Abstract
Free full text

A survey of current trends in computational drug repositioning
Abstract
Computational drug repositioning or repurposing is a promising and efficient tool for discovering new uses from existing drugs and holds the great potential for precision medicine in the age of big data. The explosive growth of large-scale genomic and phenotypic data, as well as data of small molecular compounds with granted regulatory approval, is enabling new developments for computational repositioning. To achieve the shortest path toward new drug indications, advanced data processing and analysis strategies are critical for making sense of these heterogeneous molecular measurements. In this review, we show recent advancements in the critical areas of computational drug repositioning from multiple aspects. First, we summarize available data sources and the corresponding computational repositioning strategies. Second, we characterize the commonly used computational techniques. Third, we discuss validation strategies for repositioning studies, including both computational and experimental methods. Finally, we highlight potential opportunities and use-cases, including a few target areas such as cancers. We conclude with a brief discussion of the remaining challenges in computational drug repositioning.
Introduction
Over the past decades, de novo drug discovery has grown to be time-consuming and costly, despite the advances in genomics, life sciences and technology. Investments in pharmaceutical R&D have steadily increased, while the number of new drug approvals has stagnated [1]. Indeed, failures are spread throughout the drug development pipeline, and it takes billions of investment dollars and an average of about 9–12 years to bring a new drug to the market [2]. Improving R&D productivity remains the most important priority for pharmaceutical industry [3]. In light of these challenges, drug repositioning, which concerns the detection and development of new clinical indications for those existing drugs, or for those that are in the development pipeline, has emerged as an increasingly important strategy for the new drug discovery. It could substantially reduce the risks of development and the costs, and shorten the lag between drug discovery and availability [4]. Among the 84 drug products introduced to market in 2013, new indications of existing drugs accounted for 20% [5]. Drug repositioning has played a key role in drug discovery and precision medicine paradigm [6].
In recent years, drug repositioning is becoming strongly supported by governments, non-trading organizations and academic institutions. For example, both the United States (National Center for Advancing Translational Sciences) and the United Kingdom (Medical Research Council) have launched large-scale funding programs in this area with a goal to extend molecules that already have undergone significant research and development by the pharmaceutical industry to more new indications. Furthermore, the US Food and Drug Administration (FDA) is also enabling drug repositioning, with the creation of several public databases specifically for computational drug repositioning. There are also substantial economic incentives to reposition marketed drugs for the treatment of orphan and rare disorders [7]. All of these efforts significantly promoted drug repositioning research.
Historically, the discovery of new uses of old drugs is mostly through serendipity [8] or resulted from a better understanding of the drugs’ mechanism of action. For example, the monoclonal antibody bevacizumab, originally developed to treat patients with metastatic colon cancer and non-small cell lung cancer by inhibiting angiogenesis, is now being used to slow or reverse abnormal vascularization of the retina in exudative (wet) macular degeneration [9]. With the accumulation of the large volumes of omics data, bioinformatics plays an increasingly important role in the discovery of new drug indications [4]. Depending on where the discovery comes from, these newly proposed computational methods can be categorized as either ‘drug based’ or ‘disease based’ [10]. Traditional studies mostly focus on exploring the shared characteristics among drug compounds such as chemical structures [11, 12] and side effects [13]. Other methods include rescreening the existing pharmacopeia against new targets to uncover the novel drug indications [14], looking for similarities of molecular activities [15], or exploring the relationships between drugs and diseases.
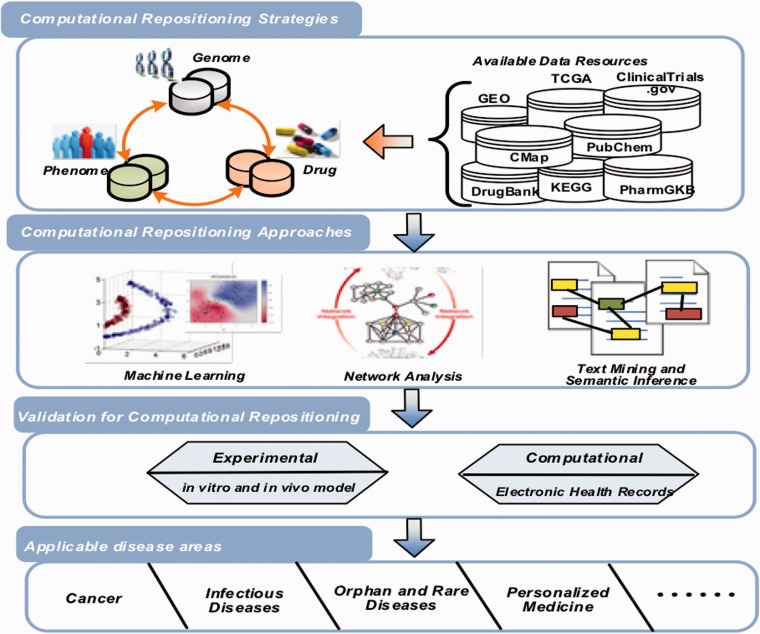
With the drug-related data growth and open data initiatives, a set of new repositioning strategies and techniques has emerged with integrating data from various sources, like pharmacological, genetic, chemical or clinical data. These methods can accumulate evidence supporting discovery of new uses or indications of existing drugs. In this review, we summarize recent progress in computational drug repositioning as the following four parts (see Figure 1): repositioning strategies (with available data sets), computational approaches, validation methods and application areas.
