Abstract
Free full text

Role of Substitutions in the Hemagglutinin in the Emergence of the 1968 Pandemic Influenza Virus
Abstract
Hemagglutinin (HA) of H3N2/1968 pandemic influenza viruses differs from the putative avian precursor by seven amino acid substitutions. Substitutions Q226L and G228S are known to be essential for adaptation of avian HA to mammals. We found that introduction of avian-virus-like amino acids at five other HA positions (positions 62, 81, 92, 144, and 193) of A/Hong Kong/1/1968 virus decreased viral replication in human cells and transmission in pigs. Thus, substitutions at some of these positions facilitated emergence of the pandemic virus.
TEXT
Pandemic influenza viruses originate from the natural reservoir of influenza A viruses in wild aquatic birds by various mechanisms, including adaptation of avian precursors in intermediate host species and reassortment with other animal and human viruses (1,–4). Because pandemic viruses usually contain an antigenically novel hemagglutinin (HA) of an animal precursor, it is essential to understand what changes in the HA are required for successful animal-to-human adaptation. Current knowledge in this field is limited. It is generally believed that HAs of pandemic viruses differ from their avian precursors by at least one or two mutations in the receptor-binding site (RBS) that alter viral receptor specificity from preferential binding to 2-3-linked sialic acids (viral receptors in birds) to preferential binding to 2-6-linked sialic acids in humans. In the case of previously characterized pandemic viruses, HA mutations G228S and/or Q226L (H2N2/1957 and H3N2/1968 viruses) and G225D and/or E190D (H1N1/1918 and H1N1/2009 viruses) were responsible for the switch in receptor specificity (for reviews, see references 3, 5, 6, 7, and 8).
The distribution of 2-3-linked and 2-6-linked receptors in the respiratory tract of pigs is similar to that in humans (9, 10). Accordingly, enzootic H1 and H3 swine influenza viruses and H2, H4, and H9 avian viruses isolated from pigs carry the same adaptive mutations in the HA RBS and show the same receptor specificity as human viruses (11,–15). Furthermore, human viruses replicate and transmit in pigs more efficiently than avian viruses, pigs can transmit influenza viruses to humans, and the pathogenesis and characteristics of influenza are very similar in humans and in pigs (16, 17). These features make swine a unique intermediate host in zoonotic transmission (18, 19) and a useful experimental model for human influenza infection (20, 21).
Recent gain-of-function studies on H5N1 avian influenza viruses showed that four amino acid substitutions in the HA, namely, two mutations in the RBS that switched receptor specificity (Q226L and either G228S or N224K), one substitution that altered HA glycosylation and receptor binding avidity, and one substitution that lowered the optimum pH of HA-mediated membrane fusion, were needed for the virus to become transmissible in ferrets by airborne droplets (22,–24). These results suggest that at least four substitutions in the HA may be required for the emergence of mammal-transmissible virus from its avian precursor. Here we aimed to test whether this notion applies to viruses that caused human pandemics in the past.
The HA of the H3N2/1968 “Hong Kong” pandemic influenza virus differed from the HA of the closest avian viruses by the aforementioned two mutations (G228S and Q226L) and by 5 additional substitutions (reference 25 and Fig. 1a). All 5 substitutions reside in the HA1 subunit of the HA; one of them (D81N) generates a new glycosylation site. Two substitutions (A144G and N193S) are located in the receptor-binding domain in the vicinity of the RBS (Fig. 1b). Three other substitutions (R62I, D81N, and N92K) are in the vestigial esterase domain, which interacts with the HA2 subunit and undergoes structural rearrangements during fusion (26, 27). Thus, based on their location, these five mutations could potentially affect the receptor-binding and fusion activities of the HA. To study combined phenotypic effects of these mutations, we prepared the recombinant pandemic virus A/Hong Kong/1/1968 (rHK) and its variant R5, which carried five amino acid substitutions in the HA restoring the ancestral avian sequence (Fig. 1a). The viruses were generated using the 8-plasmid reverse genetics system (28), and viral stocks were prepared in MDCK cells.
