and CTCF/cohesin-mediated DNA-looping topology by analyzing published datasets of ChIA-PET and ChIP-seq with specific CTCF/cohesin antibodies (ENCODE Project Consortium, 2012; Handoko et al., 2011). We first determined the orientations of 88,332 CBSs and their CTCF occupancies in K562 cells (Table S1) using position weight matrices (PWM) (Schmidt et al., 2012). We then screened for ChIA-PET interactions (from a total of 24,887) in which both tethered DNA fragments contain CBSs, and identified 19,532 such interactions (Figure 4A and Table S2). We found that 76.4% of the CTCF-mediated interactions (14,928) are in the forward-reverse orientations; by contrast, only 2.3% (443) are in the reverse-forward orientations. In addition, 11.0% of the interactions (2,155) are in the forward-forward orientations and 10.3% (2,006) are in the reverse-reverse orientations. Finally, we measured the chromatin-looping strength by counting the number of overlapped looping PETs of the ChIA-PET datasets. Interestingly, the percentages of CBS pairs in the forward-reverse orientations dramatically increased with enhanced chromatin-looping strength (Figure 4B and Table S3). Similar results were observed in data collected from mouse E14 embryonic stem cells (Table S2) (Handoko et al., 2011) and human MCF-7 breast cancer cells (Tables S1 and S2). These observations clearly show that the majority of genome-wide chromatin-looping interactions correlate with directional CTCF binding to CBS pairs in the forward-reverse orientations.
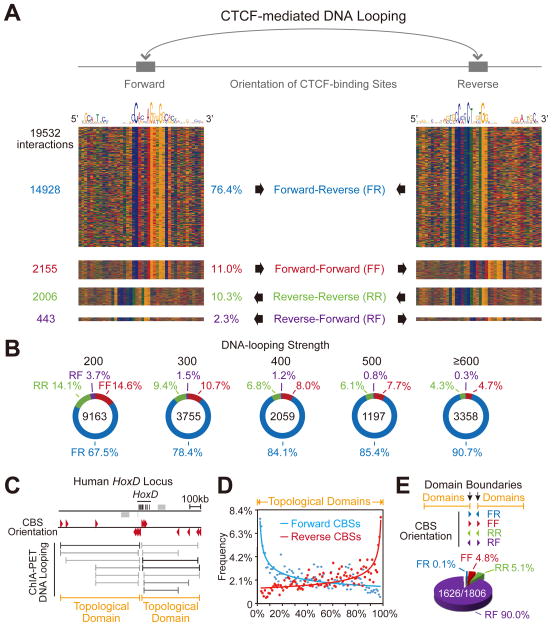
(A) Diagram of CTCF-mediated long-range chromatin-looping interactions between CBS pairs in the forward-reverse orientations. The color charts represent 19,532 interactions of CBS pairs in K562 cells. The number and percentage of CBS pairs in the forward-reverse (FR), forward-forward (FF), reverse-reverse (RR), and reverse-forward (RF) orientations are shown.
(B) The percentage of CBS pairs in the forward-reverse orientations increases from 67.5% to 90.7% as the chromatin-looping strength is enhanced.
(C) Schematic of the two topological domains in the HoxD locus. The orientations of CBSs are indicated by arrowheads. CTCF/cohesin-mediated looping interactions and the two resulting topological domains are also shown.
(D) Cumulative patterns of CBS orientations of topological domains in the human genome.
(E) Distribution of genome-wide orientation configurations of CBS pairs located in the boundaries between two neighboring domains in the human K562 genome. Note that the vast majority (90.0%) of boundary CBS pairs between two neighboring domains are in the reverse-forward orientation.
