Abstract
Free full text

Pancreatic β-cell Enhancers Regulate Rhythmic Transcription of Genes Controlling Insulin Secretion
Abstract
The mammalian transcription factors CLOCK and BMAL1 are essential components of the molecular clock that coordinate behavior and metabolism with the solar cycle. Genetic or environmental perturbation of circadian cycles contributes to metabolic disorders including type 2 diabetes. To study the impact of the cell-autonomous clock on pancreatic β-cell function, we examined islets from mice with either intact or disrupted BMAL1 expression both throughout life and limited to adulthood. We found pronounced oscillation of insulin secretion that was synchronized with the expression of genes encoding secretory machinery and signaling factors that regulate insulin release. CLOCK/BMAL1 co-localized with the pancreatic transcription factor PDX1 within active enhancers distinct from those controlling rhythmic metabolic gene networks in liver. β-cell clock ablation in adult mice caused severe glucose intolerance. Thus cell-type specific enhancers underlie the circadian control of peripheral metabolism throughout life and may help explain its deregulation in diabetes.
INTRODUCTION
The mammalian circadian system is organized hierarchically and is driven by cellular transcriptional oscillators that coordinate behavior and metabolism with the light-dark cycle. CLOCK/BMAL1 within the forward limb of the clock induces the expression of repressors (PERs/CRYs) in the negative limb and stabilizing factors (ROR/REV-ERB) in a cycle that repeats itself every 24 hrs (1, 2). A transformation in our understanding of clock function emerged from the discovery of autonomous circadian oscillation within individual tissues, and even in fibroblasts, ex vivo (3). Demonstration that disruption of central and peripheral clocks alters body weight and glucose homeostasis indicates that molecular rhythms play a critical role in systemic health. However, there has been a major gap in our understanding of how the molecular clock synchronizes transcription in distinct peripheral tissues to maintain overall physiological homeostasis (4–8).
Genome-wide analyses in liver indicate extensive rhythmicity of processed RNAs and non-coding enhancer RNAs (eRNAs) that are dependent upon temporal binding of circadian transcription factors to both promoters and enhancers (9–11). Yet the circadian clock exerts different effects on glucose metabolism within liver and other peripheral tissues, thus we sought to examine the genomic mechanism of clock control of pancreatic β-cell insulin secretion (5, 6). Here we define the targets of clock transcriptional regulation within the β-cell, and the impact of clock disruption on the temporal control of insulin secretion and glucose homeostasis.
RESULTS
The β-cell clock produces rhythmic insulin secretion and secretory gene transcription
First, to determine whether transcriptional oscillations in pancreatic islets give rise to rhythmic islet physiology, we examined the phase-dependence of pancreatic islet function by analyzing nutrient-induced insulin secretion in parallel with live-cell clock oscillation in islets from Per2Luc reporter mice (12). Following synchronization with forskolin (6, 13), we assessed insulin secretion every 4 hrs in individual groups of 5 islets at each time point over the ensuing 72 hr window (see schematic in Fig. S1A and Materials and Methods) and observed a striking self-sustained, time-of-day-dependent variation in the magnitude of response to stimulatory concentrations of both glucose and KCl, an insulin secretagogue that triggers exocytosis through direct depolarization of the β-cell (Fig. 1A). Intracellular insulin content did not cycle (Fig. S1B) despite rhythmic glucose-stimulated insulin secretion (GSIS) (Fig. 1A), consistent with circadian regulation at a step following translation of insulin. We further confirmed that GSIS rhythms were autonomous by monitoring insulin secretion following forskolin synchronization at times corresponding to the nadir (36 hr post-forskolin shock) and zenith (48 hr post-forskolin shock) of the WT GSIS rhythm in islets isolated from PdxCreER;Bmal1flx/flx mice (see Fig. 1B and schematic in Fig. S1C), which when treated with tamoxifen ex vivo display >60% reduction in Bmal1 expression (Fig. S1D). Vehicle-treated islets displayed significantly higher GSIS at the zenith than at the nadir, whereas tamoxifen-treated islets exhibited constitutively low levels of insulin secretion (Fig. 1B). Together, these data suggest that the islet molecular clock gates the rhythmic secretory response downstream of membrane depolarization.
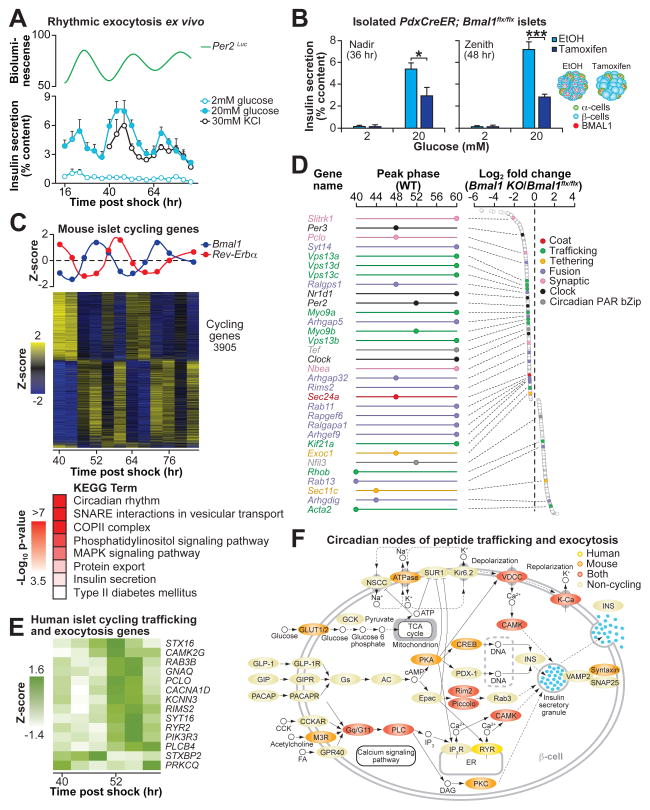
(A) Glucose and KCl-stimulated insulin secretion in synchronized WT mouse islets across either 3 or 2 consecutive days, respectively (n=3 replicate sets of islets pooled from 6–9 mice each) (bottom). Bioluminescence monitoring (counts/sec) in islets from Per2Luc reporter mice was performed in parallel (top). (B) Glucose-stimulated insulin secretion in ethanol- or tamoxifen-treated islets from PdxCreER;Bmal1flx/flx mice at the nadir (36 hr post-forskolin shock) and zenith (48 hr post-forskolin shock) of cyclic insulin secretion in wild-type islets from Fig. 1A (n=4 islet pools per time point, 3 replicates per islet pool). Of note, ethanol-treated islets displayed significant difference in GSIS comparing 36- to 48-hrs (p=0.038), whereas tamoxifen-treated islets did not (p=0.974). (C) Bmal1 and Rev-erbα RNA expression (top) and heatmap of all cycling genes identified by eJTK_CYCLE analysis (middle). Significantly enriched KEGG ontology pathways shown within the cycling gene set (bottom). (D) Peak phase expression (hrs post-forskolin shock) of cycling genes in synchronized WT islets (left) that were also altered in PdxCre;Bmal1flx/flx islets at ZT2. Log2 fold change in expression in PdxCre;Bmal1flx/flx (KO) islets compared to Bmal1flx/flx (control) at ZT2 (right) for subset of genes relevant to insulin secretion. (E) Heatmap showing expression patterns of cycling trafficking and exocytosis genes in synchronized human islets. (F) Mapping of cycling RNAs in both human and mouse islets onto the “Insulin Secretion” KEGG pathway. All values represent mean ± SEM. *p<0.05, ***p<0.001.
