Abstract
Free full text

Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing
Abstract
Exosomes are vesicles which have garnered interest due to their diagnostic and therapeutic potential. Isolation of pure yields of exosomes from complex biological fluids whilst preserving their physical characteristics is critical for downstream applications. In this study, we use 100 nm-liposomes from 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and cholesterol as a model system as a model system to assess the effect of exosome isolation protocols on vesicle recovery and size distribution using a single-particle analysis method. We demonstrate that liposome size distribution and ζ-potential are comparable to extracted exosomes, making them an ideal model for comparison studies. Four different purification protocols were evaluated, with liposomes robustly isolated by three of them. Recovered yields varied and liposome size distribution was unaltered during processing, suggesting that these protocols do not induce particle aggregation. This leads us to conclude that the size distribution profile and characteristics of vesicles are stably maintained during processing and purification, suggesting that reports detailing how exosomes derived from tumour cells differ in size to those from normal cells are reporting a real phenomenon. However, we hypothesize that larger particles present in most purified exosome samples represent co-purified contaminating non-exosome debris. These isolation techniques are therefore likely nonspecific and may co-isolate non-exosome material of similar physical properties.
Exosomes are a class of membranous extracellular vesicles which originate from inward budding of the endosomal compartment within a cell, forming a multivesicular body which subsequently fuses with the plasma membrane to release the contents1. The capacity of exosomes to transfer mRNA, miRNA and protein from their cell of origin to a recipient cell has implicated them in cell-to-cell communication2,3 and they are present in various circulating bodily fluids including blood4, urine5 and saliva6, which endear them as potential non-invasive sources for surveying the presence of a variety of diseases3,7,8,9.
The use of exosomes for biomarker analysis first requires their isolation from complex biological fluids, which is a critical step for downstream diagnostic and therapeutic applications. To this end, numerous protocols and commercially available reagents have been designed to exploit the physical properties of these vesicles to purify exosomes from heterogeneous, biological samples. For example, differential ultracentrifugation is one of the more widely cited isolation methods and comprises a series of high speed spins (~100,000 × g) to selectively sediment exosomes from solution10, although the presence of contaminating cellular and protein debris has been noted within these isolates11. Similarly, several commercially available reagents such as the Invitrogen Total Exosome Isolation Kit (Life Technologies, USA) and ExoSpin Exosome Purification Kit (Cell Guidance Systems, USA) can facilitate sedimentation of exosomes from solution during low speed centrifugation (10,000–20,000 × g) by inducing precipitation of vesicles with poly-ethylene glycol12 or similar substances; however, while these kits are less user intensive than ultracentrifugation it has similarly been noted that they may also precipitate non-exosome debris13. Finally, exosomes have also been isolated based on their buoyant density in viscous fluids, wherein samples are layered onto discontinuous sucrose or iodixanol gradients and subjected to high speed centrifugation (100,000 × g with exosomes recovered from the 1.10–1.20 g/mL fraction/s11). The advantage of this method is that it is less prone to capture contaminating cellular debris, although this method is also highly user intensive and is not suited for high-throughput applications14.
Based on purification methods such as the ones detailed above, exosomes have been described as being 30–150 nm in range15,16, with an approximate density of 1.10-1.20 g/mL11,14. The size of exosomes in particular has been reported as an important factor for in vivo vesicle behavior and localisation17, and it has been suggested that exosomes derived from tumour cells may differ in size to those from normal cells6. However, reports in the literature have also indicated that isolation methods may affect exosomal protein and RNA yield18,19 and integrity20, suggesting exosome integrity and physical characteristics may be affected by these isolation methods. As such, it is important to ensure that these observations reflect inherent differences in vesicular biology and are not simply an artefact of processing. However, to our knowledge no study has been undertaken which evaluates the effect of isolation protocols on vesicle size and aggregation.
The lack of reports on this subject are due in part to the difficulty of performing pre-isolation measurements (e.g., size, and concentration characterisation) on exosomes within heterogeneous biological fluids, which makes the effects of isolation protocols difficult to differentiate from inherent sample-to-sample variation. As well, such an analysis would ideally take place in a single-particle analysis method which maintains the sample in a physiological buffer.
In response to this, here we report on the use of liposomes as a model vesicle system for evaluation of different exosome isolation methods. To characterise these vesicles and isolation methods we employed Tunable Resistive Pulse Sensing (TRPS), a platform capable of single-particle measurements of size and concentration21. This technique has previously been used to characterise exosomes and microvesicles22,23 in an in vitro setting. Here, the system was employed to compare the model liposome system both prior and post isolation using four exosome isolation techniques: ultracentrifugation, two sedimentation reagents (the Invitrogen Total Exosome Isolation Kit [Invitrogen] and ExoSpin Exosome Purification System [ExoSpin]), and a density gradient method (PureExo Exosome Isolation Kit [PureExo]).
Results
Comparison of exosome isolation methods using four techniques
To evaluate kit performance the purification of vesicles from serum-free cell culture media was first assessed. 2 mL of BT474 serum-free cell culture media was processed using each of the four techniques, followed by measurement of the isolated vesicles by TRPS. Vesicle yield varied between methods (Figure 1a), with the recovered content of the ExoSpin and Invitrogen kits generating the highest yields (between 2 × 1011 and 3.5 × 1011 particles/mL). By comparison the recovered yields for both the PureExo and Ultracentrifugation methods were approximately two orders of magnitude lower, with recovered concentrations between 1 × 109 and 1.5 × 109 particles/mL. All samples had a modal particle diameter of approximately 60–70 nm and a mean diameter of 70–80 nm, with these particle size distributions concordant with previously reported exosome size distributions16. No differences between processing methods were observed (Figure 1b), and no particle greater than 350 nm was measured in any sample. To ensure that contaminating particles present in the media or supplement were not being introduced during the purification method and giving a false indication of recovery, negative controls using serum-free media were processed and characterised, and there was negligible particle content in these negative control samples (data not shown).