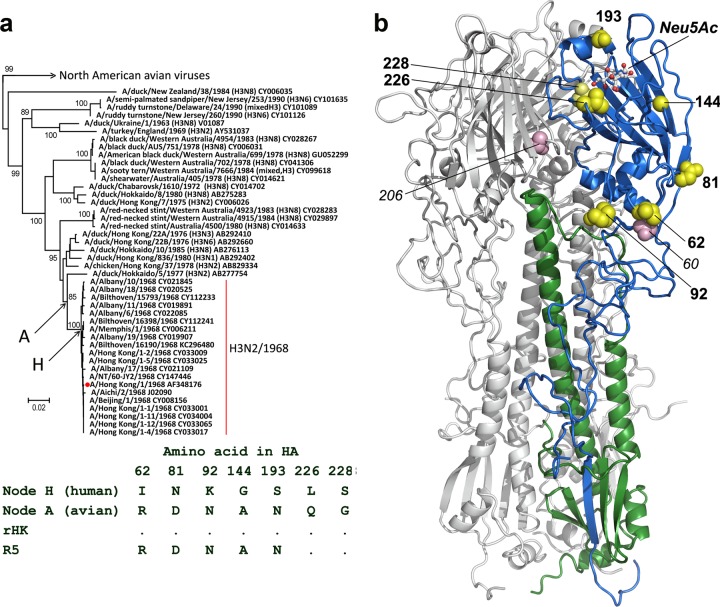
Seven amino acid substitutions in the HA separating H3N2/1968 pandemic influenza viruses from their putative avian precursors. (a) All full-length nonredundant HA sequences of avian H3 viruses isolated before 1990 (149 sequences) and of pandemic virus strains isolated in 1968 (20 sequences) were obtained from GenBank through the NCBI Influenza Virus Resource (39). The tree was generated using MEGA6 (40) with the minimum evolution method. The branch containing North American viruses is collapsed for clarity. The scale bar represents 0.02 units of nucleotide substitutions per site. Numbers depict bootstrap values (100 replications). Accession numbers are shown next to the strain names. Red dot depicts virus strain A/Hong Kong/1/1968 used to generate recombinant viruses rHK and R5 in this study. Amino acid sequences of the HAs of the putative avian precursor of the H3N2/1968 viruses (node A) and of the putative first human virus (node H) were inferred using MEGA6 with the maximum likelihood method. Amino acid differences between the avian precursor, the first human virus, and the recombinant viruses rHK and R5 are shown beneath the tree. Dot depicts amino acid identity with the top sequence. (b) Locations of amino acids shown as yellow space-filling models on the X-ray structure of H3 HA (5HMG). Two HA monomers are colored gray, and the third monomer is colored blue (HA1) or green (HA2). A ball-and-stick model depicts sialic acid in the receptor-binding site (Neu5Ac). Pink spheres depict amino acids in positions 60 and 206 which mutated during the virus transmission in experimentally infected pigs (see the text).
To infer the effect of substitutions in the HA on viral fitness in humans, we compared the replication efficiencies of R5 and rHK in differentiated cultures of human tracheobronchial epithelial (HTBE) cells, which mimic structural and functional features of the human airway epithelium in vivo (29,–32). Fully differentiated 6- to 7-week-old cultures grown on 12-mm-diameter membrane supports (Transwell-Clear; Corning Inc.) were washed with Dulbecco's modified Eagle medium (DMEM) to remove mucus. Four replicate cultures were infected from the apical side with 200 PFU of either rHK or R5 in 0.2 ml of the growth medium (GM). Following 1 h of incubation at 35°C, we removed the inoculum and continued incubation of the cultures under air-liquid interface conditions. In the first experiment, we harvested released viruses on a daily basis by washing the apical sides twice with 0.2 ml of GM with 30-min incubations. Samples were stored at −80°C and titrated using a focus-forming assay in MDCK cells (30). In this experiment, R5 grew to significantly lower titers than did rHK at the early time points postinfection (p.i.) (Fig. 2a). In the second experiment, we used HTBE cultures prepared from cells of a different human donor and harvested the viruses once at 3 days p.i. Again, the yield of R5 was lower than that of rHK (Fig. 2b). These results indicated that introduction of the 5 avian-virus-like amino acids into the HA of recombinant pandemic virus reduced its replication efficiency in HTBE cultures.