Computational repositioning strategies
Genome
Rapid advances in genomics have led to the generation of large volumes of genomic and transcriptomic data for a diverse set of disease samples, normal tissue samples, animal models and cell lines. Much of these data are publicly available. Together with other phenotypic, and clinical database, these data sets provide a unique opportunity to understand disease mechanism, elucidate drug mechanism of actions and identify new use of old drugs. Among those, transcriptomic profiles, such as gene expression data are most widely used, while other genomic and genetic profiles have been explored for drug repositioning as well.
One key source of data behind several repurposing efforts is the Connectivity Map (CMap) [16] project and its extended project Library of Integrated Network-Based Cellular Signatures (LINCS) [17], which produced large-scale gene expression profiles from human cancer cell lines treated with different drug compounds under different conditions. CMap aims to construct a detailed map for functional associations among diseases, genetic perturbations and drug actions. By integrating with other functional genomics databases [e.g. NCBI Gene Expression Omnibus (GEO)] [18], its data have been extensively explored in drug repositioning studies. One approach using these data is to look for inverse drug–disease relationships by comparing drug gene expression profiles and disease gene expression profiles. This approach is also referred as ‘signature reversion’. For example, by systematically comparing gene expression signatures of inflammatory bowel disease (IBD) derived from GEO against a set of drug gene expression signatures comprising 164 drug compounds from CMap, Dudley et al. [19] inferred several new interesting drug–disease pairs and validated one pair in IBD preclinical models. In another case, Jahchan et al. [20] used a similar systematic drug-repositioning bioinformatics approach to query a large compendium of gene expression profiles to identify antidepressant drugs for the treatment of small cell lung cancer.
Another approach, ‘guilt-by-association’, looks for drugs that provoke similar transcriptional responses, positing that they could share similar mode of action (MoA) [21]. The availability of many public repositories, such as the Drug versus Disease (DvD) [22], the database for Annotation, Visualization and Integrated Discovery (DAVID) [23] and the Gene Set Enrichment Analysis (GSEA) [24], provides such an opportunity for the comparison of drug and disease signatures from gene expression profiles.
Recently, noncoding RNAs, especially the microRNA (miRNAs), have been shown in regulating kinds of cell activities [25, 26], thus becoming promising therapeutic targets for drug repositioning [27]. For example, Liu et al. [28] developed an in silico drug repositioning strategy based on miRNA-TF feed-forward loops (FFLs). miRNAs and transcript factors (TFs) were found to be significantly enriched in cystic fibrosis (CF) associated gene regulations from public available data sources. Then they constructed FFLs in CF by defining specific TFs and miRNAs as two regulatory elements. Forty-eight existing drugs showing ability to influence the expression of miRNA that are part of FFLs were repurposed for the treatment of CF patients. Jiang et al. [29] predicted new indications for existing drugs by constructing small molecule-miRNA network for each cancer. Rukov et al. [30] developed a web server that links miRNA expression and drug function by combining data on miRNA targeting and protein–drug interactions. SM2miR is a database containing manually curated relationships between experimentally validated molecules and miRNA [31].
In addition to transcriptomic data, other genomic profiles (e.g. genetic mutations) can be applied to drug repositioning. For instance, Garnett et al. [32] carried out a large-scale screening of human cancer cell lines with 130 clinical/preclinical drugs. A multivariate analysis of genetic and gene expression profiles of cancer cell lines showed that a few mutated cancer genes that are associated with drug sensitivity may serve as potential biomarkers of drug response. To some extent, these mutations reflect the molecular activity of drugs, and can be regarded as drug signatures during the repositioning process. In another case, Okada et al. [33] performed a three-stage genome-wide association study (GWAS) meta-analysis of rheumatoid arthritis (RA) patients and linked the risk loci to known RA drug targets. In their study, logistic regression models assuming additive effects of the allele dosages were used to assess the relationship of the single-nucleotide polymorphisms (SNPs) and RA. In total, 101 RA risk loci were identified (e.g. 42 are novel), and they showed significant overlapping with approved RA drug target genes. Furthermore, several drugs approved for other diseases were connected to RA risk genes, indicating they could be repositioned for RA. In another GWAS [34], a catalog of disease-associated genes from published genome-wide associations studies were further integrated with targets of drugs from pharmaceutical projects. In this way, the drugs with targets mapped to the disease-associated genes from GWAS data may be repositioned.