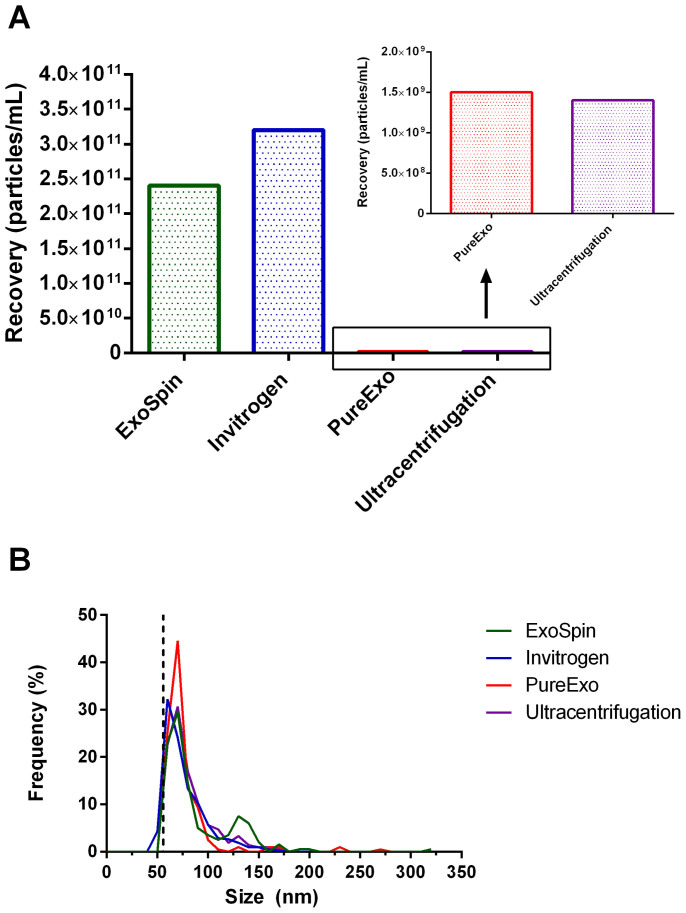
(A) Recovery (particles/mL) of exosomes from 2 mL of BT474 cell culture media via four isolation methods, as measured by TRPS with an NP100 rated pore. (B) Frequency (%) by size (nm) of recovered exosomes for each respective isolation method, measured by TRPS with a NP100 rated pore. The dotted line indicates 56 nm lower detection limit.
Liposomes as a Model System
To evaluate the fidelity of each of the four methods as relates to their recovery and potential impact on vesicle integrity, liposomes were selected as a model system. Liposomes are synthetic lipid vesicles that can be designed to share many of the physical properties of exosomes24 such as density (between 1.00 and 1.15 g/mL25,26,27), size25 and composition27, making them amenable to isolation via the exosome isolation techniques described above. Critically liposomes can be characterised before and after isolation to determine the effect of purification on vesicle size distribution and determine the percent recovery of each method. In the current study, the liposomes used were comprised of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), cholesterol and fluorescein-DHPE in a 50:45:5 mol/mol ratio. These were prepared by the manufacturer using an extrusion method to form a monodisperse vesicle population approximately 100 nm in diameter (FormuMax, USA).
To first assess the validity of liposomes as a surrogate model for exosome isolation liposome size distribution and ζ-potential was determined and compared to cell-media derived exosomes; size distributions as measured by TRPS using an NP100-pore rated for ~100 nm particles are shown in Figure 2. For liposomes, a mean particle diameter of 72 nm and a modal diameter of 63 nm were measured. For exosomes, the measured mean diameter was 78 nm and the mode 68 nm, demonstrating that the physical size of the two sample were highly similar. However, since larger particles could have potentially been gated out by the smaller pore (as outlined in Roberts et al.28) the measurements were then repeated using an NP150 pore, rated for ~150 nm particles (inset Figure 2) to assess whether larger particles were also present in the sample. This larger pore was found to have a lower sensitivity cutoff of 163 nm, compared to the NP100- pore which was 55 nm; methodology for how sensitivity cutoff limits were defined in the TRPS system is outlined in the methods section. The detection of larger particles in the NP150 pore indicated some gating of particles from the sample in the smaller NP100- pore, however this also indicated that larger particles were present in the original liposome samples and were not caused by aggregation during the extraction processes. For this reason, all further experiments were performed with an NP150 pore which was tuned to a measurement range (100–400 nm) that lay between those shown in Figure 2. This allowed detection of liposomes in the size range reported by the manufacturer (~100 nm) as well as detection of larger particles known to be present in these samples. Additionally, any changes in size distribution towards larger particles due to aggregation caused by extraction techniques were readily detectable and not truncated by the pore itself. Zetasizer measurements were also performed to assess the ζ-potential of the processed liposomes and compared to processed exosomes, to determine the similarities in surface charge between the two sample types. Processed liposomes had a ζ-potential of +7.2 mV (±1.2 mV), while the processed exosome sample was +8.0 mV (±0.5 mV) in PBS + 0.05% Tween-20.