Replication of rHK and R5 in HTBE cultures. Cultures were infected with 200 PFU of the viruses, and viral yields (in focus-forming units [FFU] per culture) were determined by titration in MDCK cells. Panels a and b depict results of independent experiments; in the second experiment, the viruses were harvested once at 72 h postinfection. Results show mean values and standard deviations from four replicates. *, P < 0.05; ***, P < 0.0005 (unpaired two-tailed Student t test).
We next compared efficiencies of replication and transmission of rHK and R5 in pigs as a model for humans using the experimental protocol described previously (33). The experiments were authorized by the Ethical and Animal Welfare Committee of the Faculty of Veterinary Medicine, Ghent University. Six 6- to 8-week-old pigs that were serologically negative for influenza were obtained from a commercial herd. Groups of six pigs housed in separate high-efficiency particulate-air-filtered isolation units were inoculated intranasally with 106 PFU of either rHK or R5. Two days later, 6 contact pigs were introduced into each group. The housing allowed both direct contact and airborne contact between inoculated and sentinel animals. Virus shedding was monitored by collecting nasal swabs daily for 9 days postinoculation (p.i.) or postcontact (p.c.) and endpoint titrating in MDCK cells. No major differences in virus shedding of rHK and R5 between groups of directly inoculated pigs were observed (Fig. 3), and the differences in titers were statistically significant only on day 1 p.i. (P = 0.0007 [unpaired Student's t test]). Transmission of rHK occurred to all contact animals, starting as early as day 1 p.c. Excretion in contact animals lasted for 7 to 8 days, reaching peak titers similar to those observed in the inoculated group but with a somewhat slower starting profile. In contrast, the transmission of R5 was highly inefficient (Fig. 3). Only 5 of 6 contact pigs were shedding virus at some time point. The transmission was strongly delayed, with the first positive nasal virus excretion detected at 6 days p.c. Of the 5 virus-positive contact pigs, only 2 pigs (pig 4 and pig 6) were reaching peak titers similar to those seen with the inoculated animals.
Replication and transmission of rHK and R5 in pigs. Groups of six pigs were infected with 106 PFU of rHK (top panel) and R5 (bottom panel), and 6 contact pigs were introduced into each group at day 2 postinfection. Nasal swabs were collected daily and stored frozen until the end of the experiment. Viral titers in the swabs of inoculated pigs (left bars) and contact pigs (right bars) were assayed in MDCK cells. Horizontal dotted lines show the detection limit (10 50% tissue culture infective doses [TCID50]/ml). Colors indicate mutations in the HA identified by sequencing of virus-positive samples collected from contact pigs in the R5 group on days 6 to 9 as follows: blue, T206I; red, D60G; red with black stripes, D60D/G. Gray depicts viral samples without mutations. #, the virus-positive sample was not sequenced.
To detect potential changes in the HA of the viruses in pigs, we isolated viral RNA from the nasal swabs and performed Sanger sequencing. No mutations in the HA of rHK were observed in the swabs of inoculated and contact pigs, respectively, at 5 days p.i. and 5 days p.c. The virus shed by R5-inoculated pigs at 4 and 6 days p.i. also contained no mutations with respect to the inoculum. In contrast, 4 of 5 contact pigs in the R5 group contained one additional mutation (Fig. 3), either T206I (pig 2) or D60G (pigs 4, 5, and 6). The first mutation was located in the interface between the HA trimers, whereas the second mutation occurred in the vestigial esterase domain in the close vicinity of the I62R mutation separating R5 from rHK (see Fig. 1). The results of these experiments show that the avian-virus-like amino acids hampered transmission of R5 in pigs and suggest that transmissibility was facilitated by adaptive mutations in the HA.