Phenome
The phenome, defined as the comprehensive collection of phenotypic information, was emerged as a new source for drug repositioning. In recent years, the phenome-wide association study (PheWAS) has become increasingly popular as a systematic approach to identify important genetic associations with human diseases [35]. For instance, Denny et al. [36] performed a large-scale application of the PheWAS using electronic medical records (EMRs), and demonstrated that PheWAS is a useful tool to enhance the analysis of the genomic basis and to detect novel associations between genetic markers and human diseases.
Meanwhile, clinical side effects are shown to be capable of profiling drug-related human phenotypic information and can subsequently help discover new therapeutic uses. For example, Yang et al. [37] used drug side effects as features to predict its indications. Ye et al. [38] identified novel indications based on the hypothesis that similar side-effect profiles may share similar therapeutic properties. Bisgin et al. [39] developed a Latent Dirichlet Allocation model for drug repositioning that adopted the phenome information from the Side Effect Resource (SIDER) [40]. Using drug side-effect profiles to suggest its novel indications has shown to be attractive but its practical use would require deep understanding of the underlying molecular/pathological mechanisms.
Finally, the phenome can be incorporated with other kinds of data for drug repositioning. As an example, Hoehndorf et al. [41] developed an integrated system to predict novel drug–disease associations by linking genotype-disease associations with drug–gene associations. In this model, beginning with integrating phenotype ontologies for disease and gene or genotype, they derived a semantic similarity-based score to measure genotype–disease associations. With this approach, most of the known drug–disease associations have been retrieved and the new associations may indicate a new repositioning opportunity. Although some researchers have demonstrated the potential correlations between genome and phenome [42], there is still an urgent need to understand these correlations better and turn them into disease treatment or personalized health care. For example, the BRCA mutation (mutation in either BRCA1 or BRCA2 genes) was found to be associated with the risk of getting breast cancer for ovarian cancer patients [43]. Because BRCA mutations are clinically significant [44], a deeper understanding of the relationship between BRCA mutation status and cancer phenotype will be important for making precise treatment decisions for patients.
Drug chemical structures
The drug chemical structures can also point toward repositioning opportunities. Moreover, publicly available databases of chemical structures, high-throughput screening data and literature-derived biochemical data containing massive amounts of information useful for repositioning [45–48]. The key insight behind these approaches is that the molecules with similar chemical structures often affect proteins and biological systems in similar ways. Similarity may be measured in many different methods using different structural features, including 2D topological fingerprints or 3D conformations, and is an active area of research. The way to incorporating similarity between chemicals into repositioning inferences is also an active area of research.
For example, Swamidass et al. [46] proposed to use chemical structure to infer which targets modulate disease-relevant phenotype. Knowing which targets modulate disease-relevant phenotypes is a signal that can indicate what other drugs might work to treat the disease.
Recently, chemical structure information turned to be integrated with other types of data for computational repositioning. Wang et al. [49] proposed an integrated repositioning model that incorporated drug chemical structure, molecular activity and side effect. All three different types of data were integrated to define a kernel function used by a support vector machine (SVM) classifier. This method was further compared with other methods, and showed high efficiency. Similarly, Tan et al. [50] incorporated drug chemical structure similarity and gene semantic similarity to construct a drug similarity network, which was further used to extract novel indications. Ng et al. [51] proposed a novel algorithm called ‘ligENTS’ to define novel drug–target associations by mapping the drug to its global pharmacological space according to its chemical information. Then structural systems biology platform was integrated to reposition approved drugs for malaria. A full review of chemical structure-based approaches to repositioning is beyond our scope, but these studies show a trend toward including chemical structures alongside other types of data.
Drug combinations
As many diseases are driven by complex molecular and environmental interactions, targeting a single component may not be sufficient to disrupt those mechanisms, and interest in early drug discovery stages has increasingly evolved to target multiple molecules using combined drugs or multi-target inhibitors. For example, the activated B-cell–like (ABC) subtype of diffuse large B-cell lymphoma (DLBCL) is a malignant cancer with poor prognosis. Constitutive activation of the NF-kB by IkB kinase (IKK) has shown to be a key pathogenic factor. Ceribelli et al. [52] screened 466 drugs that have been approved or in early stage for cancer therapy and found that ibrutinib, a kinase inhibitor that can block B-cell receptor signaling pathway to activate IKK, shows a significant synergistic effect with JQ1 in killing ABC DLBCL cells both in vitro and in vivo, suggesting that the combination of JQ1 and ibrutinib might be a new effective therapy.