We next sought to examine the genome-wide effect of rhythmic transcription on insulin secretory dynamics by performing RNA-sequencing (RNA-seq) over two circadian cycles in RNA isolated from wild-type islets synchronized ex vivo (see schematic in Fig. S1A and Materials and Methods). We analyzed polyadenylated RNAs using eJTK_CYCLE (14), a modified non-parametric algorithm with increased sensitivity for detecting cycling transcripts. We detected a total of 3,905 cycling transcripts (Bonferroni-corrected p-value <0.05) which accounted for ~27% of all expressed transcripts within the islet that met a minimum mean expression threshold of 10 normalized counts (Fig. 1C). As expected, we observed high amplitude rhythms for the core clock transcription factors, including Bmal1, Clock, Npas2, Per2, Cry1, Rev-Erbα, and Rorα, with Bmal1 and its repressor Rev-Erbα displaying anti-phasic expression (Fig. 1C) (15).
To determine the identity of functional circadian gene networks in the islet, we tested for overrepresentation of defined KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways amongst rhythmic RNAs. We observed enrichment of factors mediating vesicle exocytosis, suggesting that daily variation in insulin secretory capacity arises from genomic regulation of the transport and release of peptidergic hormone (Fig. 1C and Table S1). Over-represented pathways in the circadian transcriptome included factors involved in i) vesicle budding, including genes encoding the COPII coat proteins (Sec24a and Sec31a), which mediate vesicle budding from the endoplasmic reticulum (16, 17); ii) cargo trafficking, specifically the motor proteins (Kif1b, Myo9a, and Dync2h1) that enable vesicle transport along cytoskeletal filaments (18); and iii) vesicle tethering and fusion to the plasma membrane, including v- and t-SNAREs such as Vamp1, Vamp5, Vamp8, Stx1a, Stx4a, and Stx8 (19, 20). In addition to the cycling of RNAs encoding factors involved in insulin exocytosis, we also identified rhythmic RNA expression of insulinotropic signals involved in vesicle movement and membrane fusion, including i) targets of cAMP/EPAC signaling (Pclo, Rims2, Rab3b, Rap1a, Rap1b, Rapgef2, Rapgef6), which mediate vesicle docking and fusion to the plasma membrane (21, 22); ii) Ca2+-sensing synaptotagmins (Syt11, Syt14, Syt16), which stimulate membrane fusion of synaptic vesicles (23, 24); and iii) calmodulin-dependent protein kinases (Camk1, Camk4, Camkk2, Camk2g), which regulate vesicle exocytosis and recycling (25). Lastly, we detected significant oscillation in targets of phosphoinositide signaling, including the protein kinase C (Prkca, Prkcb) (26), exocyst actin interacting factors including Exoc1/Sec3 (27), and the cytoskeletal filament remodeling Rho GTPases Rho, RhoA, RhoB, RhoC (18). Collectively, cycling of RNAs encoding factors involved in insulin exocytosis and signaling components reveals a genomic basis for circadian variation in insulin secretion.
To further understand the physiologic function of tissue-specific rhythmic gene transcription, we next compared genome-wide rhythms of RNA expression in WT islets to those in pancreas-specific clock mutant mice (PdxCre;Bmal1flx/flx) which exhibit severe hypoinsulinemic diabetes due to defects downstream of glucose metabolism and mitochondrial respiration (Fig. S2) (6). We performed RNA-seq using RNA immediately isolated from PdxCre;Bmal1flx/flx and control littermate islets at the start of the light phase (ZT2), the time of maximal GSIS impairment (Fig. S3A) (6). We identified changes in the expression of 1,757 genes in islets isolated from clock mutant animals relative to littermate controls (Bmal1flx/flx), including transcripts that were both decreased (1,074) and increased (683) in expression, consistent with actions of the clock as both an activator and repressor of gene expression (FDR-adjusted p-value <0.05) (Fig. S3B–C).
Importantly, many of the RNAs that were altered in islet-clock knockout mice were identified as cycling RNAs in WT islets - overall, a total of 720 oscillating genes exhibited altered expression in animals with disrupted pancreatic clock function (Fig. S3C), indicating an autonomous role of the islet clock in the rhythmic transcriptional regulation of insulin secretion. Among the most significantly changed RNAs were factors in the negative limb of the core clock containing the canonical E-box transcription motif, in addition to circadian PAR bZip transcription factors, including Per2, Rev-Erbα (Nr1d1), Tef, and E4bp4 (Nfil3) (Fig. 1D). We also found a broad range of alterations in cycling genes that are circadian outputs and grouped into similar exocytosis networks by KEGG annotation as described for the WT islets, including genes encoding factors involved in trafficking, such as the vacuolar protein sorting factors Vps13b and Vps13c, the motor protein involved in vesicular transport Myo9, the kinesin transport factor Kif21, and the small GTPase Rab11, a factor in trans-golgi vesicular biogenesis (28) (Figs. 1D, S3E–F, and Table S1). Ontology analysis also identified genes related to vesicle tethering and fusion as altered in clock deficient islets, including the conserved exocyst component Exoc1/Sec3, cAMP-EPAC controlled Rims2 and Pclo, and the synaptogamnin Syt14 (Fig. 1D), while islet genes involved in glucose sensing were unchanged (Table S2). Interestingly, whereas the complete set of cycling RNAs display broadly distributed peak phases (Fig. S3D), the majority of exocytosis-related RNAs that were differentially expressed in clock mutants exhibited peak expression at two distinct phases (48 and 60 hrs post forskolin shock) (Fig. 1D). Although this suggests that these genes may represent direct targets of CLOCK/BMAL1 and/or a clock repressor (REVERBα/β or E4BP4), nascent RNA-seq studies indicate that peak circadian mRNA phases are not directly correlated with nascent transcription (11). Collectively, sequencing results indicate that secretory pathway genes represent a major output of the islet clock.