We previously demonstrated that CTCF and the cohesin complex colocalize to promoters and enhancers in the Pcdh clusters (Monahan et al., 2012; Guo et al., 2012). In order to investigate the relationship between the binding of CTCF and cohesin, CBS orientation, and DNA looping, we identified chromatin-looping interactions containing CTCF/cohesin co-occupied CBSs in K562 cells. We found 16,610 such interactions, in which the majority (78.7%) occur between CBS pairs in the forward-reverse orientations (Table S4). In addition, we found that the tethered CBSs have a higher occupancy of CTCF and cohesin than the non-tethered CBSs (Figure S4A), suggesting that high levels of CTCF/cohesin co-occupancy at CBSs are required for establishing these long-range chromatin-looping interactions. Thus, in addition to the location and orientation of CBSs, levels of their CTCF/cohesin occupancy are also an important determinant for directional chromatin looping.
Interestingly, in the K562 cell genome, 46% of the p300 enhancer marks (Heintzman et al., 2007) have at least one CBS located within 2 kb (Figure S4B). On the other hand, 54% of the marks of the silencer factor REST/NRSF (Johnson et al., 2007) have at least one CBS located within 2 kb (Figure S4C). These observations suggest that CTCF/cohesin-mediated DNA-looping interaction may enhance or inhibit gene expression, depending on its proximity to p300 or REST/NRSF. This possibility is consistent with the observation that a REST/NRSF binding site in HS5-1 is required for repression of the α cluster in non-neuronal cells (Kehayova et al., 2011).
We next identified genome-wide overlapping CTCF/cohesin-mediated chromatin-looping interactions, and merged clusters of the overlapping interactions as single CCDs. The two CCDs in the HoxD locus are shown as examples in Figure 4C. The cumulative features of CBSs in the looping PETs of all human CCDs demonstrate that most CBSs are located near the boundaries (Figure 4D and Tables S4 and S5). By analyzing the orientations of the boundary CBS pairs between neighboring CCDs, we found that the vast majority (90.0%) of the boundary CBS pairs between neighboring CCDs in K562 cells are in the reverse-forward orientations (1,626) (Figure 4E and Tables S4 and S6). Similar results were obtained for MCF-7 cells (Tables S4, S5 and S6). Taken together, these genome-wide data suggest that directional CTCF binding to CBS pairs in the reverse-forward orientations at the boundary between neighboring CCDs is important for establishing distinct topological domains.
Finally, because CBSs are enriched at the boundaries of TADs (Dixon et al., 2012; Shen et al., 2012), we analyzed the orientations of CBSs of the TAD boundaries identified in H1-hESC and IMR90 cells. We found that CBS pairs in the reverse-forward orientation exist in >60% neighboring TAD boundaries (Figure S4D), suggesting that the boundary reverse-forward CBS pairs play an important role in the formation of most of TADs. For example, there is a CBS pair in the reverse-forward orientation in a Chr12 genomic region of H1-hESC cells, located at or very close to each of the six TAD boundaries (boundaries 1–6), except for boundary5, which has only one closely-located CBS in the forward orientation (Figure S4E). These data, taken together, strongly suggest that directional binding of CTCF to boundary CBS pairs in the reverse-forward orientations causes opposite topological looping, and thus appears to function as insulators.