Current combination strategies rely mainly on clinical and empirical experience, computational prediction is thus highly demanded [53]. Huang et al. [54] used side effects as features for drug–drug combinations (DDCs), and further classify the safe DDCs from the unsafe ones. Sun et al. [55] developed a model to predict effective drug combinations by integrating gene expression profiles of multiple drugs. Existing drug combinations were extracted from the Drug Combination Database (DCDB) [56]. They started to identify important features related to drug combinations by using statistical methods that look for side effects, genes or disease pathways that could be affected by drugs in the combination. Those features were further used to construct a machine-learning classifier for predicting effective drug combinations.
Other than in silico methods, experimental characterizations of drug efficiency (e.g. library screening and cell viability assays) were also adopted to identify new drug combinations. Kang et al. [57] identified antileukemic drugs that could be combined with imatinib to overcome drug resistance in BCR-ABL +
+ leukemia. They first used library screening, literature search and correlation analysis to select 11 candidate drugs that might be combined with imatinib. Dose responses for these candidates with/without imatinib were applied in an iterative search algorithm to identify effective combinations that can overcome drug resistance. These predicted combinations were further confirmed in preclinical models.
leukemia. They first used library screening, literature search and correlation analysis to select 11 candidate drugs that might be combined with imatinib. Dose responses for these candidates with/without imatinib were applied in an iterative search algorithm to identify effective combinations that can overcome drug resistance. These predicted combinations were further confirmed in preclinical models.
Available data sources
Currently, many different kinds of data sources (e.g. genetic, pharmacogenomics, clinical, chemical agent) are available for supporting computational drug repositioning, and to some extent they have promoted the development of various repositioning strategies. In Table 1, we summarize a brief list of those frequently used data sources.
Table 1.
Data resources supporting computational repositioning strategies
| Repositioning strategies | Database | URL | Brief description |
|---|---|---|---|
| Genome | ArrayExpress | http://www.ebi.ac.uk/arrayexpress | Public repositories of functional genomics data. |
| Gene Expression Omnibus (GEO) | http://www.ncbi.nlm.nih.gov/geo | ||
| Gene Expression Atlas | http://www.ebi.ac.uk/gxa | Gene expression patterns under different biological conditions. | |
| The Cancer Genome Atlas (TCGA) | http://cancergenome.nih.gov | Genomic data (e.g. Exome, SNP, Methylation, mRNA, miRNA, Clinical) of >10 000 patient tissue samples across > > 30 common cancers. 30 common cancers. | |
| Cancer Cell Line Encyclopedia (CCLE) | http://www.broadinstitute.org/ccle | Genomic data (e.g. DNA copy number, mRNA expression, mutation data) of > > 1000 cancer cell lines. 1000 cancer cell lines. | |
| International Cancer Genome Consortium | https://icgc.org | A comprehensive description of genomic, transcriptomic and epigenomic changes. | |
| The Connectivity Map (CMap) | http://www.broadinstitute.org/cmap | Gene expression profiles of >1000 drugs across three primary cell lines. | |
| Library of Integrated Network-based Cellular Signatures (LINCS) | http://www.lincsproject.org | Aim to produce >1 million gene expression profiles of drugs and genetic perturbagens across >15 cell lines. | |
| Molecular Signature Database (MsigDB) | http://www.broadinstitute.org/gsea/msigdb | Collections of annotated gene signatures from different sources. | |
| Gene Signature Database (GeneSigDB) | http://compbio.dfci.harvard.edu/genesigdb | ||
| Database for Annotation, Visualization and Integrated Discovery (DAVID) | http://david.abcc.ncifcrf.gov | Functional annotation tools. | |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | http://www.genome.jp/kegg | Resource for understanding high-level functions and utilities of the biological system from molecular-level information. | |
| Gene Set Enrichment Analysis (GSEA) | http://www.broadinstitute.org/gsea | Tool to determine if an a priori defined gene signature shows statistically significant, concordant differences between two biological states. | |
| Drug versus Disease (DvD) | www.ebi.ac.uk/saezrodriguez/dvd | Computational pipeline for comparing disease and drug-response gene expression signatures from publicly available resources. | |
| Genome/Phenome | The Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB) | http://www.pharmgkb.org | Genetic variation on drug response. |
| Online Mendelian Inheritance in Man (OMIM) | http://www.omim.org | Relationship between genes and genetic phenotypes, particularly disorders. | |
| Phenome | Side Effect Resource (SIDER) | http://sideeffects.embl.de | Adverse drug reactions of >900 drugs. |
| ClinicalTrials.gov | http://www.clinicaltrials.gov | A registry and result database of publicly and privately supported clinical studies. | |
| Phenome/Drug | Drugs@FDA Database | http://www.fda.gov/Drugs/InformationOnDrugs/ucm135821.htm | Information about FDA approved drugs, such as brand name, therapeutic products. |
| Drug | Drug Combination Database (DCDB) | http://www.cls.zju.edu.cn/dcdb | Known examples of drug combinations, models for drug combinations. |
| The NCGC Pharmaceutical Collection (NPC) | http://tripod.nih.gov/npc/ | A comprehensive, publically accessible collection of approved and investigational drugs. | |