To determine whether the rhythmic islet transcriptome is conserved from mouse to humans, we performed RNA-seq in RNA isolated from synchronized human islets (Fig. S4A). Human islets displayed characteristic circadian patterns in the expression of core clock components BMAL1 and REV-ERBα (Fig. S4B) (29) and genome-wide rhythmic patterns in the transcriptome with 1,800 cycling RNAs (Bonferroni-corrected p-value <0.05) (Fig. S4B). While striking differences have been described between mouse and human islet cell composition and cytoarchitecture (30), the expression of key genes involved in insulin release are conserved between species (30). Remarkably, 481 of the rhythmic human islet genes were orthologous to those in mouse islets (Fig. S4C), including factors involved in exocytosis, trafficking, and fusion (Figs. 1E–F and S4C). Mapping cycling human islet RNAs onto KEGG-curated human insulin secretion pathways revealed regulation of GPCR, cAMP, Ca2+ and phosphoinositide-responsive signaling molecules important in nutrient responsive and hormone release (Fig. 1E–F). Specifically, these included the Gq protein GNAQ, insulin vesicle associated RIMS2 and PCLO, calmodulin-activated protein kinase CAMK2G, and phospholipase C PLCB4, all of which were also rhythmic in mouse islets (Figs. 1E–F and S4D). Circadian gene regulation in the endocrine pancreas of both mice and humans thus converges on the late secretory pathway, demonstrating conservation of clock control of rhythmic tissue function across species.
BMAL1 and CLOCK bind near cell-type specific enhancers in pancreatic β-cells
Since our genome-wide RNA sequencing studies indicate that genomic regulation by the clock gives rise to rhythmic insulin secretion, we next sought to analyze how core circadian transcription factors (TFs) regulate this process by analyzing the extent of binding by BMAL1 and CLOCK to rhythmically expressed genes. In this context, cistrome studies have recently characterized β-cell transcriptional hubs encoding genes that program both development and function (31), revealing co-localization within regions of accessible chromatin (H2A.Z) and active enhancers (H3K4Me1 co-localized with H3K27Ac) containing binding sites for lineage-determining transcription factors (PDX1, MAFB, FOXA2, NKX6-2, and NKX2-2) (31–33). To determine the intersection between circadian TF regulation and genomic binding at regulatory loci, we performed chromatin immunoprecipitation sequencing (ChIP-seq) in the mouse β-cell line Beta-TC6 (Fig. 2A). As expected, we found that both BMAL1 and CLOCK physically bound to sites at core clock and other gene targets in β-cells that were enriched for the canonical CACGTG E-box motif, often occurring in tandem, as previously reported at BMAL1 binding sites in liver (Fig. S5A) (p=10−38 and p=10−91, respectively) (9, 34). Moreover, we also observed significant overlap in the genome-wide binding of BMAL1 and CLOCK (Fig. S5B). A representative UCSC genome browser track at the Rev-erbα (Nr1d1) locus is shown in Fig. 2A, revealing co-localization of BMAL1 and CLOCK binding sites at three distinct regulatory regions at the Nr1d1 locus, including within both the promoter region (shaded light orange and defined as within 2kb of the transcription start site (TSS)) and within intragenic and intergenic distal enhancer regions (shaded light green and defined as binding regions greater than 2kb from the TSS) (Fig. 2A). Of importance, histone markers representing active and accessible chromatin (H3K27Ac and H2A.Z, respectively) localize to the same promoter and enhancer regions within the Nr1d1 locus, indicating active transcriptional regulation by BMAL1 and CLOCK (Fig. 2A).

(A) Model of transcriptional targets and chromatin modifications for ChIP-seq experiments (top). UCSC genome browser tracks at Nr1d1 (Rev-erbα) locus in β-cells. Maximum track heights within viewable window are indicated to the right of each factor (bottom). (B) Distribution of BMAL1 and CLOCK peaks at cycling and non-cycling gene targets. Binding sites at cycling genes are separated into promoter proximal and distal sites (< and >2kb from TSS of nearest gene, respectively). (C) KEGG ontology terms enriched in cycling and non-cycling BMAL1 and CLOCK target genes.
To determine whether BMAL1 and CLOCK directly regulate the oscillating transcripts identified in the synchronized WT islets (Fig. 1C), we evaluated the overlap between the BMAL1 and CLOCK cistromes with genes oscillating in the WT islets. Among binding sites localized to expressed RNAs, 30% (862 binding sites) and 29% (330 binding sites) of the BMAL1- and CLOCK-targets, respectively, exhibited rhythmic transcription in synchronized WT islets (Fig. 2B), which collectively accounted for 742 cycling direct target genes of which 165 were differentially expressed in Bmal1 knockouts (Fig. S5C), suggesting direct BMAL1 and CLOCK regulation. Moreover, KEGG analysis of the direct gene targets in mouse islets that were present in BMAL1 and CLOCK cistromes revealed enrichment in pathways related to protein export, COPII-mediated vesicle budding from the endoplasmic reticulum, and SNARE vesicular transport and membrane fusion, in the cycling compared to non-cycling set of BMAL1 and CLOCK-controlled transcripts (KEGG pathways listed in order of descending −log10 p-values, Fig. 2C, Table S1). Together, these findings identify direct transcriptional targets of CLOCK/BMAL1 that mediate rhythmic islet physiology.
BMAL1 binds to distinct enhancers in liver and pancreatic β-cells
Given evidence for tissue-specific regulation at enhancers as a predominant mode of circadian regulation in liver (10), we next analyzed the binding position of core clock TFs BMAL1 and CLOCK in relation to the transcription start site (TSS) of rhythmic genes in β-cells. We classified binding events occurring within 2kb of the nearest annotated gene TSS as promoter-proximal since genome-wide promoter activity studies and epigenetic characterization of mammalian regulatory regions have indicated that the majority of core promoter activity is localized within 2kb of the TSS (35–37). Surprisingly, we found that BMAL1 and CLOCK bind predominantly at distal sites (defined as greater than 2kb from the TSS) rather than at proximal promoter sites (defined as less than 2kb from the TSS) of rhythmically regulated genes (Fig. 2B and Fig. S5D), suggesting that the islet clock TFs impact rhythmic physiology through binding to distal regulatory sites, an observation concordant with the general finding that TFs exert physiologic effects through regulation within tissue-specific enhancers (31).
While clock factors have been shown to exert distinct physiologic functions across tissues, a major gap remains in understanding the underlying genomic mechanisms accounting for these tissue-specific functions. To determine whether BMAL1 regulates rhythmic genes through unique sites in the β-cell compared to liver, the tissue in which the circadian cistrome has been best characterized (9–11, 34, 38), we compared sites of BMAL1 occupancy in the β-cells to a published set of liver BMAL1 peaks (9). Unexpectedly, although there was a significant overlap of genes identified as direct BMAL1 binding targets in β-cells and liver (40%, 1063 genes out of 2660 total β-cell target genes) (Fig. 3A), BMAL1 binding at the regulatory regions of those shared gene sets localizes to distinct sites (Fig. 3A). Remarkably, in comparing genome-wide binding patterns, we only observed common locations of binding in 4% of these instances; thus BMAL1 binding at all β-cell and liver-defined sites are uncorrelated (Fig. 3A, Fig. S5E) (R2=0.01874 and 0.03286 for BMAL1 binding at β-cell and liver sites, respectively), while binding at canonical E-box sites in Per2, Cry1, and Dbp was similar between tissues (Fig. S5F). Furthermore, when we compared the shared set of BMAL1 target genes that were rhythmic in islets and also reported to be rhythmic at the mRNA level in liver, BMAL1 likewise bound to unique sites (9) (Fig. 3B). These data suggest convergent regulation of BMAL1-targets in β-cell and liver through divergent regulatory elements.