The Human β-globin Locus Provides an Additional Example of CBS Orientation-dependent Topological Chromatin Looping
Based on the location and orientation of CBSs as well as their CTCF/cohesin occupancy, we identified four CCDs (domains 1–4) in the well-characterized β-globin cluster (Figure 5A). The β-globin gene cluster is located between CBS3(5′HS5) and CBS4(3′HS1) in domain1 (Figure 5A) (Hou et al., 2010; Splinter et al., 2006). We generated a series of CBS4/5 mutant K562 cell lines using CRISPR/Cas9 with one or two sgRNAs (Li et al. 2015) (Figures S2B and S2C). In the CRISPR cell lines D3, D7 and D19 (out of 38 clones screened) in which the internal CBS4(3′HS1) was deleted (Figure S2B), chromatin-looping interactions between CBS3(5′HS5) in the forward orientation and the boundary CBS5 in the reverse orientation in domain1 persisted, although its interaction with the CBS4(3′HS1) region was abolished (Figures S5A and S5B). As expected, the interactions between CBS6/7 and CBS8/9 in domain2 were unchanged (Figure S5C). Strikingly, however, in the CBS4(3′HS1) and CBS5 double-knockout CRISPR cell lines C2, C4 and C14 (out of 49 clones screened) (Figure S2C), novel chromatin-looping interactions between CBS3(5′HS5) in the forward orientation of domain1 and CBS8/9 in the reverse orientation of the neighboring domain2 were observed, suggesting that these two domains merge as a single domain in CRISPR cell lines with CBS4/5 double-knockout (Figure S5B). Similarly, when CBS8 was used as an anchor, this reverse-oriented CBS in domain2 establishes new long-range chromatin-looping interactions with CBS1-3 in the forward orientation of domain1 in the CBS4/5 double-deletion CRISPR cell lines (Figure S5C). We conclude that cross-domain interactions can be established after deletion of CBSs up to the boundary of topological domains, but not after deletion of the internal CBS in the β-globin locus.
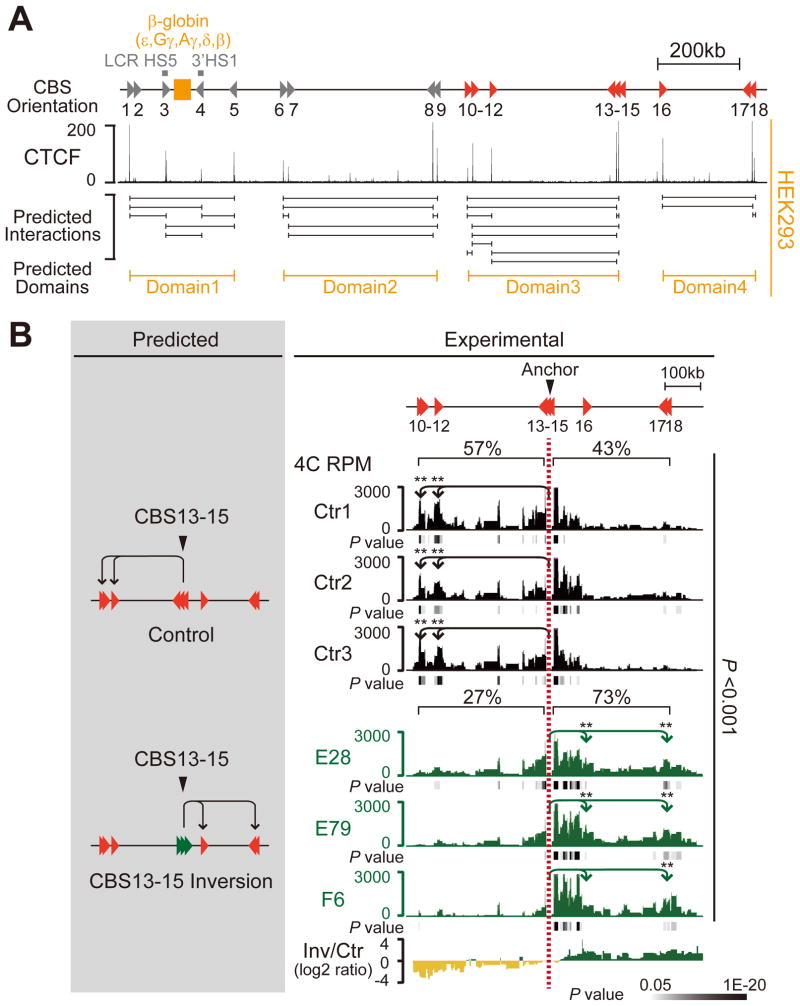
(A) Diagram of the human β-globin region. Predicted looping interactions and topological domains are shown, based on CTCF occupancy in HEK293 cells.