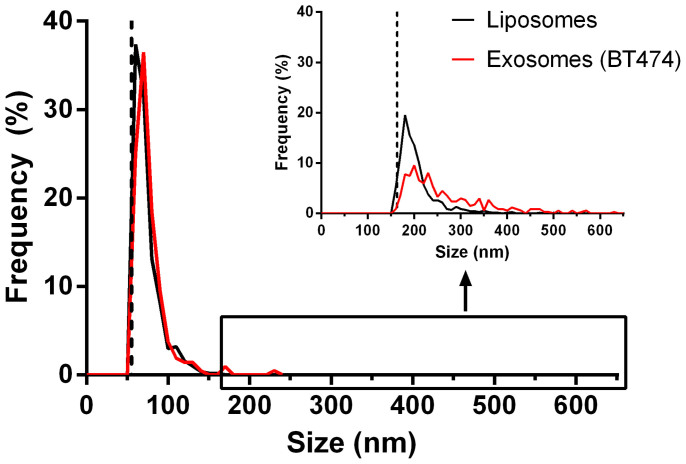
Size profile and frequency (%) by size (nm) of fluorescin-labelled liposomes (n = 503) and BT474 cell line-derived exosomes (n = 211) as measured by TRPS with an NP100 rated pore. Dotted line indicates 55 nm lower sensitivity limit. Inset) Frequency (%) by size (nm) of of the same fluorescin labeled liposomes (n = 686) and BT474 cell line-derived exosomes (n = 334) as measured by TRPS with an NP150 pore. Dotted line indicates 163 nm lower sensitivity limit.
Exosome isolation protocols do not induce changes to the size distribution of a model vesicle system
To determine the effect of isolation on vesicle size distribution and yield, measurements were first made of diluted unprocessed liposomes, followed by liposomes processed in triplicate using each of the four isolation techniques (ultracentrifugation, ExoSpin Exosome Purification System, Invitrogen Total Exosome Isolation Kit, PureExo Exosome Isolation Kit). Notably, it was observed that purification of liposomes using these methods required re-suspension of liposomes in serum free media (Medium 171, Life Technologies, USA) together with the addition of Mammary Epithelial Growth Supplement (MEGS, Life Technologies, USA); without MEGS supplement liposomes could not be purified from serum-free media. To ensure that contaminating particles present in the media or supplement were not being introduced during the purification method and giving a false indication of recovery, negative controls of cell-media without spiked liposomes were processed and characterised, and there was negligible particle content in these negative control samples (data not shown).
Post-processing characterisation measurements for intensity (%) and size (nm) were obtained for purified liposomes using dynamic light scattering (DLS), and representative data from one replicate of each processing method is shown in Figure 3a. The shape of the particle size distribution differed between sample processing measurements for DLS, with the size distributions for ExoSpin (z-ave = 97.6 ± 9.7 nm) and Invitrogen (z-ave = 95.1 ± 3.1 nm) samples similar to that of the unprocessed control (z-ave = 144.8 ± 6.2 nm), with an intensity peak centered around 150 nm. However, distributions for ultracentrifugation (z-ave = 108.8 ± 3.3 nm) and PureExo (z-ave = 125.7 ± 15.8 nm) samples were both shifted towards larger particles (>200 nm) relative to the control.
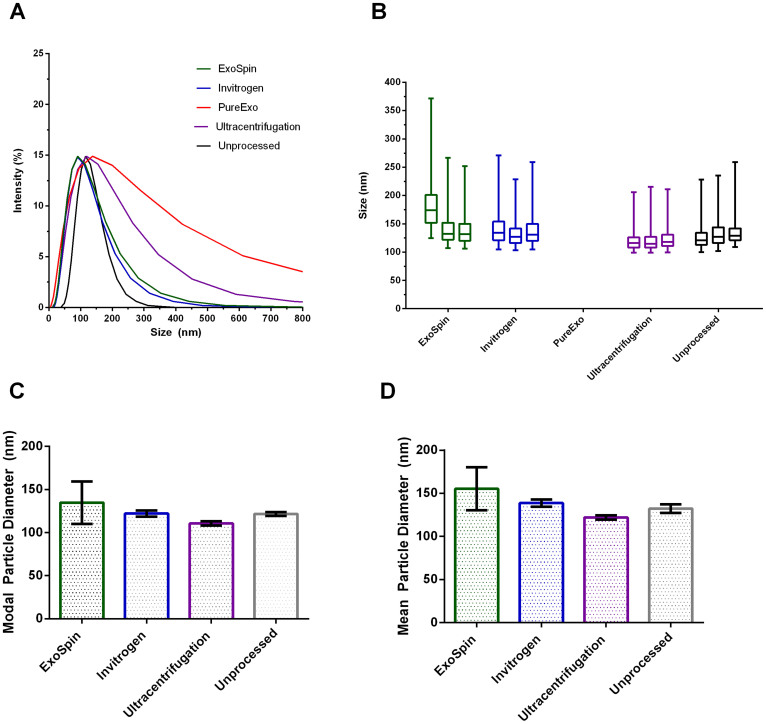
(A) DLS percent intensity by size (nm) for ExoSpin, Invitrogen, PureExo, Ultracentrifugation and Unprocessed control. (B) TRPS measurements with an NP150 pore of particle size distributions for ExoSpin (n = 1777), Invitrogen (n = 3302), PureExo (n = 0), Ultracentrifugation (n = 923) and unprocessed (n = 1312) samples respectively. Boxes indicate 25th, 50th and 75th percentile values, whiskers encapsulate 1st and 99th percentile values. (C) Modal ± SD particle diameter for ExoSpin (n = 3), Invitrogen (n = 3), ultracentrifugation (n = 3) treated samples and the unprocessed negative control as measured by TRPS with an NP150 pore. There was no significant difference in size between any of the processed samples and the unprocessed control (p>0.05). (D) Mean ± SD particle diameter for ExoSpin (n = 3), Invitrogen (n = 3), ultracentrifugation (n = 3) treated samples and the unprocessed negative control as measured by TRPS with an NP150 pore. There was no significant difference between any of the processed samples and the unprocessed control.