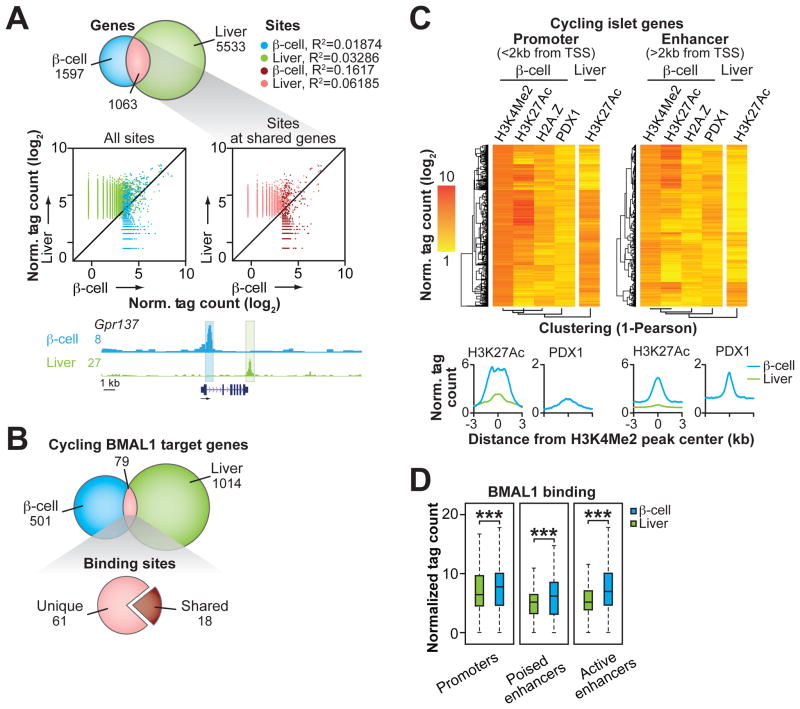
(A) Overlap of genes identified at BMAL1 binding sites in β-cells and liver (top). Scatter plots show BMAL1 binding in liver (y-axis) versus β-cells (x-axis) within 500bp windows surrounding peaks identified in each tissue (middle). Browser track view of BMAL1 binding in β-cells and liver at the Gpr137 locus (bottom). (B) Overlap of cycling and direct BMAL1 target genes in β-cells that have been reported to cycle in liver (top). Cycling BMAL1 direct target genes containing shared or unique binding sites in β-cells and liver (bottom). (C) Heatmaps comparing binding of indicated factors within 1kb windows surrounding promoter (3,492) and enhancer (5,771) localized H3K4Me2 peaks annotating to genes containing cycling RNAs in WT islets. Histograms summarizing normalized tag counts for H3K27Ac (in β-cells and liver) and PDX1 (in β-cells) across 6kb span centered at all β-cell H3K4Me2 peaks (bottom). (D) Box and whiskers plots (whiskers represent IQR 1.5) comparing BMAL1 binding in β-cells and liver at loci corresponding to H3K4Me2 peaks defined in heatmaps. Poised enhancers refer to H3K4Me2 sites that do not co-localize with H3K27Ac, whereas active enhancers are defined as H3K4Me2 sites co-localized with H3K27Ac. ***p<0.0001 by Mann-Whitney non-parametric, unpaired t-test. All reported ChIP-seq tag counts normalized per 107 reads.
Because BMAL1 predominantly bound at distal regulatory regions in islets that were divergent from liver, we next sought to examine the chromatin regulatory context at all cycling genes in β-cells. To do so, we defined all regulatory regions at cycling loci using H3K4Me2 peaks within 2kb of the TSS (promoter) and >2kb from the TSS (enhancer) (Fig. 3C). The binding patterns of the histone marks H3K4Me2, H2A.Z, and H3K27Ac (which represent promoter/enhancer regulatory regions, chromatin accessibility, and enhancer activity, respectively), as well as binding of the lineage-determining TF for β-cells PDX1 (39) at promoters and enhancer regions are displayed in heatmaps in Fig. 3C. By performing hierarchical clustering, we found that all epigenetic and PDX1 signals at promoter and distal enhancer regions at cycling genes more frequently displayed correlated binding than did H3K27Ac at these loci in liver, as indicated by the clustering dendogram (Fig. 3C). Accordingly, the genomic coordinates in liver corresponding to enhancers defined in β-cells displayed markedly reduced H3K27Ac, indicating that these enhancers defined specific loci of β-cell regulation (Fig. 3C). Frequent binding of PDX1 at distal enhancer loci suggested that tissue-specificity arose from early events in islet cell development (Fig. 3C) (40). Consistent with tissue-specific clock TF regulation at β-cell regulatory regions, BMAL1 displayed a greater degree of binding to promoter and enhancer regions at cycling genes in β-cells than in liver, particularly at active enhancers containing both H3K4Me2 and H3K27Ac (Fig. 3D). These results indicate that clock TFs generate unique patterns of rhythmic RNA expression across tissues according to the pattern of cell-specific enhancer repertoires and provide a molecular basis for the distinct and opposing effects of the clock in pancreas and liver, which primarily affect post-prandial and fasting glucose metabolism, respectively (5, 6).
β-cell clock disruption during adulthood impairs insulin secretion and causes diabetes
To test the hypothesis that clock genes modulate genome-wide transcription on a daily basis throughout adult life, we examined the impact of acute clock inhibition on glucose metabolism in PdxCreER;Bmal1flx/flx mice at 2–3 months of age following administration of tamoxifen, which abrogates BMAL1 expression exclusively within the β-cell (Fig. S6) (41). While these mice displayed normal wheel running rhythms, period length, food intake, and body weight (Fig. S7) compared to littermate tamoxifen-treated PdxCreER and Bmal1flx/flx animals, they developed significant hyperglycemia, impaired glucose tolerance, and hypoinsulinemia within 10–14 days following tamoxifen administration during both the day (ZT2) and night time (ZT14) (Figs. 4A–B and S8), despite no differences in islet mass (Fig. S9A). These results establish that circadian disruption in fully differentiated cells is sufficient to induce metabolic disease independent of effects on early development.