(B) The predicted interactions (left) and the altered looping directions (right) in the three subcloned CRISPR cell lines with inversion of CBS13-15 (E28, E79 and F6) are confirmed by 4C with CBS13-15 as an anchor. The looping interactions of three mock controls are also shown. The average log2 ratios of interactions between inversions and controls are also indicated. **P < 0.01.
See also Figures S2 and S5.
To further test the functional significance of this organization of CBSs, we again performed CRISPR/cas9-mediated DNA-fragment editing in the HEK293T cells and screened 198 CRISPR cell clones for inversions of CBS13-15, which is located at the boundary of domain3 (Figure 5A), and obtained three CRISPR inversion cell clones (E28, E79 and F6) (Figure S2D). We then performed 4C using CBS13-15 as an anchor. Strikingly and similar to the inversion of the Pcdh domain boundary, we observed a significant increase of chromatin-looping interactions with the downstream domain containing CBS16-18 (from 43% to 73%) and a corresponding decrease of chromatin-looping interactions with the upstream domain containing CBS10-15 (from 57% to 27%) (Figure 5B). These observations, taken together, clearly show that the orientations of CBSs determine the directionality of topological DNA looping.
In summary, we find that changing the relative orientations of CBS elements at domain boundaries by CRISPR/Cas9 alters the direction of CTCF/cohesin-mediated topological chromatin looping, which consequently leads to the establishment of new chromatin-looping interactions with CBS targets located in neighboring topological domains.
DISCUSSION
The diverse neuronal cell-surface PCDH repertoires, encoded by more than 50 clustered mammalian Pcdh genes, provide individual neurons with “identity tags” that engage in highly specific combinatorial homophilic interactions (Chen and Maniatis, 2013; Hirayama et al., 2012; Schreiner and Weiner, 2010; Thu et al., 2014; Wu, 2005; Wu and Maniatis, 1999). The functional significance of these interactions, based on direct evidence and by analogy to the Dscam system of invertebrates, is that they are required for the normal assembly of neural circuits during brain development (Chen et al., 2012; Garrett et al., 2012; Lefebvre et al., 2012; Suo et al., 2012; Thu et al., 2014; Wu and Maniatis, 1999). Therefore, understanding how PCDH diversity is generated in individual neurons is of fundamental importance.
The architecture rule of Pcdh CBSs provides interesting insights into their insulator functions. Rather than, or in addition to, blocking the cross-domain activities of enhancers as generally thought, the location and relative orientation of CBSs in enhancers determine the direction of looping therefore indirectly “insulate” one expression domain from another. This perspective may explain seemingly contradictory data previously obtained from reporter gene assays or transgenic mice experiments that addressed whether insulators function in an orientation-dependent manner. The enhancer activity of HS5-1 was demonstrated both with reporter genes in transgenic mice (Ribich et al., 2006) and by targeted deletion (Kehayova et al., 2011). The presence of both oriented CBSs (Guo et al., 2012) and a functional REST/NRSF binding site (NRSE) in the HS5-1 enhancer regulates the directional looping and neuronal cell-specific activity of the enhancer (Guo et al., 2012; Kehayova et al., 2011). Analysis of HS5-1 reporter constructs revealed that the NRSE functions as a silencer in transfection experiments, and deletion of the HS5-1 enhancer in mice resulted in an increase in Pcdhα gene expression in the kidney (Kehayova et al., 2011). The computational analyses presented here revealed that 46% of the potential enhancers genome-wide have a nearby CBS (Figure S4B), and 54% of genome-wide REST/NRSF sites (Figure S4C) have a close CBS, suggesting that CTCF functions as an activator or a silencer of transcription by controlling directional looping in different genomic contexts or specific cell types.