| Protein Data Bank (PDB) | http://www.rcsb.org/pdb/home/home.do | 3D structure of proteins, nucleic acids. | |
| SWEETLEAD | https://simtk.org/home/sweetlead | A database containing chemical structures representing approved drugs, chemical isolates from traditional medicinal herbs and regulated chemicals. | |
| DrugBank | http://www.drugbank.ca/ | Detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. | |
| ChEMBL | https://www.ebi.ac.uk/chembl/ | A large literature-derived database of molecule structures and molecule-protein interactions. This includes a catalog of approved drugs. | |
| PubChem | https://pubchem.ncbi.nlm.nih.gov/ | A repository of biological assay results for hundreds of thousands of molecules. |
Computational repositioning approaches
Machine learning
For the various data resources that support the exploration of repositioning opportunities, machine learning-based models can leverage the data to study the underlying systems for the prediction of novel associations between drugs and diseases. In recent years, an increasing number of machine learning methods have been proposed when coupled with the elucidation of various features.
Menden et al. [58] developed machine-learning models to predict the response of cancer cell lines to drug treatment, quantified through IC50 values. In the model, cancer genomic features of the cell lines (e.g. mutation status of 77 oncogenes and microsatellite status) and chemical properties (e.g. structural fingerprint) were used to build a feed-forward perceptron neural network model and a random forest regression model. The predicted IC50 values were further validated by a cross-validation and an independent blind test. Napolitano et al. [59] focused on a drug-centered approach to predict drug therapeutic class by using drug-related features (e.g. drug chemical structure similarity, drug molecular target similarity and drug gene expression similarity). They merged these features into a single drug similarity matrix, which was used as a kernel for SVM classification. Other than drug-related features, Gottlieb et al. [60] integrated various disease-related features (e.g. phenotypic and genetic features). Based on these features, drug–drug and disease–disease similarity measures were computed to construct classification features. They further used a logistic regression classifier to predict novel drug indications.
Moreover, there exist machine learning algorithms that use collaborative filtering techniques to predict unknown drug–disease associations. For instance, Zhang et al. [61] proposed a unified computational framework for integrating multiple aspects of drug similarity and disease similarity. Briefly, genome (e.g. drug target protein, disease gene), phenome (e.g. disease phenotype, drug side effect) and chemical structure (e.g. drug chemical structure) were integrated to extract drug similarity matrix and disease similarity matrix. Based on all this information, the authors turned the drug–disease network analysis into an optimization problem. This computational framework shows high efficiency in exploring novel drug indications. Yang et al. [62] used a causal inference-probabilistic matrix factorization approach to infer drug–disease associations. In this model, they integrated multilevel relations to construct causal networks connecting drug–target–pathway–gene–disease, PMF modes were learned based on known interactions. This approach can predict novel drug–disease associations, thus providing potential values for drug repositioning.
Network analysis
Network-based analysis is another widely used strategy for computational drug repositioning. With the advances of high-throughput technology and bioinformatics methods, molecular interactions in the biological systems can be modeled by networks. Previous studies have suggested that drug–target network, drug–drug network, drug–disease network, protein interaction network, transcriptional networks and signaling networks are useful in the identification of therapeutic targets or characteristics of drug targets [63–65], thus providing new opportunities for drug discovery or repositioning.
Li et al. [66] developed a bipartite drug–target network method to identity potential new indications of an existing drug through its relation to similar drugs. In the bipartite network model, drug pair similarity integrated drug chemical structure similarity, common drug targets and their interactions. Built on their past success, the same authors recently built a causal network (CauseNet) [67]—a multilayered pathway of gene, disease and drug target—to identify new therapeutic uses of existing drugs. In the causal network, the transition likelihood of each chain is estimated on the basis of known drug–disease treatment association. Wu et al. [68] applied network clustering to a drug–disease heterogeneous network to identify closely connected modules of disease and drugs, which can be used for extracting possible drug–disease pairs for drug repositioning. In the network, two nodes (one drug or one disease) with shared genes/targets and enriched features (biological process, pathway and phenotypes) were connected and the connection was weighted by a Jaccard score. Jin et al. [69] developed a novel method to repurpose drugs for cancer therapeutics by leveraging off-target effects (OTEs) that may affect important cancer cell signaling pathways. The OTEs of drugs on signaling proteins were recognized by a hybrid model composed by a network component called cancer-signaling bridges and a regression model called Bayesian Factor Regression Model.