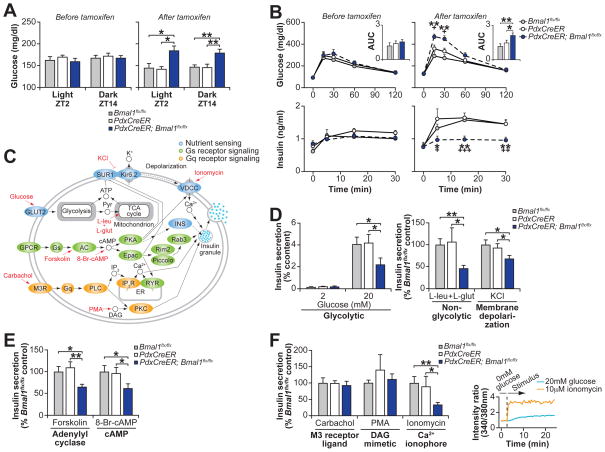
(A) Blood glucose levels in ad libitum fed mice before and after tamoxifen administration (n=6–12 mice per genotype). (B) Glucose tolerance and insulin secretion at ZT2 following intraperitoneal glucose administration in PdxCreER;Bmal1flx/flx mice and littermate controls before and after tamoxifen treatment (n=4–11 mice per genotype). Inset represents area under the curve (AUC) for glucose (104 mg/dl/120min). (C) Model of intersecting pathways driving insulin exocytosis highlighting nutrient, Gs, and Gq receptor signaling that are used to stimulate insulin secretion in (D–F). (D) Glucose and nutrient-stimulated, (E) cyclase-pathway, and (F) catecholamine-stimulated insulin secretion in islets isolated from tamoxifen-treated PdxCreER;Bmal1flx/flx and control mice (n=3–8 mice per genotype, 3 repeats per mouse). Inset is ratiometric determination of intracellular Ca2+ using Fura2-AM dye in Beta-TC6 cells in response to insulin secretagogues (n=3 replicates per condition). All values represent mean ± SEM. For (B), asterisks denote significance between Bmal1flx/flx and PdxCreER;Bmal1flx/flx, while plus symbols denote significance between PdxCreER and PdxCreER;Bmal1flx/flx. *p<0.05, **p<0.01, ***p<0.001
We further found that islets isolated from tamoxifen-treated PdxCreER;Bmal1flx/flx mice secreted significantly less insulin compared to littermate controls when exposed to (i) 20mM glucose, (ii) 10mM leucine combined with 2mM glutamine, which bypasses glycolysis to trigger mitochondrial ATP production, or (iii) 30mM KCl, which chemically closes the KATP channel, thus inducing membrane depolarization distal to glucose metabolism and an increase in cytosolic calcium (Fig. 4C–D), while glucose-stimulated calcium influx was unchanged (Fig. S9B). Remarkably, this data is consistent with our observation that circadian oscillation in insulin secretory capacity is regulated downstream of KATP channel closure. Consistent with impaired heterotrimeric G protein-coupled receptor (GPCR) signaling, PdxCreER;Bmal1flx/flx islets also secreted significantly less insulin than controls in response to glucose together with the cyclase agonist forskolin and the non-hydrolyzable cAMP analogue 8-br-cAMP (Fig. 4C, E).
Finally, we tested the response to Gq-type GPCR signaling by stimulating islets with the muscarinic agonist carbachol, the DAG mimetic PMA, and Ca2+ ionophore ionomycin. Surprisingly, carbachol and PMA restored insulin secretion in PdxCreER;Bmal1flx/flx islets (Fig. 4C, F), while the response to ionomycin, which raised intracellular Ca2+ in β-cells (Fig. 4F), was significantly reduced in mutants, indicating that the DAG-arm of the Gq pathway restored second messenger signaling. DAG regulates exocytosis in β-cells and other neurosecretory cells by acting as a ligand for the vesicle priming protein Munc13-1 (42) and PKC, which phosphorylates and activates SNAP25 and MUNC18-1 to initiate vesicle fusion (43). We observed rhythmic RNA expression of the PKC activating Rho and Rap GTPases Rho, Rhoa, Rhob, and Rap1a in WT islets, raising the intriguing possibility that elevated DAG concentrations in carbachol or PMA treated islets pharmacologically bypassed a deficiency in Rho- and Rap-mediated signaling. Together, these results demonstrate that pharmacologic Gq agonism reverses the insulin secretory blockade induced by clock disruption, indicating convergence of cholinergic and phosphoinositol signaling within the β-cell in temporal homeostasis.
DISCUSSION
We have established the genome-wide basis of coordinated cross-tissue circadian oscillation through integrated studies of β-cell physiology and cistrome regulation. We focused on pancreatic β-cells as a paradigm of peripheral clock regulation of metabolism since clock disruption in the islet leads to severe hypoinsulinemic diabetes and has direct application to understanding human tissue rhythms and disease. Although the circadian system functions as a hierarchy in the intact animal, our results reveal organ-autonomous cycles of nutrient-coupled insulin secretion in isolated islets ex vivo that result in a significant amplitude in maximal glucose responsiveness—suggesting that the clock primes insulin secretion within limited windows each day. We further find that circadian-driven transcriptional oscillation within pancreas drives daily waves of expression of genes involved in the biogenesis, transport, and signal-induced activation of peptide exocytosis, indicating that genomic rhythmicity gives rise to tissue-specific function of the clock. Our observations suggest that autonomous transcription cycles enable islet cells to anticipate diurnal changes in the demand for insulin.
Cistromic profiling within the β-cell provides further insight into the regulation of tissue-specific genome oscillation. We found that CLOCK/BMAL1 bind predominantly within distal tissue-specific enhancers rather than the promoters of cycling genes in proximity to H3K4Me2-, H2A.Z-, and H3K27Ac-modified nucleosomes that are co-occupied by PDX1. Consistent with tissue specificity in enhancer selection across cell types, BMAL1 binding in islet cells were highly divergent from liver, even within shared cycling target genes across the two tissues. These findings suggest that the establishment of accessible chromatin domains during development is a critical determinant of the available regulatory sites for clock-mediated transcription across distinct cell types. Further studies will be needed to define the underlying mechanisms through which divergent tissue-specific regulation gives rise to convergent oscillation of rhythmic genes. Finally, our studies using chemically-inducible genetic clock inactivation demonstrate that inhibition of circadian signaling in differentiated β-cells acutely blocks peptide exocytosis and leads to hypoinsulinemic diabetes, providing evidence that clock function throughout adult life is necessary for glucose constancy. An intriguing possibility is that cell-autonomous genomic rhythms may regulate peptidergic secretion across diverse secretory and neuronal cell types, coordinating the availability of signaling molecules with the sleep/wake cycle each day. Furthermore, given the association between circadian and sleep disruption with human metabolic disease in both clinical (44, 45) and genetic (46) studies, the finding that circadian transcription is conserved human islets suggests that clock deregulation in β-cells may contribute to the pathogenesis of human diabetes. The demonstration of coordinated circadian genomic and physiologic rhythms in pancreatic β-cell insulin exocytosis and its control by enhancers provides a new window to understand the transcriptional coupling of geophysical and physiologic time, and how errors in this process may contribute to diabetes and other metabolic disorders.
MATERIALS AND METHODS
Animals
Male wild-type C57BL/6J mice were purchased from The Jackson Laboratory. Per2Luc (12) and PdxCre;Bmal1flx/flx (6) mice were produced and maintained on a C57BL/6J background at the Northwestern University Center for Comparative Medicine. Bmal1flx/flx mice (47) were crossed with PdxCreER transgenic mice (kindly provided by Dr Douglas Melton, Harvard University) (48) to generate PdxCreER;Bmal1flx/flx mice, as well as Bmal1flx/flx and PdxCreER littermate controls. Unless otherwise stated, mice were maintained on a 12:12 light:dark (LD) cycle with free access to regular chow and water. All animal care and use procedures were conducted in accordance with regulations of the Institutional Animal Care and Use Committee at Northwestern University.