Given the difference in size measurements between samples using DLS, the same samples were then assayed by single-particle TRPS, and Figure 3b shows the size distribution of each replicate as measured by TRPS. The particle size distributions for ExoSpin (n = 3), Invitrogen (n = 3) and ultracentrifugation (n = 3) are comparable to that of the unprocessed control (n = 3), however there was negligible measurable particle recovery for the PureExo treated samples (n = 3) when measured by TRPS, in contrast to the measurement by DLS (Figure 3a). A one-way ANOVA was performed to test for significant differences in the mean and modal values for each of the processed samples relative to the control, but no significant difference was found between any of the methods for either analysis (Figure 3c & 3d); as PureExo recovery was insufficient for TRPS analysis this was not tested for significance.
ExoSpin and Invitrogen Purification does not cause loss of liposome yield
To assess the total recovery of each of the methods the yield of recovered particles for each of the isolation technique was quantified with TRPS. Samples were measured directly after isolation, alongside an unprocessed liposome sample, and a standard set of conditions was maintained across all measurements. As explained previously, these measurements were performed using a larger nanopore (i.e., NP150 verses NP100 than the previous exosome experiments (see Figure 1 and and2)2) to assay whether large aggregates were formed during purification. The use of the larger pore allowed us to detect large aggregates but does result in a lower sensitivity for smaller particles, as explained earlier, thus an artificial shift towards a mean/modal size distribution of approximately 125 nm has appeared to occur. The lack of resolution of smaller particles does not limit our ability to compare the data from these different sets of samples, as inter-measurement variables are conserved, as detailed in the following work (paper in review). Figure 4 shows the concentration (particles/mL) measured for each of the processed samples directly after the isolation process. To account for differing resuspension volumes between processed samples, all concentration values were normalised to the negative control for comparison. Particles recovered from ExoSpin (mean concentration = 1.58 × 1012 ± 3.079 × 1011 particles/mL), Invitrogen (mean concentration = 1.51 × 1012 ± 4.067 × 1011 particles/mL) and ultracentrifugation (mean concentration = 4.03 × 1011 ± 4.978 × 1010 particles/mL) processed samples were not significantly different (p>0.05) from the particle content of the unprocessed control (mean concentration = 1.22 × 1012 ± 3.05 × 1011 particles/mL). PureExo processed samples were excluded from analysis, as there was no measurable particle content in any of the three replicate measurements. Negative control samples consisting of cell culture media without liposomes added were processed via each respective method and measured. None of the control samples contained a measurable quantity of particles (data not shown).
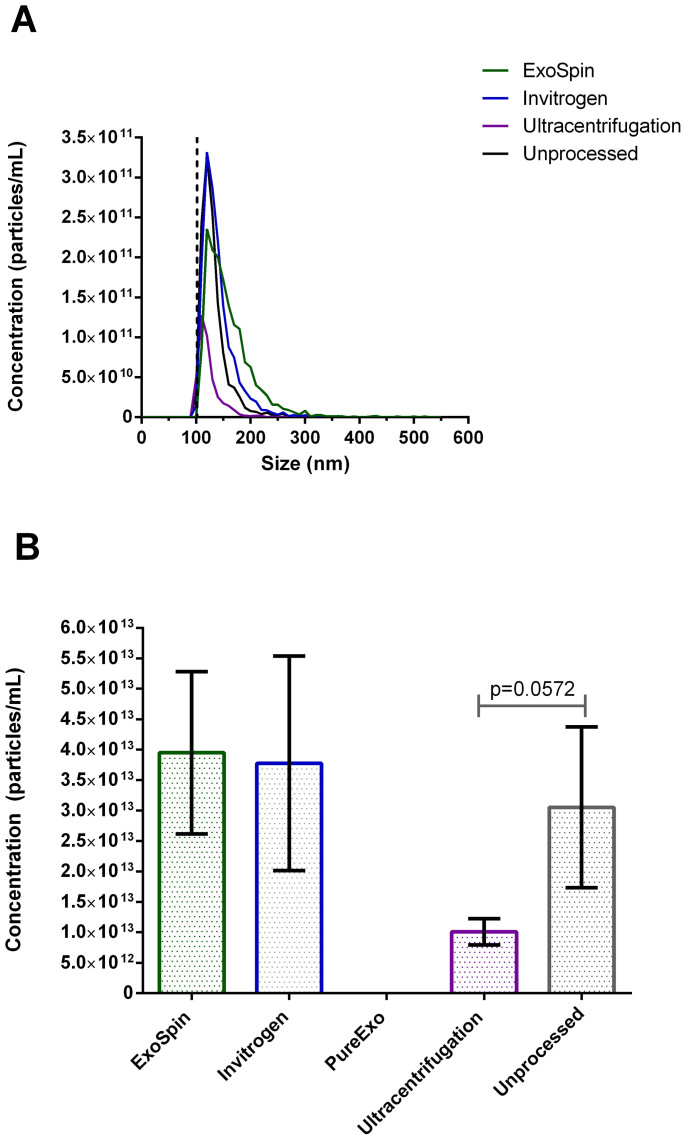
(A) Concentration (particles/mL) by size (nm) for each of the four processed samples and the unprocessed control as measured via TRPS with an NP150 pore. Dotted line indicates 102 nm lower detection limit. (B) Mean ± SD (n = 3) concentration (particles/mL) for ExoSpin, Invitrogen, PureExo and Ultracentrifugation processed samples and unprocessed control. There was no significant difference in concentration the unprocessed control and each of the processed samples respectively, although the difference in recovered yield between the unprocessed sample and ultracentrifugation purification approached significance (p = 0.0572).