Insulators function to ensure proper interactions between remote enhancers and cognate promoters in vivo by blocking enhancers from targeting non-cognate promoters (Ong and Corces, 2014). Considering that CTCF and cohesin plays a pivotal role in the enhancer-blocking activity of insulators, in conjunction with the striking switching of looping directions with in-situ CRISPR inversion, as well as biophysical and computational analyses, we propose that directional CTCF/cohesin recognition of CBS pairs in the forward-reverse orientations establishes topological domains in mammalian genomes, resulting in a boundary element with CBS pairs in the reverse-forward orientations between adjacent domains (Figure 6). In particular, the directional CTCF binding to forward-reverse CBS sites and the asymmetric recruitment of cohesin through the CTCF C-terminal domain (Xiao et al., 2011) determine the looping direction with adjacent CBS sites (Figure 6). The reverse-forward boundary element between neighboring topological domains functions as an insulator to ensure the proper targeting of cognate promoters by distal enhancers (Inset, Figure 6). This model provides a molecular explanation for the pivotal role of CTCF in organizing chromatin during higher-order chromosome folding, and defines a unifying mechanism for the multivalent and seemingly conflicting functions of CTCF in the regulation of gene expression. Interestingly, computational simulation suggests that chromatin loops can “facilitate” or “insulate” enhancer-promoter interactions, depending on their locations outside or inside of the loops (Doyle et al., 2014). We note, however, that additional levels of control over directional DNA looping must exist, as the orientations of CBSs alone could not explain the specificity of DNA looping at such long distances since the chromatin fiber is likely to be sufficiently flexible to allow the DNA to be positioned to bring enhancers into proper orientation to interact with promoters.
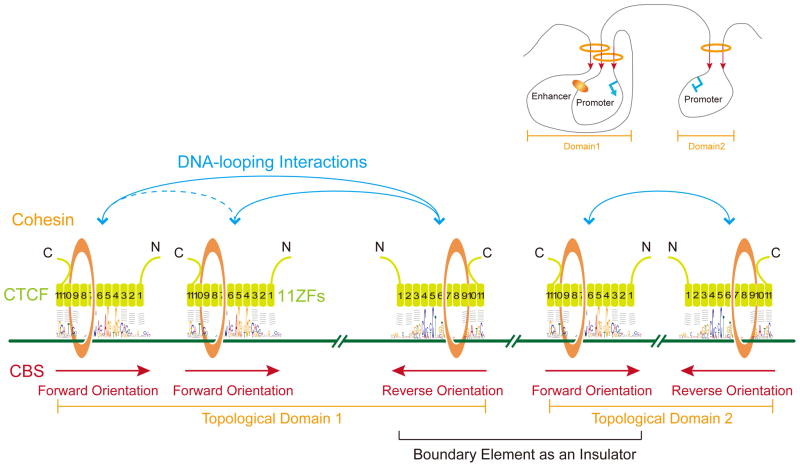
In mammalian genomes, CTCF directionally recognizes CBSs by distinct combinations of its 11 ZF domains, and asymmetrically recruits the cohesin complex to CBS sites through its C-terminal domain (Xiao et al., 2011). CTCF together with the cohesin complex establishes specific long-range chromatin-looping interactions between CBS pairs in the forward-reverse orientations to form distinct topological domains (domains 1 and 2, see the upper right inset). The weak interactions between the two CBSs in the same forward orientation in topological domain1 may be the consequence of their looping interactions with a common CBS in the reverse orientation (Guo et al., 2012). The two CBSs in the reverse-forward orientations form a boundary insulator element between the two neighboring domains 1 and 2, blocking remote enhancers located within one domain from aberrantly activating promoters located in the neighboring domain, and thus “indirectly” ensuring proper activation of cognate promoters by distal enhancers within the same topological domain (see Inset).