Islet isolation, insulin secretion assays, and in vitro islet synchronization
Mouse pancreatic islets were isolated via bile duct collagenase digestion (Collagenase P, Sigma) and Ficoll gradient separation and left to recover overnight (16 hrs) at 37°C in RPMI 1640 with 10% FBS, 1% L-glutamine, and 1% penicillin/streptomycin. For standard insulin release assays, 5 islets were statically incubated in Krebs-Ringer Buffer (KRB) and stimulated for 1 hr at 37°C with various glucose concentrations, 30mM KCl, 2.5μM forskolin, 1mM 8-Br-cAMP, 10mM L-leucine + 2mM L-glutamine, 1mM carbachol, 10μM PMA, or 10μM ionomycin. Supernatant was collected and assayed for insulin content by ELISA (Crystal Chem Inc). Islets were then sonicated in acid-ethanol solution and solubilized overnight at 4°C before assaying total insulin content by ELISA. For rhythmic insulin release assays, islets were first synchronized with 10μM forskolin (Sigma) for 1 hr and allowed to recover for 16 hrs. Insulin secretion assays were then performed as above in individual groups of 5 islets every 4 hrs for 72 hrs (Fig. S1A). Human islets (obtained from IIDP) were cultured in RPMI 1640 with 10% human AB serum, 1% L-glutamine, and 1% penicillin/streptomycin (see table in Fig. S4A for details re: sex, age, BMI, and IIDP ID numbers of the three donors). For the rhythmic analysis of RNAs in murine and human islets, RNA was isolated (described below) in groups of 200 islets every 4 hrs for 48 or 24 hrs, respectively, starting 40 hrs following forskolin synchronization (Fig. S1A).
Lumicycle analysis
Approximately 2 hrs prior to the start of the dark period (i.e., lights off), ~100–150 pancreatic islets were isolated from Per2Luc mice as described above. Islets were cultured on tissue culture membranes (Millipore) in 1.2ml DMEM media (Gibco) containing 352.5mg/ml sodium bicarbonate, 10mM HEPES (Gibco), 2mM L-glutamine, 2% B-27 serum-free supplement (Invitrogen), 25units/ml penicillin, 20mg/ml streptomycin (Gibco), and 0.1mM luciferin sodium salt (Biosynth AG). Sealed cultures were placed at 37°C in a LumiCycle luminometer (Actimetrics) and bioluminescence from tissues was recorded continuously. After several days in culture, islets were synchronized by 10μM forskolin (Sigma) treatment for 1 hr followed by incubation in fresh media. Period was calculated via a modified best-fit sine wave analysis using Lumicycle Analysis software (Actimetrics).
Measurement of islet oxygen consumption
Following bile duct collagenase digestion, 40 purified pancreatic islets were plated in wells of a 96-well respirometry plate (Seahorse Bioscience) and cultured overnight in complete medium. The next day culture medium was replaced with assay buffer containing 3mM glucose, 0.8mM Mg2+, 1.8mM Ca2+, 143mM NaCl, 5.4mM KCl, 0.91mM NaH2PO4, and Phenol red 15mg/ml (Seahorse Bioscience) and allowed to equilibrate at 37°C in a CO2-free incubator for 1–2 hrs. The plate was then loaded into a Seahorse XF96 instrument, and the oxygen consumption rate (OCR) was measured for 4 sequential 3-minute intervals at basal conditions and following injection of glucose (20mM final concentration), oligomycin (F1FOATPsynthase inhibitor) (5μM final concentration), and antimycin A (complex III inhibitor) (5μM final concentration). OCR values presented represent the average of 4 sequential measurements. Mitochondrial oxygen consumption was calculated by subtracting OCR values following antimycin A treatment (representing non-mitochondrial oxygen consumption).
RNA isolation and qPCR mRNA quantification
Islets were added to microfuge tubes containing Tri Reagent (Molecular Research Center, Inc) and frozen at −80°C. RNA was isolated according to the manufacturer’s protocol and purified using RNeasy columns (Qiagen). cDNAs were then synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative real-time PCR analysis was performed with SYBR Green Master Mix (Applied Biosystems) and analyzed using an Applied Biosystems 7900 Fast Real-Time PCR System. Relative expression levels were determined using the comparative CT method to normalize target gene mRNA to Gapdh. Exon-specific primer sequences for qPCR as follows: Bmal1 Exons 5–7 Forward: 5′-ATCGCAAGAGGAAAGGCAGT-3′, Reverse: 5′-ATCCTTCCTTGGTGTTCTGCAT-3′; Bmal1 Exons 7–9 Forward: 5′-AGGCCCACAGTCAGATTGAA-3′ Reverse: 5′-TGGTACCAAAGAAGCCAATTCAT-3′; Bmal1 Exon 8 Forward: 5′-GGCGTCGGGACAAAATGAAC-3′ Reverse: 5′-TCTAACTTCCTGGACATTGCAT-3′; Bmal1 Exons 8–9 Forward: 5′-TGCAATGTCCAGGAAGTTAGAT-3′ Reverse: 5′-TGGTGGCACCTCTCAAAGTT-3′; Bmal1 Exons 10–12 Forward: 5′-TAGGATGTGACCGAGGGAAG-3′ Reverse: 5′-AGCTCTGGCCAATAAGGTCA-3′.
RNA-sequencing and analysis
Following RNA isolation (described above), RNA quality was assessed using a Bioanalyzer (Agilent), and sequencing libraries were constructed using an Illumina TruSeq Stranded mRNA sample prep kit LT (Illumina, RS-122-2101) according to the manufacturer’s instructions. Libraries were quantified using both a Bioanalyzer (Agilent) and qPCR-based quantification (Kapa Biosystems) and sequenced on either an Illumina HiSeq 2000 or NextSeq 500 instrument to a depth of at least 30 million reads using 100bp or 75bp paired-end reads, respectively. Differential Expression Analysis: For differential expression comparison between PdxCre;Bmal1flx/flx and Bmal1flx/flx islets, RNA raw sequence reads were aligned to the reference genome (mm10) using STAR version 2.3.1s_r366 (49). Differentially expressed RNAs were identified using DESeq2 version 1.6.3 (50) (FDR-adjusted p-value <0.05). Cycling RNA Analysis: For cycling RNAs, raw sequence reads were similarly aligned using STAR (mm10 index for mouse and hg19 for human) and uniquely mapped reads (tags) were normalized using the algorithm employed in DESeq2 (50). The geometric mean of the raw read counts was calculated for each gene. A normalization factor was calculated for each sample using the median of the raw read counts of each gene divided by the geometric mean of the gene. The normalized read counts were computed by dividing the raw read counts by the normalization factor. The normalized tags for the mouse and human time series were separately concatenated and z-scored within each gene (14). Rhythm detection of the z-scored and normalized counts was performed with empirical JTK_CYCLE with asymmetry search, which increases sensitivity to detecting cycling transcripts by extending cosine wave-fiting to include asymmetric waveforms, which better represents expression patterns seen in biological data. Rhythmic time series were examined with reference waveforms with a period of 24 hr, a phase of either 0, 4, 8, 12, 16, or 20, and an asymmetry of either 4, 12, or 20. Due to the low number of waveforms compared, the Bonferroni correction was used instead of the empirical p-values. Genes with a Bonferroni-adjusted p-value below 0.05 were considered to be rhythmic. Ontology Analysis: For Kyoto Encyclopedia of Genes and Genomes (KEGG) ontology term enrichment (51, 52), Ensembl gene IDs were supplied and analyzed using Homer (version 4.7.2) command “findGO” (53). Cycling Genes in Liver: Genes exhibiting rhythmic mRNA accumulation in vivo in liver were derived from reported “exon cycling” transcripts (9).