Discussion
Over the past 5 years interest in the field of exosome research has increased dramatically, but methods to assess the potential impact of exosome purification protocols on vesicle recovery and size distribution have been limited. In this work we demonstrate that liposomes possess similar physical characteristics to exosomes, including size distribution (Figure 1), zeta potential (Figure 1) and density11,27, making them a useful tool to assay exosome purification techniques. Using liposomes as a model system together with a particle-by-particle analysis platform we have assessed the performance of four different purification methods, and conclude that liposomes are amenable to isolation via conventional exosome purification techniques such as precipitation reagents (ExoSpin and Invitrogen Total Exosome Isolation) and ultracentrifugation. Notably, vesicle recovery was found to vary between isolation methods, with the ExoSpin and Invitrogen kits recovering substantially more particles than either the PureExo or ultracentrifugation methods for both exosomes and liposomes. The difference in recoveries could potentially be due to the more user-intensive nature of the PureExo and ultracentrifugation protocols, which could cause an increased sample loss due to handling during the isolation process and result in a lower end yield. This lower yield also partly explains the difference in size profiles for the PureExo and ultracentrifugation methods as measured by DLS (Figure 3a), as the low yield results in a noisier signal due to the higher attenuator strength required to detect the low concentration of particles present in the sample29. It should also be noted that as DLS is based on light scattering, this technique is poor at resolving polydispersity which may skew measurement results towards larger particles21. Alternately the ExoSpin and Invitrogen kits may isolate non-exosome debris to a higher degree than the PureExo or ultracentrifugation methods, but as the size distributions for each of the recovered exosome samples were concordant with previously reported observations16 this seems unlikely.
In examining these isolation techniques we also sought to determine whether any significant impact on particle size or propensity to aggregation as a result of purification occurred. This question reflects reports in the literature indicating that purification methods may affect exosomal protein and RNA yield18,19 and integrity20, suggesting potential changes in vesicle integrity or aggregation. To address this question, we employed a model system of liposomes spiked in serum-free media. This meant that our system was free of vesicular and protein contaminants which could confound analysis of size distributions pre and post processing, thus allowing a direct assessment of purification techniques. Both DLS and TRPS data (Figure 3) demonstrated that the particle size distribution of processed liposome samples was comparable to that of the unprocessed control, and using the TRPS single particle analysis system we did not observe any substantial changes in liposome size distribution. While there was a slightly elevated level of larger liposome particles (i.e., >200 nm) in the processed samples relative to the unprocessed control, this is likely to be an artefact of instability of the resuspended liposomes in PBS, rather than a large-scale aggregation induced by the isolation methods. A one-way analysis of variation (ANOVA) for differences in the mean and modal particle diameter between the processed samples and the negative control did not reach significance at the 5% level. Overall, we conclude that the size distribution profile and characteristics of vesicles is stably maintained during processing.
However, while we observed no changes in the size distribution of the recovered liposome particles post-processing, we did measure particles larger than the reported 30–150 nm size range in processed exosome samples (Figure 1B). As no induced aggregates were observed in our model liposome system this suggests these larger particles in exosomes samples most likely represent non-exosome debris which has been co-purified, a hypothesis which is concordant with previous studies that observed the presence of contaminating membranous and protein material in exosome preparations11,14. It is possible that differences in exosome phospholipid content and protein composition could possibly lead to different outcomes with respect to aggregation and changes in exosome integrity, but applications which require pure exosomes may benefit from additional affinity purification steps to eliminate contamination by microvesicles and other non-exosomal membranous material. Moreover, the finding that liposomes can be purified using these kits further suggests that these isolation methods are not selective for exosomes and will likely facilitate sedimentation of other cellular material of similar size and density.
An unexpected finding was that liposome purification required the serum-free cell media used in this analysis (Medium 171, Life Technologies, USA) to be supplemented with Mammary Epithelial Growth Supplement (MEGS, Life Technologies, USA) to facilitate liposome recovery. Without MEGS supplement, liposomes remained visibly suspended in the serum free media after centrifugation step, with an absence of the expected pellet. This suggests that the additional protein content of the supplement is essential for liposome sedimentation, perhaps through adsorption of protein onto the surface of the liposome, though the exact mechanism of this is still under investigation.
In conclusion our results demonstrate that the isolation protocols assessed in this study do not substantially alter vesicle size distribution during purification, and are able to robustly isolate both exosomes and synthetic liposomes using three of the four methods investigated. This conclusion is of particular importance in the context of exosome purification and analysis, since the size of exosomes has been reported to be an important factor in in vivo vesicle behavior and localisation17, and reports have suggested that exosomes derived from tumour cells may differ in size to those from normal cells6. Moreover, the finding that exosome isolation protocols do not induce aggregation or alter the size profile is also relevant for those studies which have reported a difference in size and propensity towards aggregation6 between vesicles released from cancerous and healthy cells. Our findings suggest that such observations are not artefacts of sample preparation, however, more rigorous characterization methodologies are required before firm conclusions can be made on this issue30. However, the robust purification of both exosomes and liposomes using these techniques also demonstrates that these methods will co-isolate material of similar physical properties, and we therefore suggest an additional immunoaffinity purification step be included where highly pure exosomes are required for downstream analysis or applications to ensure that microvesicles and other cellular debris is not co-isolated. The current study therefore presents an important step to verify that vesicle size distributions remain unaffected by processing and preserves functionality for downstream applications.
