Abstract
Free full text

Optimization of RGD containing cyclic peptides against αvβ3 integrin
Abstract
We have previously reported the use of one-bead-one-compound (OBOC) combinatorial technology to develop a disulfide cyclic, Arg-Gly-Asp containing octapeptide LXW7 (cGRGDdvc), that targets αvβ3 integrin with high affinity and specificity (Mol Cancer Ther, 9:2714–23, 2010). αvβ3 integrin is known to be over-expressed in many cancers and in tumor vasculature, and it has been established as a cancer therapeutic target. To further optimize LXW7, we have performed systematic structure activity relationship (SAR) studies. Based on the results, two highly focused OBOC peptide libraries were designed, synthesized and screened against αvβ3 integrin transfected K562 cells. One of the best ligands, LXW64 was found to have 6.6-fold higher binding affinity than LXW7, and showed preferential binding to cells expressing αvβ3 integrin. In addition to binding strongly to U-87MG glioblastoma cells in vitro, LXW64 also targets U-87MG xenografts implanted in nude mice, indicating that it is an excellent vehicle for delivery of cytotoxic payload to tumors and tumor blood vessels that overexpress αvβ3 integrin.
Introduction
Integrins are heterodimeric transmembrane receptors of the extracellular matrix consisting of non-covalently associated α- and β-subunit. So far 24 distinct integrins have been identified in mammals, resulting from different combination among 18 α- and 8 β-subunits. Integrins mediate cell-cell, cell-extracellular matrix (ECM) interaction and regulate key biological processes including migration, proliferation, differentiation, and apoptosis (1–4). It is well known that integrin signaling plays an important role in tumor progression and metastasis (5, 6). Integrin αvβ3 is a known receptor for a wide variety of ECM ligands, including vitronectin, fibronectin, fibrinogen, laminin and proteolyzed collagen. Up-regulation of intergrin αvβ3 has been identified in both tumor cells and angiogenic endothelial cells. It has received increasing attention as cancer therapeutic and imaging target, because of its critical role in tumor-induced angiogenesis and tumor metastasis (7, 8). In vivo imaging methods to depict and characterize angiogenesis are becoming increasingly important to study this process in both clinical and research settings. Several classes of inhibitors against αvβ3 integrin have been developed (9–12). Many antibodies, cyclic peptides, disintegrins, peptidomimetics and small molecules antagonists against integrins are currently in preclinical and clinical development.(8, 13–15).
Utilizing the one-bead-one-compound (OBOC) combinatorial library technology, which was first reported by Lam et al (16) over 20 years ago, we have identified a number of peptides and peptidomimetic ligands against various cell surface receptors including integrins (16–20). Using this approach, we had reported a novel RGD containing cyclic octapeptide LXW7 (cGRGDdvc) (Figure 1), which preferentially targets αvβ3 integrin. LXW7 exhibited excellent tumor uptake and low liver uptake in in vivo and ex vivo optical imaging studies using glioblastoma U-87MG or melanoma A375M xenograft models (21). In order to further reveal structure-activity relationship (SAR) and develop novel LXW7 analogues with higher binding affinity, we designed and synthesized a series of LXW7 analogues and analyzed their binding affinity using competition binding assay on αvβ3 integrin transfected cells (K562/αvβ3+). Based on the results of these studies, two OBOC focused libraries were designed and synthesized. The screening of these OBOC focused libraries resulted in the development of several cyclic peptides with higher affinities than LXW7. LXW64 was identified as the lead compound showing preferential binding to αvβ3 integrin expressing cells. We have also used near infra-red optical imaging to demonstrate that this peptide was able to target implanted xenograft in vivo with high efficiency and specificity.
Materials and Methods
Materials
TentaGel S NH2 resin (90 μm diameter, 0.27 mmol/g) was purchased from Rapp Polymere GmbH (Tübingen, Germany). Rink amide MBHA resin (0.59 mmol/g), amino acid derivatives, HOBt, and DIC were obtained from GL Biochem (Shanghai, China), Anaspec (Fremont, CA), and Chem-Impex (Wood Dale, IL). All organic solvents and other chemical reagents were purchased from Aldrich (Milwaukee, WI). The solvents were directly used in the synthesis without further purification unless otherwise noted. A Perkin-Elmer/Applied Biosystems Protein Sequencer (ABI Procise 494) was used for library bead decoding. Analytical HPLC analysis were performed on a Water 2996 HPLC system equipped with a 4.6 x 150mm Waters Xterra MS C18 5.0μm column and utilized a 20 min gradient from 100% aqueous H2O (0.1% TFA) to 100% acetonitrile (0.1% TFA) at a flow rate of 1.0 mL/min. Preparative HPLC purification was performed on a System Gold 126NMP solvent module (Beckman) with a C18 column (Vadac, 20mm x 250 mm, 5 m, 300 Å, C18, 7.0 ml/min) 45 min gradient from 100% aqueous H2O (0.1% TFA) to 100% acetonitrile (0.1% TFA), 214nm. Mass spectra (MALDI-TOF-MS) were measured on an ABI 4700 TOF/TOF instrument. This instrument employs an Nd: Yag laser (352nm) at a repetition rate of 200 Hz. The applied accelerating voltage was 20kV. Spectra were recorded in delayed extraction mode (300 ns delay). All spectra were recorded in the positive reflector mode. Spectra were sums of 1000 laser shots. Matrix alpha-cyano-4-hydroxycinnamic acid was prepared as saturated solutions in 0.1% TFA in 50% CH3CN.