β-cell culture
Beta-TC6 cells were purchased from ATCC (CRL-11506) and cultured in DMEM supplemented with 15% FBS, 1% L-glutamine, and 1% penicillin/streptomycin. All cells used in experiments were at fewer than 15 passages.
Mouse BMAL1 and CLOCK polyclonal antibody generation
Guinea pig anti-mouse BMAL1 and CLOCK polyclonal antibodies were generated using a 37- and 39-amino acid peptide fragment of the mouse BMAL1 and CLOCK proteins, respectively (RS synthesis). Guinea pigs were immunized with KLH-conjugated peptides (Pocono Farms), and BMAL1- and CLOCK-specific antibodies were affinity-purified from whole serum using resin cross-linked with antigen peptides (Pierce).
Chromatin immunoprecipitation (ChIP)
Beta-TC6 cells (~40–160 million) were fixed for 30 minutes in 2mM DSG and for 10 minutes in 1% formaldehyde and then either frozen at −80°C or processed immediately. Nuclei were isolated in buffer containing 1% SDS, 10mM EDTA, 50mM Tris-HCl pH 8.0, and protease inhibitors and sonicated using a Diagenode Bioruptor to shear chromatin to 200–1000bp fragments. Protein-DNA complexes were incubated with antibodies against BMAL1 and CLOCK (affinity-purified guinea pig IgGs as described above), H3K4Me2 (Abcam), H3K27Ac (Active Motif), H2AZ (Active Motif), or PDX1 (Novus Biologicals) and immunoprecipitated with IgG paramagnetic beads (Invitrogen). Eluted chromatin was isolated using MinElute PCR purification columns (Qiagen).
ChIP-sequencing and analysis
Sequencing libraries were generated using KAPA DNA Library Preparation kits (Kapa Biosystems, KK8504) according to manufacturer’s instructions. Library concentrations were assessed by both a Bioanalyzer (Agilent) and qPCR-based quantification (Kapa Biosystems). Libraries were sequenced using 75bp single-end reads on an Illumina Next-Seq 500 instrument to a depth of > 10 million mapped reads. Alignment and Peak Finding: Raw sequence reads were aligned to the mm10 reference genome and displayed using UCSC annotated genes using bowtie version 1.1.1 (54) with parameters “--best” and “-m 1” to ensure reporting of uniquely mapped reads (tags). ChIP-Seq peaks were designated as regions with 4-fold enrichment over both the input sample and the local background and were normalized to 10 million reads using default parameters for the Homer “findPeaks” command (53) and specifying “-style factor” for BMAL1, CLOCK, and PDX1 and “-style histone” for H2A.Z, H3K4Me2, and H3K27Ac. For BMAL1 and CLOCK peaks, promoter binding was defined as peaks occurring within 2kb of the nearest gene TSS, and distal binding was defined as those occurring greater than 2kb from a nearest TSS. Motif Analysis: To identify consensus motifs for BMAL1 and CLOCK, we scanned 50bp windows surrounding TF peaks using “findMotifsGenome.pl” with standard background (random genomic sequences sampled according to GC content of peak sequences). We determined the occurrence of tandem E-boxes with variable length spacing by generating synthetic canonical E-box motifs separated by the indicated number of random spacers (i.e. CACGTGNNNCACGTG = 3 spacers) using “seq2profile.pl” allowing for 2 mismatches and testing for their occurrence at BMAL1 and CLOCK peaks using “annotatePeaks.pl”. Analysis of Liver ChIP-Seq Data: Fastq files for all BMAL1 and H3K27Ac ChIP-Seq were downloaded from the ENA server (Study Accession Number: SRP014752) and raw sequence reads for 12 sequential time points were concatenated into a single file. Alignments and peak calling were performed using bowtie and Homer as described above. Shared BMAL1 binding sites were identified by comparing binding locations between β-cells and liver using the Homer command “mergePeaks” and specifying “-d 200” which identified peaks occurring within 200bp as shared across tissues.
Tamoxifen treatment
For in vivo delivery of tamoxifen (Sigma, dissolved in corn oil), mice received three IP injections of 200μg tamoxifen/g body weight, administered every other day. Subsequent experiments were conducted 10–14 days following tamoxifen treatment. For in vitro administration of tamoxifen, isolated islets were incubated for 24 hrs with 1mM tamoxifen (dissolved in ethanol) prior to transfer to complete media for 24 hrs to recover. Islets were then synchronized with forskolin prior to insulin secretion assays as described above.
Immunohistochemical analysis
Mice were anesthetized with intraperitoneal injection of phenobarbital (Nembutal, 50mg/ml) and perfused with heparinized saline, followed by 4% paraformaldehyde (PFA) (Sigma) in PBS. Brain and pancreas were removed and post-fixed with 4% PFA overnight at 4°C. Brain tissues were then cryoprotected in 30% sucrose (Sigma), frozen in O.T.C. (Tissue Tek), and 30μm brain sections collected for antibody staining. Pancreata were embedded in paraffin, and blocks of 6μm sections were mounted on slides. The following primary antibodies were used for staining: guinea pig anti-insulin (1:500, DAKO), mouse anti-glucagon (1:500, Sigma), and rabbit anti-BMAL1 (1:500, Novus Biological). Triple staining was visualized with the following secondary antibodies: AMCA goat anti-guinea pig (1:400, Jackson ImmunoResearch), Alexa Fluor 488-conjugated goat anti-mouse (1:400, Invitrogen), and Alexa Flour 546-conjugated goat anti-rabbit (1:400, Life Technologies). Nuclei were counterstained with DAPI as indicated. Images were acquired with PictureFrame 1.0 using a Zeiss Axioskop 50. β–cell mass was assessed by morphometric analysis of insulin immunostained pancreatic sections (DAKO; Histomouse™Plus kit, Life Technologies). Four pancreatic sections, spaced 50μm apart, were stained for each animal, and endocrine versus total pancreas area was calculated using Image-Pro Premier software (Media Cybernetics) using the smart segmentation feature.
Glucose and insulin measurements and glucose tolerance tests
Blood glucose and plasma insulin levels in ad libitum fed mice were assessed at ZT2 and ZT14 from tail vein bleeds. Glucose tolerance tests were performed in mice following a 14 hr fast, and blood glucose and plasma insulin levels were measured at the indicated times following intraperitoneal (IP) glucose injection of either 2 or 3g/kg body weight, respectively. Plasma insulin levels were measured by ELISA.