Methods
Preparation of Samples
Serum Free Media (Medium 171, Life Technologies, USA) was obtained and passed through a 0.22 µm filter to eliminate contaminating debris. Fluorescin-labelled liposomes (Formumax, USA) were spiked into the filtered media at a dilution of 1:500, and dispensed into 2 mL aliquots. Negative controls consisting of filtered media without liposomes added were also separated into 2 mL aliquots. For exosome samples, BT474 cell culture media was collected and dispensed into 2 mL aliquots for processing.
ExoSpin Exosome Purification Kit
2 mL aliquots of sample were processed according to manufacturer instructions. To remove cellular debris, BT474 cell culture media was centrifuged at 300 × g for 10 minutes, and the resulting supernatant centrifuged for a further 30 minutes at 20,000 × g 0.5 volumes of Buffer A was added to each sample and vortexed to mix. Samples were incubated at 4°C for a minimum of 1 hour, and then centrifuged for 1 hour at 20,000 × g Following centrifugation, the supernatant was discarded and each pellet resuspended in 100 µL PBS. The resuspended pellets were applied to ExoSpin columns, and centrifuged at 50 × g for 60 seconds. The eluates were discarded and a further 200 µL of PBS applied to each column. This was centrifuged at 50 × g for 60 seconds and the eluate containing exosomes was stored at 4°C. The purification process for liposomes was identical with omission of the initial centrifugation steps for removal of debris.
Invitrogen Total Exosome Purification Kit
2 mL aliquots were processed according to manufacturer instructions. To remove cellular debris, BT474 cell culture media was centrifuged at 2,000 × g for 20 minutes and the supernatant transferred to a new microfuge tube. 0.5 volumes of the Invitrogen reagent were added to the sample and vortexed to mix. Samples were incubated overnight at 4°C, and subsequently centrifuged at 10,000 × g for 1 hour at 4°C. The supernatants were discarded and each pellet resuspended in 100 µL PBS respectively. The purification process for liposomes was identical with omission of the initial centrifugation steps for removal of debris.
PureExo Exosome Isolation Kit
2 mL aliquots were processed according to manufacturer instructions To remove cellular debris, BT474 media was centrifuged at 2,000 g for 10 minutes and the pellet discarded. In order, solutions A B and C were added to a glass tube in a 1:1:4 ratio and vortexed to mix. Sample was added to this mixture, vortexed and incubated for 30 minutes at 4°C. Of the resulting three-phase separation, the top and bottom layers were discarded, preserving the middle ‘fluff’ layer. This layer was centrifuged at 1,000 × g for 3 minutes forming a new three-phase separation. The top and bottom layers were discarded and the centrifugation repeated. The ‘fluff’ layer was gently dried with nitrogen gas and resuspended in 100 µL PBS. The resuspension was placed on a horizontal shaker at 450 rpm for 10 minutes. The resuspension was then centrifuged at 5,000 × g for 5 minutes and the pellet discarded. The supernatant was applied to a PureExo spin column and centrifuged at 1,000 g for 5 minutes. The flow through was collected and stored at 4°C. The purification process for liposomes was identical with omission of the initial centrifugation steps for removal of debris.
Ultracentrifugation
2 mL aliquots were processed according to a protocol adapted from Théry et al.10. To remove large debris, BT474 media was centrifuged at 2,000 × g for 20 minutes and the pellet discarded. The supernatant was centrifuged at 10,000 × g for a further 30 minutes and the resulting pellet discarded. Samples were transferred to 3.5 mL polycarbonate ultracentrifuge tubes (Beckman Coulter, USA) and centrifuged for 70 minutes at 4°C and 100 000 × g in a TLA100.3 fixed angle rotor (Beckman Coulter, USA). The resulting pellets were resuspended in 1 mL PBS and transferred to 1 mL polycarbonate ultracentrifuge tubes (Beckman Coulter, USA). To concentrate the exosomes, samples were centrifuged at 4°C and 100 000 × g for 60 minutes in a TLA120.2 fixed angle rotor (Beckman Coulter, USA). The resulting pellet was resuspended in 100 µL PBS and stored at 4°C. The purification process for liposomes was identical with omission of the initial centrifugation steps for removal of debris.
Dynamic Light Scattering
DLS measurements were performed with a ZetaSizer 3000-HA (Malvern Instruments, UK). Samples were diluted 1:1000 in PBS + 0.05% Tween-20 to a total volume of 1.5 mL. 3 × 10 measurement runs were performed, with standard settings (Refractive Index = 1.331, viscosity = 0.89, temperature = 25°C).
Measurement of ζ-Potential
ζ-Potential measurements were performed with a Zetasizer 3000-HA (Malvern Instruments, UK). Exosomes isolated via the ExoSpin kit (see above) were 1:100 in PBS + 0.05% Tween-20 for measurements. Liposomes isolated via the ExoSpin kit (see above) were diluted 1:1000 in PBS + 0.05% Tween-20 for measurements. 5 mL of each sample was applied to a flow cell, and 50 ζ-potential measurements taken. Standard settings were used (viscosity = 0.89, dielectric constant = 80, temperature = 25°C).