We next compared receptor binding and membrane fusion activities of rHK, R5, and the D60G mutant of R5 (R5mut), which we isolated from pig nasal swab material in MDCK cells (Fig. 3). The receptor-binding properties were characterized using desialylated/resialylated preparations of peroxidase-labeled fetuin containing either 2-3-linked or 2-6-linked sialic acid moieties (34). We determined the binding of the viruses to fetuin in a solid-phase assay and calculated association constants of virus-fetuin complexes as previously described (24, 34). R5 bound to both fetuin preparations with higher avidity than did rHK (Fig. 4a). Comparison of R5mut and R5 indicated that the D60G mutation reduced the binding avidity to the levels of avidity of rHK, although the effect was statistically significant only in the case of 2-3-fetuin. To corroborate these findings, we compared viruses with respect to their ability to infect MDCK cells that were treated with Vibrio cholerae sialidase and as a result contained reduced levels of sialic acid receptors on the surface (Fig. 4b). rHK and Rmut infected partially desialylated cells less efficiently than did R5, confirming the higher receptor-binding avidity of the latter virus. We speculate on the basis of these experiments that reduced transmissibility of R5 in pigs was caused, at least in part, by excessive binding avidity of the HA and that the D60G mutation alleviated this problem.
Phenotypic analyses of rHK, R5, and a swine-derived mutant (R5mut). (a) Association constants of viral complexes with peroxidase-labeled fetuin carrying either 2-3-linked or 2-6-linked sialic acids were determined in a solid-phase binding assay. Data represent combined results from two experiments performed on different days with a total of 6 replicates for each virus. AU, arbitrary units. (b) Inhibition of infection by Vibrio cholerae sialidase. MDCK cells were incubated with the indicated concentrations of sialidase for 30 min at 37°C. Viruses (200 FFU) were added without removing sialidase, and incubation continued for 16 h. To avoid multicycle replication, no trypsin was added to the medium. The cells were fixed and immunostained for viral nucleoprotein. Numbers of infected cells were counted and expressed in percentages with respect to the control cultures that were incubated with the culture medium instead of sialidase. Four independent experiments were performed on different days with the same results. Data show results of one experiment performed with 2 to 3 replicates for each virus. (c) pH-dependent conformational transition of the HA. Viruses adsorbed in the wells of microtiter plate were incubated in either phosphate-buffered saline (PBS) or 0.1 M sodium acetate low-pH buffers for 15 min at 37°C, followed by washing with PBS and incubation with 0.1 mg/ml of proteinase K in PBS for 1 h at 37°C. After washing, binding to the viruses of peroxidase-labeled fetuin was determined and expressed in percentages of low-pH-exposed virus with respect to the PBS-exposed control. Three experiments were performed on different days with the same results. The data in the panel represent results of one experiment performed with 4 replicates for each virus. (d) Inhibition of virus entry into cells by the lysosomotropic agent ammonium chloride. MDCK cells were infected with 200 FFU of the viruses in the presence of the indicated concentrations of NH4Cl. The cells were fixed and immunostained 16 h postinfection. Percentages of infected cells with respect to the mock-treated control are shown for one experiment performed with 4 replicates for each virus. The results were reproduced in two additional independent experiments. In all panels, error bars show standard deviations and stars depict P values calculated using an unpaired two-tailed Student t test (*, P < 0.05; **, P < 0.005; ***, P < 0.0005).
The fusogenic activity of influenza viruses depends on the extensive low-pH-triggered conformational transition of the HA that makes HA sensitive to protease digestion (35, 36). We used this effect to study the pH dependence of the transition (Fig. 4c). R5mut was significantly less sensitive to low-pH treatment than were rHK and R5, indicating that the conformational change of R5mut occurs at a lower pH. The difference between R5 and rHK was less pronounced and significant only at pH 5.1. To further compare the fusogenic characteristics of the viruses, we studied inhibition of viral infection in MDCK cells by the lysosomotropic agent ammonium chloride, which counteracts the acidification of cellular endosomes (37, 38) (Fig. 4d). R5mut was significantly more sensitive to neutralization by ammonium chloride than were R5 and rHK, leading to the conclusion that R5mut relies on a stronger acidification of endosomes for fusion and uncoating than do R5 and rHK. These results imply that nonoptimal conformational stability and/or membrane fusion activity of the R5 HA could play a role in the inefficient transmission of the virus in pigs.