K562 and U-87MG cells were purchased from American Type Culture Collection (Oct 2007; no further authentication was done). The αvβ3-K562 cells (May 2006; tested by immunostaining) were furnished by Dr. Yoshikazu Takada. The αIIbβ3-K562 (Jun 2009; tested by immunostaining) and αvβ5-K562 (Mar 2010; tested by immunostaining) were a gift from Dr. Jennifer Cochran (Stanford University) and Dr. Scott D. Blystone (SUNY Upstate Medical University).
Synthesis of analogues of LXW7
Peptides were synthesized using Fmoc chemistry. Peptide cyclization was completed in the 50 mM NH4HCO3 buffer with activated charcoal. The crude product was purified by RP-HPLC. The final products were characterized by MALDI-TOF-MS and the purity was determined by analytical HPLC (Supplementary data Table S1).
Synthesis of Cyclic LXW7 Derivatives with N-Terminal Modification
The synthesis of linear peptide cGRGDdvc was done on Rink amide resin using Fmoc chemistry. After Fmoc deprotection, the resin were split into 14 portions, 14 different carboxylic acids were coupled to each portion of resin using DIC/HOBt in DMF. The reaction mixture was shaken overnight at room temperature. The resin was then completely washed and dried under vacuum. After cleavage from the beads, the crude products were cyclized and purified to give peptide 18–31 (Supplementary data Table S1).
Synthesis of Focused “One-Bead-One-Compound” Peptide Libraries
The OBOC libraries were synthesized on TentaGel S NH2 resin using the bilayer bead encoding strategy and “split-and-pool” method. A biphasic solvent and topologic segregation approach was used to create bilayer beads. The library compounds are displayed on the outer layer (4% down substitution) and the coding peptide tags on the bead interior. The two focused OBOC peptide libraries were constructed on bilayer beads using Fmoc chemistry. The synthesis details are presented in the supplementary data “synthesis of focused libraries”, Table S2, S3 and S4.
On-Bead Whole Cell Screening Assay
U-87 MG cells adherent to the bottom of a T75 flask were trypsinized with 0.05% trypsin-EDTA and neutralized with culture medium. Floating cells were collected, spun down, and resuspended in 10 mL of culture medium in a 10 cm Petri dish. The library beads were washed sequentially with ethanol, water, and PBS. The beads were then incubated with suspended U-87 MG cells, and the entire dish was swirled at a speed of 40 rpm inside an incubator at 37°C under 5% CO2. The plate was then examined under an inverted microscope every 15 min.
Flow Cytometry. K562 and K562/αvβ3+ cells were cultured in RPMI 1640 medium
To demonstrate the peptides binding affinity, the cells (3 × 105) in each sample were incubated with 0.5μM biotinylated peptides in 50 μL of PBS containing 10% FBS and 1 mM MnCl2 for 30 min on ice. Then each sample was washed three times with 1 mL PBS containing 1% FBS. Samples thereafter were incubated with a 1:500 dilution of streptavidin-PE (1 mg/mL) for 30 min on ice followed by a single wash with 1 mL of PBS containing 1% FBS. Finally, the samples were analyzed via flow cytometry (Coulter XL-MCL). To determine the IC50 of the peptides, series of diluted peptides were incubated with 0.5 μM biotinylated LXW7 first, then the mixture was incubated with cells and followed by streptavidin-PE incubation. Samples were run with flow cytometry and Mean Fluorescence Intensity was decided for each individual sample. IC50s were calculated based on the readings from each sample.
Data Processing and Statistics
Histogram analysis with determination of mean fluorescence intensity (MFI) was conducted after running Flowcytometry. The IC50 of peptides were calculated using Graph Prism software. All of the data are shown as mean (SD of n independent measurements).