Computational analyses reveal vast majority of genome-wide chromatin loops occur between forward-reverse CBSs and minority of loops between forward-forward or reverse-reverse clusters of CBSs. Together with CRISPR and conformation capture evidence, the genome-wide architectural mechanism of CTCF/cohesin-mediated chromosome topology (Figure 6) has important implications not only for long-distance chromatin-looping contacts ranging from several kb to several Mb but also for the enhancer insulation functions of insulators to ensure proper promoter activation by distal enhancers. We suggest that genome-wide topological chromatin looping can be predicted based on CTCF/cohesin directional binding and its controlling elements can be engineered by CRISPR genome editing. Thus, our findings reveal how nonlinear 3D genome topology could be encoded by linear 1D genomic sequences.
EXPERIMENTAL PROCEDURES
CRISPR/Cas9 System
The templates for producing target sgRNAs were constructed in pLKO.1 or pGL3-U6-sgRNA-PGK-Puro plasmids (Li et al., 2015). All constructs were confirmed by sequencing. To screen for inversion cell clones, cells cultured to about 80% confluence were transfected with Lipofectamine 2000 (Invitrogen) in a 6-well plate with 6 μg of plasmid DNA, including 2 μg of pcDNA3.1-Cas9 and 4 μg of sgRNA constructs (2 μg each). One day after transfection, puromycin was added to a final concentration of 2 μg/ml. Ten to twelve days later, the cells were serially diluted and plated in 96-well plates to isolate clonal CRISPR cell lines. The primer sets used are shown in Table S7.
Circularized Chromosome Conformation Capture (4C)
The 4C-seq libraries were constructed as described (Guo et al., 2012; Jia et al., 2014). A series of 4C-seq libraries were generated by inverse PCR using a high-fidelity DNA polymerase. High-throughput sequencing was performed using 49 bp single-end reads on the Illumina HiSeq 2000 platform. The sequenced reads were mapped to reference genomes using the Bowtie program (version 1.0.0). The r3Cseq program in the R/Bioconductor package was used to detect statistical significance. All 4C-seq experiments were performed with at least two biological replicates.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (Guo et al., 2012; Jia et al., 2014). Briefly, HEC-1B cells were cross-linked with 1% formaldehyde for 10 min at 37°C. The lysate was immunoprecipitated with antibodies against CTCF (07-729; Millipore) or RAD21 (ab992; Abcam). The DNA was purified for real-time PCR. Statistical analysis was performed using the Student’s t test.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed using LightShift Chemiluminescent EMSA reagents as described (Guo et al., 2012). The probes were incubated with in-vitro-synthesized proteins in binding buffer containing 10 mM Tris, 50 mM KCl, 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM dithiothreitol, 0.1% Nonidet P-40 (NP-40), 50 ng/μl poly (dI-dC), and 2.5% (v/v) glycerol.
Supplementary Material
Supplemental Figure Legends
Supplemental figures
Supplemental text
SupplementalTable Legends
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Acknowledgments
We thank M. Capecchi, Z. Chen, S. Lomvardas, K. Monahan, S. O’Keeffe, L. Peng, and Y. Shi for critical reading of the manuscript. X. Huang and J. Xi for providing CRISPR/Cas9 plasmids. This study was supported by grants from MOST (2009CB918700), NSFC (31171015 and 31470820), Shanghai Municipality (13XD1402000 and 14JC1403600), and State Key Laboratory of Medical Genomics to Q.W.; from NIH (NS043915) to T.M.; from a collaborative grant between NSFC (81261120390) and NCI (3P01-CA013106-41S1) to Q.W. and A.R.K.; from NIH (HG001696), MOST (2012CB316503) and NSFC (91019016) to M.Q.Z. D.C. is a Helen Hay Whitney postdoctoral fellow. Q.W. is a Shanghai Subject Chief Scientist.