In summary, our data show that a combination of five amino acid substitutions in positions 62, 81, 92, 144, and 193 acquired by the HA of the 1968 pandemic influenza virus during its evolution from an avian precursor facilitates viral replication in human airway epithelial cells and transmission in pigs. Thus, at least some of these mutations, in addition to the well-known Q226L and G228S mutations, could have been instrumental in the bird-to-human adaptation and pandemic emergence. Our analyses suggest that these mutations optimize fusogenic and/or receptor-binding activities of the HA. Further studies using single-point mutants are warranted to clarify the role of individual mutations and the underlying molecular mechanisms. Such studies will improve current knowledge on phenotypic and genotypic markers of emerging zoonotic and pandemic viruses and facilitate risk assessment and pandemic preparedness.
ACKNOWLEDGMENTS
This work was supported by the European Union's Seventh Framework Programme for Research, Technological Development, and Demonstration under grant agreement 258084-FLUPIG and the German Research Foundation (SFB 1021).
We thank Earl Brown (University of Ottawa, Canada) for A/Hong Kong/1/68 and Robert Webster (St. Jude Children's Research Hospital, Memphis, TN, USA) for pHW2000 plasmid.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.01292-15
Read article for free, from open access legal sources, via Unpaywall:
https://jvi.asm.org/content/jvi/89/23/12211.full.pdf
Citations & impact
Impact metrics
Article citations
Probing altered receptor specificities of antigenically drifting human H3N2 viruses by chemoenzymatic synthesis, NMR, and modeling.
Nat Commun, 15(1):2979, 06 Apr 2024
Cited by: 3 articles | PMID: 38582892 | PMCID: PMC10998905
Vaccination and Antiviral Treatment against Avian Influenza H5Nx Viruses: A Harbinger of Virus Control or Evolution.
Vaccines (Basel), 11(11):1628, 24 Oct 2023
Cited by: 1 article | PMID: 38005960 | PMCID: PMC10675773
Review Free full text in Europe PMC
A Single Vaccination of Chimeric Bivalent Virus-Like Particle Vaccine Confers Protection Against H9N2 and H3N2 Avian Influenza in Commercial Broilers and Allows a Strategy of Differentiating Infected from Vaccinated Animals.
Front Immunol, 13:902515, 08 Jul 2022
Cited by: 4 articles | PMID: 35874682 | PMCID: PMC9304867
Genetic and Antigenic Characteristics of Highly Pathogenic Avian Influenza A(H5N8) Viruses Circulating in Domestic Poultry in Egypt, 2017-2021.
Microorganisms, 10(3):595, 09 Mar 2022
Cited by: 12 articles | PMID: 35336170 | PMCID: PMC8948635
Epidemiology, Genetic Characterization, and Pathogenesis of Avian Influenza H5N8 Viruses Circulating in Northern and Southern Parts of Egypt, 2017-2019.
Animals (Basel), 11(8):2208, 26 Jul 2021
Cited by: 9 articles | PMID: 34438666 | PMCID: PMC8388380
Go to all (17) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(1 citation)
PDBe - 5HMGView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Characterization of changes in the hemagglutinin that accompanied the emergence of H3N2/1968 pandemic influenza viruses.
PLoS Pathog, 17(9):e1009566, 23 Sep 2021
Cited by: 2 articles | PMID: 34555124 | PMCID: PMC8491938
The avian-origin PB1 gene segment facilitated replication and transmissibility of the H3N2/1968 pandemic influenza virus.
J Virol, 89(8):4170-4179, 28 Jan 2015
Cited by: 18 articles | PMID: 25631088 | PMCID: PMC4442368
Genetic drift influenza A(H3N2) virus hemagglutinin (HA) variants originated during the last pandemic turn out to be predominant in the 2011-2012 season in Northern Italy.
Infect Genet Evol, 13:252-260, 19 Nov 2012
Cited by: 12 articles | PMID: 23174527
[Accumulation of amino acid substitutions promotes irreversible structural changes in the hemagglutinin of human influenza AH3 virus during evolution].
Uirusu, 56(1):91-98, 01 Jun 2006
Cited by: 0 articles | PMID: 17038817
Review
 a and
a and 