Tunable Resistive Pulse Sensing
TRPS measurements were performed with the qNano (Izon Science, UK). The instrument was set up and calibrated as per manufacturer recommendations. For liposome samples, a polyurethane nanopore rated for particles between 100 and 250 nm (NP150, Izon Science, UK) was used to perform all measurements, and was axially stretched to 48 mm, as measured from adjacent teeth on the qNano unit. For exosome samples, a polyurethane nanopore rated for particles <100 nm (NP100-, Izon Science, UK) was used, and was axially stretched to 48 mm, as measured from adjacent teeth on the qNano unit. 40 µL of sample diluted to an appropriate particle content in PBS + 0.05% Tween-20 was measured with this system. Optimally, measurement durations were greater than two minutes except where system instability limited this. All measurements were calibrated with 115 nm (NP100-) or 212 nm (NP150) polystyrene beads appropriately diluted (Izon Science, UK). Data processing and analysis were carried out on the Izon Control Suite software v2.2 (Izon Science, UK). Pore sensitivity was defined by the smallest possible particle able to be seen above system noise (i.e. pulse magnitudes >0.05 nA), as detailed in the following work (paper in review).
Statistical Analysis
All analyses were performed using Prism 6.0 (GraphPad Software, UK).
Cell culture
Breast cancer BT-474 cell lines were maintained in serum free Media 171 (Gibco, UK) supplemented with Mammary Epithelial supplement (Gibco, UK), 1% Pencillin/streptomycin and grown in 5% CO2 at 37°C. The conditioned medium from 106 cells was collected after 60 h after which exosomes were purified and analyzed as described above.
Acknowledgments
The authors wish to acknowledge the National Breast Cancer Foundation (NBCF) and Australian Research Council (ARC) for ongoing financial support of our group's research activities. Authors W.A. and M.T. are pleased to acknowledge a scientific collaboration with Izon Science Ltd. that was in place at the time that the manuscript was prepared. In this collaboration Izon Science Ltd. provided the researchers with equipment so that fundamental investigations into the qNano platform could be made. The authors gratefully acknowledge the receipt of this equipment. Izon Science Ltd. had no intellectual input into the work presented in this manuscript.
Footnotes
M.T and W.A. acknowledge a scientific collaboration with Izon Science where research equipment is provided for fundamental investigations into the qNano platform. The webpage http://www.aibn.uq.edu.au/aibn-izon outlines an agreement between Izon, M.T. and his research group in the middle of 2012. In addition to this, W.A. through a small student contract (12 days work over a total of 2 years), was contracted to perform qNano training for some of Izon's prospective customers. Apart from this, he is a full-time PhD student at the University of Queensland. R.L, D.K and R.V have no competing financial interests to declare.
Author Contributions D.K. and W.A. conceived and oversaw the research. R.L. performed the experiments with assistance from R.V. The manuscript was prepared by R.L., D.K. and W.A. All authors (R.L., D.K., W.A., R.V. and M.T.) contributed to discussions and manuscript review.
References
- Beaudoin A. R. & Grondin G. Shedding of vesicular material from the cell surface of eukaryotic cells: different cellular phenomena. Biochim Biophys Acta 1071, 203–219 (1991). [Abstract] [Google Scholar]
- Valadi H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659, 10.1038/ncb1596 (2007). [Abstract] [CrossRef] [Google Scholar]
- Skog J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10, 1470–1476, 10.1038/ncb1800 (2008). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Orozco A. F. & Lewis D. E. Flow cytometric analysis of circulating microparticles in plasma. Cytometry A 77, 502–514, 10.1002/cyto.a.20886 (2010). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Gonzales P. A. et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol 20, 363–379, 10.1681/ASN.2008040406 (2009). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Sharma S., Gillespie B. M., Palanisamy V. & Gimzewski J. K. Quantitative nanostructural and single-molecule force spectroscopy biomolecular analysis of human-saliva-derived exosomes. Langmuir 27, 14394–14400, 10.1021/la2038763 (2011). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lv L. L. et al. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. Am J Physiol Renal Physiol 305, F1220–1227, 10.1152/ajprenal.00148.2013 (2013). [Abstract] [CrossRef] [Google Scholar]
- Alvarez S. et al. Urinary exosomes as a source of kidney dysfunction biomarker in renal transplantation. Transplant Proc 45, 3719–3723, 10.1016/j.transproceed.2013.08.079 (2013). [Abstract] [CrossRef] [Google Scholar]
- Duijvesz D., Luider T., Bangma C. H. & Jenster G. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol 59, 823–831, 10.1016/j.eururo.2010.12.031 (2011). [Abstract] [CrossRef] [Google Scholar]
- Thery C., Amigorena S., Raposo G. & Clayton A. Isolation and characterizationof exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 30, 3.22, 10.1002/0471143030.cb0322s30 (2006). [Abstract] [CrossRef] [Google Scholar]
- Tauro B. J. et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 56, 293–304, 10.1016/j.ymeth.2012.01.002 (2012). [Abstract] [CrossRef] [Google Scholar]
- van der Pol E. et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost 12, 1182–1192, 10.1111/jth.12602 (2014). [Abstract] [CrossRef] [Google Scholar]
- Oosthuyzen W. et al. Quantification of human urinary exosomes by nanoparticle tracking analysis. J Physiol 591, 5833–5842, 10.1113/jphysiol.2013.264069 (2013). [Abstract] [CrossRef] [Google Scholar]