Text mining and semantic inference
The biomedical and pharmaceutical knowledge available in literature [70, 71] or databases contains vast amount of information for drugs and diseases, which can be automatically mined [72–74] and retrieved [75], thanks to the recent advances in text mining research [76]. Therefore, it is possible to detect novel indications for existing drugs by finding relevant knowledge through text mining approaches [77]. One important basis rooted in this method is the occurrence of biological ontology, which makes it possible for the comparison and analysis of biological information from different sources.
A recent study by Andronis et al. [78] summarized several literature mining approaches and sources for drug repurposing. For example, if one study finds that disease A was caused by the lack of nutrition B while another study reports that drug C used for another disease was an activator of nutrition B, then drug C might be repurposed for disease A through literature mining.
Moreover, semantic technologies have facilitated the integration of various data sources and the discovery of new drug indications. For instance, Zhu et al. [79] developed an ontology to model FDA-approved breast cancer drugs and their relations with pathways, drugs, genes, SNPs and diseases. New drug–disease pairs were inferred from the ontology-based knowledge base. Chen et al. [80] developed a statistical model to assess drug–target associations from a semantic linked network comprised by drugs, chemical compounds, protein targets, diseases, side effects and pathways and their relations. The model considered the topology and semantics of the subgraph between a drug and a target. The similar drug–drug pair from different disease areas may indicate a potential repositioning opportunity.
Validation for computational drug repositioning
Computational models often predict a handful interesting hits; however, the ultimate goal of drug repositioning is to move only one or two hits into clinical application to benefit patients. Therefore, experimental validation of these computer-generated hits becomes important. Despite some known limitations, in vitro and in vivo models (e.g. cell-based targeted assays and mouse models) have been increasingly used to validate the candidate hits for preclinical drug evaluation. For example, Zerbini et al. [81] identified several FDA-approved drugs showed sensitivity for clear cell renal cell carcinoma (ccRCC). To confirm the drug efficacy, they further used apoptosis assays and xenograft mouse models and demonstrated pentamidine as a potential therapeutic agent for ccRCC, as it can significantly induce apoptosis in tumor cells and slow tumor growth. Kang et al. [57] used cell viability assays to validate the drug combinations generated from a computational search algorithm. Subsequently, drug combinations with significant efficacy in cancer cell killing were confirmed. Végner et al. [82] performed an experimental validation study for a previous reported computational drug repositioning strategy where they confirmed the efficacy of top-predicted drugs through ACE inhibition assay, COX inhibition assay, dopaminergic agonist and antagonist assay.
In addition to in vitro and in vivo models, electronic health/medical records may be helpful to validate the predictions. For example, Khatri el al. [83] identified atorvastatin as a new therapeutic for organ transplantation using meta-analysis of public microarray data sets and validated the beneficial effect of atorvastatin on graft survival by retrospective analysis of EMRs of a single-center cohort of 2515 renal transplant patients followed for up to 22 years. Xu et al. [84] used electronic health records to validate metformin, a drug for type 2 diabetes mellitus, associated with cancer mortality.
Understanding and selecting the appropriate validation model are critical for the success of the prediction, as the contexts of the validation models may be different from those used to make the prediction or some validation models are not reliable per se. For example, it is debatable that genomic responses in mouse models mimic human inflammatory diseases [85, 86]. Half of the hepatocelluar carcinoma (HCC) cell lines are not significantly correlated to the HCC tumors from TCGA, while a few rarely used ovarian cancer cell lines more closely resemble ovarian tumor profiles than common used cell lines [87]. In addition to the selection of the right model, the selection of the right hits for validation is also critical. Some drugs may not be favored by physicians or biologists owing to various reasons such as high toxicity, high cost and low bioavailability. The early involvement of all stakeholders would likely be to increase the rate of success in translational research.
Current and future target areas: cancers, infectious and orphan diseases and personalized medicine
There are several high potential applications of drug repositioning in disease or related therapeutic areas. One important application is the anticancer drug discovery. Motivated by the benefit of drug repositioning and the high demand for anticancer drugs, searching novel anticancer therapies from existing drugs has become increasingly popular. Drug repositioning also turns out to be a promising strategy for the discovery of anti-infectious drugs that can overcome drug resistance, as the emergence of drug resistance is awful to human beings and can largely reduce the drug efficacy. Alternative therapeutics for orphan and rare diseases can also be identified from approved drugs, and this strategy has gained various grants supporting research in this field. Finally, drug repositioning provides a new accessibility to personalized treatment. Table 2 shows examples of drug repositioning applied in cancer, infectious, orphan diseases and personalized medicine.