Footnotes
AUTHOR CONTRIBUTIONS
Q.W. conceived the project. Y.G. performed 3C, 4C, ChIP-qPCR, and RNA-seq. Q.X. performed computational analyses. D.C., D.U.G., and I.J. preformed Hi-C. J.S. and J.L. performed CRISPR. H.W. and Y.Z. performed EMSA. Y.T., Y.L., Y.W., Z.J., and W.L. performed cell culture and mouse work. Y.G., M.Q.Z., B.R., A.R.K., T.M., and Q.W. prepared the manuscript.Supplemental information includes Extended Experimental Procedures, seven Tables and five Figures.
References
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. [Europe PMC free article] [Abstract] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. [Abstract] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen WV, Alvarez FJ, Lefebvre JL, Friedman B, Nwakeze C, Geiman E, Smith C, Thu CA, Tapia JC, Tasic B, et al. Functional significance of isoform diversification in the protocadherin gamma gene cluster. Neuron. 2012;75:402–409. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen WV, Maniatis T. Clustered protocadherins. Development. 2013;140:3297–3302. [Europe PMC free article] [Abstract] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. [Abstract] [Google Scholar]
- de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. [Abstract] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. [Abstract] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. [Europe PMC free article] [Abstract] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. [Europe PMC free article] [Abstract] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. [Europe PMC free article] [Abstract] [Google Scholar]
- Doyle B, Fudenberg G, Imakaev M, Mirny LA. Chromatin loops as allosteric modulators of enhancer-promoter interactions. PLoS Comput Biol. 2014;10:e1003867. [Europe PMC free article] [Abstract] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [Europe PMC free article] [Abstract] [Google Scholar]
- Garrett AM, Schreiner D, Lobas MA, Weiner JA. gamma-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron. 2012;74:269–276. [Europe PMC free article] [Abstract] [Google Scholar]
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–782. [Europe PMC free article] [Abstract] [Google Scholar]
- Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci USA. 2012;109:21081–21086. [Europe PMC free article] [Abstract] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. [Europe PMC free article] [Abstract] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. [Abstract] [Google Scholar]
- Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2:345–357. [Abstract] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci USA. 2010;107:3651–3656. [Europe PMC free article] [Abstract] [Google Scholar]
- Jia Z, Guo Y, Tang Y, Xu Q, Li B, Wu Q. Regulation of the protocadherin Celsr3 gene and its role in globus pallidus development and connectivity. Mol Cell Biol. 2014;34:3895–3910. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. [Abstract] [Google Scholar]
- Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc Natl Acad Sci USA. 2011;108:17195–17200. [Europe PMC free article] [Abstract] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. [Europe PMC free article] [Abstract] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488:517–521. [Europe PMC free article] [Abstract] [Google Scholar]
- Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. [Europe PMC free article] [Abstract] [Google Scholar]
- Li J, Shou J, Guo Y, Tang Y, Wu Y, Jia Z, Zhai Y, Chen Z, Xu Q, Wu Q. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol. 2015;7:284–298. [Europe PMC free article] [Abstract] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. [Europe PMC free article] [Abstract] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [Abstract] [Google Scholar]
- Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc Natl Acad Sci USA. 2012;109:9125–9130. [Europe PMC free article] [Abstract] [Google Scholar]
- Nakahashi H, Kwon KR, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A, et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–1689. [Europe PMC free article] [Abstract] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. [Europe PMC free article] [Abstract] [Google Scholar]
- Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. [Europe PMC free article] [Abstract] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. [Europe PMC free article] [Abstract] [Google Scholar]
- Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem. 2007;282:33336–33345. [Abstract] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. [Europe PMC free article] [Abstract] [Google Scholar]
- Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci USA. 2006;103:19719–19724. [Europe PMC free article] [Abstract] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. [Europe PMC free article] [Abstract] [Google Scholar]
- Schreiner D, Weiner JA. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc Natl Acad Sci USA. 2010;107:14893–14898. [Europe PMC free article] [Abstract] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. [Europe PMC free article] [Abstract] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. [Europe PMC free article] [Abstract] [Google Scholar]
- Suo L, Lu H, Ying G, Capecchi MR, Wu Q. Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. J Mol Cell Biol. 2012;4:362–376. [Abstract] [Google Scholar]
- Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10:21–33. [Abstract] [Google Scholar]
- Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, Maniatis T. Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell. 2014;158:1045–1059. [Europe PMC free article] [Abstract] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. [Europe PMC free article] [Abstract] [Google Scholar]
- Wu Q. Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics. 2005;169:2179–2188. [Europe PMC free article] [Abstract] [Google Scholar]
- Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. [Abstract] [Google Scholar]
- Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. [Europe PMC free article] [Abstract] [Google Scholar]
- Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol. 2011;31:2174–2183. [Europe PMC free article] [Abstract] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci USA. 2007;104:7145–7150. [Europe PMC free article] [Abstract] [Google Scholar]
- Yokota S, Hirayama T, Hirano K, Kaneko R, Toyoda S, Kawamura Y, Hirabayashi M, Hirabayashi T, Yagi T. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J Biol Chem. 2011;286:31885–31895. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang T, Haws P, Wu Q. Multiple variable first exons: a mechanism for cell- and tissue-specific gene regulation. Genome Res. 2004;14:79–89. [Europe PMC free article] [Abstract] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IWF, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA. 2014;111:996–1001. [Europe PMC free article] [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.cell.2015.07.038
Read article for free, from open access legal sources, via Unpaywall:
http://www.cell.com/article/S0092867415009150/pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The chromatin tapestry as a framework for neurodevelopment.
Genome Res, 34(10):1477-1486, 29 Oct 2024
Cited by: 0 articles | PMID: 39472026 | PMCID: PMC11529992
Review Free full text in Europe PMC
Multifaceted role of CTCF in X-chromosome inactivation.
Chromosoma, 133(4):217-231, 21 Oct 2024
Cited by: 0 articles | PMID: 39433641
Review
YY1-controlled regulatory connectivity and transcription are influenced by the cell cycle.
Nat Genet, 56(9):1938-1952, 29 Aug 2024
Cited by: 2 articles | PMID: 39210046
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments.
Sci Adv, 10(37):eado1662, 13 Sep 2024
Cited by: 0 articles | PMID: 39270011 | PMCID: PMC11397430
Integration of CTCF loops, methylome, and transcriptome in differentiating LUHMES as a model for imprinting dynamics of the 15q11-q13 locus in human neurons.
Hum Mol Genet, 33(19):1711-1725, 01 Sep 2024
Cited by: 1 article | PMID: 39045627 | PMCID: PMC11413648
Go to all (531) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Three-dimensional genome architectural CCCTC-binding factor makes choice in duplicated enhancers at Pcdhα locus.
Sci China Life Sci, 63(6):835-844, 02 Apr 2020
Cited by: 3 articles | PMID: 32249388
Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection.
Genome Biol, 21(1):75, 23 Mar 2020
Cited by: 37 articles | PMID: 32293525 | PMCID: PMC7087399
CTCF Binding Polarity Determines Chromatin Looping.
Mol Cell, 60(4):676-684, 29 Oct 2015
Cited by: 346 articles | PMID: 26527277
CTCF: the protein, the binding partners, the binding sites and their chromatin loops.
Philos Trans R Soc Lond B Biol Sci, 368(1620):20120369, 06 May 2013
Cited by: 129 articles | PMID: 23650640 | PMCID: PMC3682731
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NCI NIH HHS (3)
Grant ID: P01 CA013106
Grant ID: 3P01-CA013106-41S1
Grant ID: P30 CA045508
NHGRI NIH HHS (2)
Grant ID: R01 HG001696
Grant ID: HG001696
NIGMS NIH HHS (1)
Grant ID: K12 GM068524
NINDS NIH HHS (2)
Grant ID: R01 NS043915
Grant ID: R56 NS043915