- Kalra H. et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 13, 3354–3364, 10.1002/pmic.201300282 (2013). [Abstract] [CrossRef] [Google Scholar]
- Dragovic R. A. et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine 7, 780–788, 10.1016/j.nano.2011.04.003 (2011). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- van der Pol E. et al. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J Thromb Haemost 8, 2596–2607, 10.1111/j.1538-7836.2010.04074.x (2010). [Abstract] [CrossRef] [Google Scholar]
- Alvarez-Erviti L. et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29, 341–345, 10.1038/nbt.1807 (2011). [Abstract] [CrossRef] [Google Scholar]
- Taylor D. D., Zacharias W. & Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol 728, 235–246, 10.1007/978-1-61779-068-3_15 (2011). [Abstract] [CrossRef] [Google Scholar]
- Rani S. et al. Isolation of exosomes for subsequent mRNA, MicroRNA, and protein profiling. Methods Mol Biol 784, 181–195, 10.1007/978-1-61779-289-2_13 (2011). [Abstract] [CrossRef] [Google Scholar]
- Alvarez M. L., Khosroheidari M., Kanchi Ravi R. & DiStefano J. K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int 82, 1024–1032, 10.1038/ki.2012.256 (2012). [Abstract] [CrossRef] [Google Scholar]
- Anderson W., Kozak D., Coleman V. A., Jamting A. K. & Trau M. A comparative study of submicron particle sizing platforms: accuracy, precision and resolution analysis of polydisperse particle size distributions. J Colloid Interface Sci 405, 322–330, 10.1016/j.jcis.2013.02.030 (2013). [Abstract] [CrossRef] [Google Scholar]
- Boing A. N. et al. Active caspase-3 is removed from cells by release of caspase-3-enriched vesicles. Biochim Biophys Acta 1833, 1844–1852, 10.1016/j.bbamcr.2013.03.013 (2013). [Abstract] [CrossRef] [Google Scholar]
- de Vrij J. et al. Quantification of nanosized extracellular membrane vesicles with scanning ion occlusion sensing. Nanomedicine (Lond) 8, 1443–1458, 10.2217/nnm.12.173 (2013). [Abstract] [CrossRef] [Google Scholar]
- Teng H. et al. Suppression of inflammation by tumor-derived exosomes: a kind of natural liposome packaged with multifunctional proteins. J Liposome Res 22, 346–352, 10.3109/08982104.2012.710911 (2012). [Abstract] [CrossRef] [Google Scholar]
- Alanazi F. K., Lu D. R., Shakeel F. & Haq N. Density gradient separation of carborane-containing liposome from low density lipoprotein and detection by inductively coupled plasma spectrometry. J Liposome Res 24, 53–58, 10.3109/08982104.2013.833224 (2014). [Abstract] [CrossRef] [Google Scholar]
- Versluis A. J., Rump E. T., Rensen P. C., van Berkel T. J. & Bijsterbosch M. K. Stable incorporation of a lipophilic daunorubicin prodrug into apolipoprotein E-exposing liposomes induces uptake of prodrug via low-density lipoprotein receptor in vivo. J Pharmacol Exp Ther 289, 1–7 (1999). [Abstract] [Google Scholar]
- Abe A., Miyanohara A. & Friedmann T. Enhanced Gene Transfer with Fusogenic Liposomes Containing Vesicular Stomatitis Virus G Glycoprotein. J Virol 72, 6159–6163 (1998). [Europe PMC free article] [Abstract] [Google Scholar]
- Roberts G. S. et al. Tunable nano/micropores for particle detection and discrimination: scanning ion occlusion spectroscopy. Small 6, 2653–2658, 10.1002/smll.201001129 (2010). [Abstract] [CrossRef] [Google Scholar]
- DeBlois R. W. & Bean C. P. Counting and Sizing of Submicron Particles by the Resistive Pulse Technique. Rev Sci Instrum 41, 909–916, 10.1063/1.1684724 (1970). [CrossRef] [Google Scholar]
- Zhuang X. et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol Ther 19, 1769–1779, 10.1038/mt.2011.164 (2011). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Articles from Scientific Reports are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1038/srep07639
Read article for free, from open access legal sources, via Unpaywall:
https://www.nature.com/articles/srep07639.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/srep07639
Article citations
Advancements in extracellular vesicle targeted therapies for rheumatoid arthritis: insights into cellular origins, current perspectives, and emerging challenges.
Stem Cell Res Ther, 15(1):276, 04 Sep 2024
Cited by: 0 articles | PMID: 39227964 | PMCID: PMC11373471
Review Free full text in Europe PMC
Differential extracellular vesicle concentration and their biomarker expression of integrin αv/β5, EpCAM, and glypican-1 in pancreatic cancer models.
Sci Rep, 14(1):14273, 20 Jun 2024
Cited by: 0 articles | PMID: 38902362 | PMCID: PMC11189911
TrkB phosphorylation in serum extracellular vesicles correlates with cognitive function enhanced by ergothioneine in humans.
NPJ Sci Food, 8(1):11, 06 Feb 2024
Cited by: 1 article | PMID: 38321007 | PMCID: PMC10847428
Exosomes: Methods for Isolation and Characterization in Biological Samples.
Methods Mol Biol, 2835:181-213, 01 Jan 2024
Cited by: 0 articles | PMID: 39105917
Review
Potential therapeutic effects of milk-derived exosomes on intestinal diseases.
J Nanobiotechnology, 21(1):496, 20 Dec 2023
Cited by: 4 articles | PMID: 38115131 | PMCID: PMC10731872
Review Free full text in Europe PMC
Go to all (121) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches.
Methods, 87:46-58, 02 Jun 2015
Cited by: 231 articles | PMID: 26044649
Effect of exosome isolation methods on physicochemical properties of exosomes and clearance of exosomes from the blood circulation.
Eur J Pharm Biopharm, 98:1-8, 03 Nov 2015
Cited by: 89 articles | PMID: 26545617
Optimized exosome isolation protocol for cell culture supernatant and human plasma.
J Extracell Vesicles, 4:27031, 17 Jul 2015
Cited by: 819 articles | PMID: 26194179 | PMCID: PMC4507751
Methods for the Determination of the Purity of Exosomes.
Curr Pharm Des, 25(42):4464-4485, 01 Jan 2019
Cited by: 6 articles | PMID: 31808383
Review