Table 2.
Current and future target areas for computational drug repositioning
| Target area | Key findings | Main data sources | Main methods |
|---|---|---|---|
| Cancer | Repositioning of auranofin for the treatment of gastrointestinal stromal tumors [88]. | The FDA-approved drug library | High-throughput screening |
| Repositioning of irinotecan for the treatment of breast cancer [89]. | Genome (expression, structure, activity), clinicaltrials.gov | Biomarker-guided repurposing | |
| Repositioning of the anthelmintic drug mebendazole for the treatment for colon cancer [90]. | The Pharmakon 1600 library, NCI 60 data, CMap | In silico drug screening | |
| Repositioning of metformin for various types of cancer including breast, colorectal, endometrial, esophageal, etc [91]. | Clinical trials for cancer treatment with metformin | Clinical studies | |
| Repositioning of an existing phenothiazine-like antipsychotic drug, trifluoperazine, as a potential anti-CSC (cancer stem cell) agent [92]. | CMap | In silico drug screening, in vitro and in vivo models | |
| Infectious disease | Identified novel Plasmodium falciparum targets of drug-like active compounds [51]. | DrugBank | Drug chemical structure analysis, network analysis |
| Rare and orphan diseases | Drug repositioning for orphan genetic diseases [93], such as proposing HDAC1 and TSPO as two significant targets for epileptic syndromes. | Gene Expression Omnibus, OMIM phenotypes, DrugBank, CMap, FDA | Systematic analysis of gene coexpression, text mining |
| Establishment of a new resource, the Rare Disease Repurposing Database (RDRD) [94]. | FDA orphan designation database, FDA-approved drugs | Database analysis | |
| Personalized medicine | ISM strategy for personalized AML therapy defined dasatinib and sunitinib could be repositioned for subsets of AML [95]. | Drug-response profiles, clinical responses information | Ex vivo drug sensitivity and resistance testing (DSRT) |
| Generation of a novel method, PREDICT, for inferring novel drug indications with application to personalized medicine [60]. | DrugBank, OMIM, ArrayExpress, DCDB, KEGG DRUG databases, SIDER, UniProt, clinicaltrials.gov | Text mining, machine learning, network analysis | |
| Current opportunities and existing challenges of drug repositioning for personalized treatment [96]. | Genome (expression, structure), clinical trials, Gene Expression Omnibus, DrugBank, etc. | Experimental approaches (cell-based screening, in vivo mouse studies), computational approaches (network analysis) |
Discussion
Each of the aforementioned computational repositioning strategies and approaches has their methodological advantages and limitations. A combination of these methods is often desired for achieving better results. For example, Wang et al. [49] integrated drug chemical structures, target protein sequences and phenotype data and applied machine learning approach (i.e. SVM) to identify drug–disease relationships, and further made the drug–disease network analysis. Gottlieb et al. [60] integrated chemical structures, drug side effects, drug target protein sequences and target protein interactions and phenotype data and applied network analysis for target protein distance calculation, applied text mining to identify disease phenotypes and used machine learning algorithms to classify true and false drug–disease associations. These integrative methods showed better performance in both sensitivity and specificity when comparing with individual methods.
Despite several successful use cases of computational drug repositioning, challenges remain. First, the transformation of theoretical computational models into practical use is far from straightforward, due to some inevitable factors like missing data, data bias and technical limitations of computational methods. For example, many in silico repositioning approaches search potential drug–target interactions through chemical structure information. Thus, the lack of high-resolution structural data for targets makes such methods inadequate. Large scale of drug-induced transcriptomic profiles (e.g. gene expression data in CMap) might have experimental variation across batches. Additional gene expression normalization techniques are needed to ensure the systematical genome-wide analysis in an unbiased way [97]. Second, the lack of structured gold standard for drug repositioning made it hard to compare and evaluate the performance of computational methods. In response, several recent studies focus on curating a comprehensive and public catalog of existing drug indications [98, 99]. Although common metrics (e.g. sensitivity, specificity, and area under ROC curve (AUC)) are adopted, previous studies performed evaluation on their own data sets rather than a shared gold standard for various reasons. Third, although computational drug repositioning may merely shorten the process of drug discovery in preclinical and Phase I trials, challenges may still exist after Phase II trials for the repositioned drugs. For instance, as the test population becomes larger for Phase II trials, more resources will be needed for the further drug validation [100]. Recently, some researchers have proposed the idea of repositioning the approved drugs for the treatment of Ebola, and the debate has been surprisingly intense [101]. Antiviral drugs or drugs with immune system’s modulation drugs were repositioned to treat Ebola, but World Health Organization has ignored these proposed FDA-approved drugs, owing to the deficiency of experimental tests, as well as the potential drug toxicity.