Behavioral analysis
Locomotor activity was analyzed in 2–4 month old pancreas-specific Bmal1 knockout mice and their respective littermate controls following tamoxifen treatment. All animals were individually housed in standard mouse cages equipped with running wheels and allowed free access to food and water. Mice were placed in a 12:12 LD cycle for 14 days, followed by 14 days in constant darkness (DD). Total activity data was recorded and analyzed in 6-minute bouts using ClockLab software (Actimetrics). The free-running period was determined as the duration of time between the major activity periods on consecutive days in DD. Period was calculated using a Chi-square periodogram for days 7–14 in DD. Food consumption was analyzed in pancreas-specific Bmal1 knockout mice and their littermate controls prior to and following tamoxifen treatment. All animals were individually housed with free access to water and regular chow. Day- and night-time food consumption was determined by manual measurement of food at both ZT0 and ZT12 for 3 consecutive days.
Intracellular calcium determination
BetaTC-6 cells were plated at a density of 100,000 cells per well in black 96-well plates with clear bottoms and cultured overnight at 37°C and 5% CO2. Islets were dispersed to single cells by incubating in 0.05% Trypsin-EDTA at 37°C for 3 minutes and plated at a density of 100 islets per well in laminin-treated black 96-well plates with clear bottoms and cultured in complete media for 48-hours at 37°C and 5% CO2. Cells were then washed with BSA-free KRB buffer with no glucose and loaded with 5mm Fura-2 (Invitrogen) and 0.04% Pluronic F-127 (Invitrogen) for 30 min at 37°C. Following a wash with BSA-free KRB, Fura-2 intensity was measured following injection of either glucose or ionomycin (Sigma) to final concentrations of 20mM or 10μM, respectively. Cells were alternately excited with 340 nm and 380 nm wavelength light, and the emitted light was detected at 510 nm using a Cytation 3 Cell Imaging Multi-Mode Reader (Bio Tek) at sequential 30-second intervals. Raw fluorescence data were exported to Microsoft Excel and expressed as the 340/380 ratio for each well.
Acknowledgments
This work was supported by NIDDK grant R01DK090625, NIA grant P01AG011412, Chicago Biomedical Consortium S-007, JDRF (17-2013-511, 1-INO-2014-178-A-V, 1-INO-2015-23-AV), and the University of Chicago Diabetes Research and Training Center (P60DK020595) (to J.B.), NIDDK T32 grant DK007169 (to B.M.), NIHBI T32 grant HL007909 (to M.P.), and the Defense Advanced Research Projects Agency (DARPA) (D12AP00023) to A.D.. A.H. is a trainee of the National Institutes of Health Medical Scientist Training program at the University of Chicago (NIGMS T32GM07281). We thank Elizabeth Rosenzweig for technical assistance with RNA-seq library generation, as well as all members of the Bass and Barish laboratories for helpful discussions. J.B. has financial interest in and serves as an advisor to Reset Therapeutics. ChIP-seq and RNA-seq data sets have been deposited in NCBI’s Gene Expression Omnibus accession GSE69889.
REFERENCES AND NOTES
Full text links
Read article at publisher's site: https://doi.org/10.1126/science.aac4250
Read article for free, from open access legal sources, via Unpaywall:
https://science.sciencemag.org/content/sci/350/6261/aac4250.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The metabolic and circadian signatures of gestational diabetes in the postpartum period characterised using multiple wearable devices.
Diabetologia, 12 Nov 2024
Cited by: 0 articles | PMID: 39531039
Maternal diet during pregnancy and adaptive changes in the maternal and fetal pancreas have implications for future metabolic health.
Front Endocrinol (Lausanne), 15:1456629, 23 Sep 2024
Cited by: 0 articles | PMID: 39377073 | PMCID: PMC11456468
Review Free full text in Europe PMC
Non-canonical metabolic and molecular effects of calorie restriction are revealed by varying temporal conditions.
Cell Rep, 43(9):114663, 20 Aug 2024
Cited by: 0 articles | PMID: 39167490 | PMCID: PMC11427179
Circadian disruption, clock genes, and metabolic health.
J Clin Invest, 134(14):e170998, 15 Jul 2024
Cited by: 2 articles | PMID: 39007272 | PMCID: PMC11245155
Review Free full text in Europe PMC
Personal light exposure patterns and incidence of type 2 diabetes: analysis of 13 million hours of light sensor data and 670,000 person-years of prospective observation.
Lancet Reg Health Eur, 42:100943, 05 Jun 2024
Cited by: 2 articles | PMID: 39070751 | PMCID: PMC11281921
Go to all (221) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE69889
Nucleotide Sequences
- (1 citation) ENA - SRP014752
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes.
Nature, 466(7306):627-631, 01 Jul 2010
Cited by: 885 articles | PMID: 20562852 | PMCID: PMC2920067
Bmal1 is required for beta cell compensatory expansion, survival and metabolic adaptation to diet-induced obesity in mice.
Diabetologia, 59(4):734-743, 13 Jan 2016
Cited by: 47 articles | PMID: 26762333 | PMCID: PMC4779402
Induction of Core Circadian Clock Transcription Factor Bmal1 Enhances β-Cell Function and Protects Against Obesity-Induced Glucose Intolerance.
Diabetes, 70(1):143-154, 21 Oct 2020
Cited by: 29 articles | PMID: 33087455 | PMCID: PMC7881843
Clock genes, pancreatic function, and diabetes.
Trends Mol Med, 20(12):685-693, 05 Nov 2014
Cited by: 30 articles | PMID: 25457619 | PMCID: PMC4862830
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Chicago Biomedical Consortium S-007
Defense Advanced Research Projects Agency (1)
Grant ID: D12AP00023
Juvenile Diabetes Research Foundation (3)
Grant ID: 1-INO-2015-23-A-V
Grant ID: 17-2013-511
Grant ID: 1-INO-2014-178-A-V
NHLBI NIH HHS (1)
Grant ID: T32 HL007909
NIA NIH HHS (2)
Grant ID: P01AG011412
Grant ID: P01 AG011412
NIAMS NIH HHS (1)
Grant ID: P30 AR057216
NIDDK NIH HHS (8)
Grant ID: K01 DK105137
Grant ID: T32 DK007169
Grant ID: P60 DK020595
Grant ID: P60DK020595
Grant ID: R01DK090625
Grant ID: R01 DK090625
Grant ID: R01 DK100814
Grant ID: P30 DK020595
NIDDK T32 (1)
Grant ID: DK007169
NIEHS NIH HHS (2)
Grant ID: ES05703
Grant ID: R01 ES005703
NIGMS NIH HHS (2)
Grant ID: T32GM07281
Grant ID: T32 GM007281
National Heart, Lung, and Blood Institute T32 (1)
Grant ID: HL007909
National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (1)
Grant ID: R01DK090625
National Institute of Environmental Health Science (1)
Grant ID: ES05703
National Institute of General Medical Sciences (1)
Grant ID: T32GM07281
National Institute on Aging (1)
Grant ID: P01AG011412
University of Chicago Diabetes Research and Training Center (1)
Grant ID: P60DK020595