Conclusions
In summary, we believe computational drug repositioning research is of great significance to improve human health through discovering new uses for existing drugs. In fact, a number of studies have already been carried out with various degrees of success. It has great potentials to accelerate drug discovery with interesting opportunities in several particular disease areas (e.g. cancer). As both public and private sectors from around the world are supporting drug-repositioning research with various funding opportunities and special programs, now is the time for the research community to further develop techniques and methods toward new discoveries and breakthroughs.
Biographies
Jiao Li, PhD, is a division chief of the Institute of Medical Information, Chinese Academy of Medical Sciences. Her research interests include data modeling for drug discovery and translational bioinformatics.
Si Zheng is a faculty member of the Institute of Medical Information, Chinese Academy of Medical Sciences. Her research focuses on complex disease molecular network modeling and mining.
Bin Chen, PhD, is a postdoctoral scholar at Stanford School of Medicine. He focuses on developing methods to leverage various data in translational drug discovery to help discover new therapeutics faster.
Atul Butte, MD, PhD, is an associate professor of Pediatrics and Genetics, and by courtesy, Medicine, Pathology and Computer Science, and is Chief of the Division of Systems Medicine at Stanford University and Lucile Packard Children's Hospital. He has authored nearly 200 publications.
S. Joshua Swamidass, MD, PhD. Both a physician and a computational scientist, Dr Swamidass is an assistant professor of Laboratory and Genomic Medicine in the Pathology and Immunology Department of Washington University in St. Louis.
Zhiyong Lu, PhD, is Earl Stadtman investigator at the National Center for Biotechnology Information (NCBI), National Institutes of Health (NIH) where he leads the biomedical text mining research group.
Funding
NIH Intramural Research Program, National Library of Medicine (to Z.L.), the National Key Technology Research and Development Program of China (Grant No. 2013BAI06B01), the National Population and Health Scientific Data Sharing Program of China, the Fundamental Research Funds for the Central Universities (Grant No. 13R0101).
References
Articles from Briefings in Bioinformatics are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/bib/bbv020
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/bib/article-pdf/17/1/2/6684680/bbv020.pdf
Citations & impact
Impact metrics
Article citations
Empowering Graph Neural Network-Based Computational Drug Repositioning with Large Language Model-Inferred Knowledge Representation.
Interdiscip Sci, 26 Sep 2024
Cited by: 0 articles | PMID: 39325266
Some Aspects and Convergence of Human and Veterinary Drug Repositioning.
Molecules, 29(18):4475, 20 Sep 2024
Cited by: 0 articles | PMID: 39339469 | PMCID: PMC11433938
Review Free full text in Europe PMC
DRML-Ensemble: drug repurposing method based on feature construction of multi-layer ensemble.
J Mol Model, 30(8):296, 31 Jul 2024
Cited by: 0 articles | PMID: 39083073
In silico transcriptome screens identify epidermal growth factor receptor inhibitors as therapeutics for noise-induced hearing loss.
Sci Adv, 10(25):eadk2299, 19 Jun 2024
Cited by: 2 articles | PMID: 38896614 | PMCID: PMC11186505
Drug to genome to drug: a computational large-scale chemogenomics screening for novel drug candidates against sporotrichosis.
Braz J Microbiol, 55(3):2655-2667, 18 Jun 2024
Cited by: 0 articles | PMID: 38888692
Go to all (229) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Computational and experimental advances in drug repositioning for accelerated therapeutic stratification.
Curr Top Med Chem, 15(1):5-20, 01 Jan 2015
Cited by: 45 articles | PMID: 25579574
Review
Computational Drug Repurposing: Current Trends.
Curr Med Chem, 26(28):5389-5409, 01 Jan 2019
Cited by: 30 articles | PMID: 29848268
Review
A review of computational drug repurposing.
Transl Clin Pharmacol, 27(2):59-63, 28 Jun 2019
Cited by: 83 articles | PMID: 32055582 | PMCID: PMC6989243
Review Free full text in Europe PMC
A Review of Recent Developments and Progress in Computational Drug Repositioning.
Curr Pharm Des, 26(26):3059-3068, 01 Jan 2020
Cited by: 2 articles | PMID: 31951162
Review
Funding
Funders who supported this work.
Intramural NIH HHS
NIGMS NIH HHS (1)
Grant ID: R01 GM079719