Results and Discussions
We have previously reported the identification of tumor targeting ligands through screening OBOC libraries using K562/αvβ3+ cell lines as living cell probes (21). In that study, we hypothesized that through screening OBOC libraries with RGD and related motifs against αvβ3 integrin expressing cell line, one would be able to optimize the amino acids flanking the RGD motif and discover ligands with higher affinity and selectivity to αvβ3 integrin. The screening results indicated that for αvβ3 integrin, the RGD triad flanked by a Gly at the N-terminal side and polar or negative charged amino acids at the C-terminal side was optimal, and the lead peptide sequence emerged from that study was cGRGDdvc (LXW7), which was found to bind strongly to αvβ3, weakly to αvβ5 and αIIbβ3 integrin, and had no binding to α5β1 integrin (21).
In this study, we performed systematic SAR studies to map the pharmacophore of LXW7. We substituted D-Cys1 and/or D-Cys8 with Cys or D-Penicillamine, substituted Gly2 with β-Ala or sarcosine, substituted the constituents of the RGD triad, substituted D-Val7 with other hydrophobic amino acids, capped the N-terminus with various carboxylic acids, and performed N-methylation scan. Half-maximal inhibitory concentrations (IC50) of all these LXW7 analogues were determined by their ability to compete the binding of biotinylated LXW7 to K562/αvβ3+ cells. The relative binding affinities of all these analogues were summarized in Table 1. The peptides were group together according to their chemical modifications, and they were numbered in order of the groupings (peptide #); included in the table are the peptide names (LXW series), which were numbered chronologically as they were designed and synthesized.
Table 1
Analogues of peptide 1 (LXW7) and Their IC50*
| Peptide # (Peptide name) | Sequences | IC50(μM) | Peptide # (Peptide name) | Sequences | IC50(μM) |
|---|---|---|---|---|---|
| 1 (LXW7) | cGRGDdvc-NH2 | 0.46 | 1 (LXW7) | cGRGDdvc-NH2 | 0.46 |
| 2 (LXW9) | CGRGDdvc-NH2 | 4.62 | 22 (LXW90) | R5-cGRGDdvc-NH2 | 1.4 |
| 3 (LXW10) | cGRGDdvC-NH2 | 7.45 | 23 (LXW80) | R6-cGRGDdvc-NH2 | 0.50 |
| 4 (LXW11) | CGRGDdvC-NH2 | >20 | 24 (LXW81) | R7-cGRGDdvc-NH2 | 1.36 |
| 5 (LXW20) | DPen-GRGDdvc-NH2 | 5.04 | 25 (LXW82) | R8-cGRGDdvc-NH2 | 1.14 |
| 6 (LXW21) | cGRGDdv-DPen-NH2 | 2.57 | 26 (LXW83) | R9-cGRGDdvc-NH2 | 0.74 |
| 7 (LXW19) | DPen-GRGDdv-DPen-NH2 | 1.86 | 27 (LXW85) | R10-cGRGDdvc-NH2 | 0.55 |
| 8 (LXW13) | caRGDdvc-NH2 | 3.57 | 28 (LXW86) | R11-cGRGDdvc-NH2 | 0.56 |
| 9 (LXW15) | c-βala-RGDdvc-NH2 | 5.06 | 29 (LXW92) | R12-cGRGDdvc-NH2 | 1.5 |
| 10 (LXW51) | cG-HoArg-GDdvc-NH2 | >20 | 30 (LXW89) | R13-cGRGDdvc-NH2 | 0.76 |
| 11 (LXW59) | cG-Agb-GDdvc-NH2 | >20 | 31 (LXW84) | R14-cGRGDdvc-NH2 | 0.64 |
| 12 (LXW58) | cG-Agp-GDdvc-NH2 | >20 | 32 (LXW14) | c-Sar-RGDdvc-NH2 | 1.92 |
| 13 (LXW25) | cGRGDd-DAbu-c-NH2 | 0.64 | 33 (LXW53) | cG-(NMe)Arg-GDdvc-NH2 | 0.63 |
| 14 (LXW26) | cGRGDdic-NH2 | 1.84 | 34 (LXW54) | cGR-Sar-Ddvc-NH2 | >20 |
| 15 (LXW16) | cGRGDd-DAgl-c-NH2 | 2.38 | 35 (LXW55) | cGRG-(NMe)Asp-dvc-NH2 | >20 |
| 16 (LXW17) | cGRGDd-DPra-c-NH2 | 2.46 | 36 (LXW24) | cGRGD-D(NMe)Asp-vc-NH2 | 10 |
| 17 (LXW18) | cGRGDd-DBug-c-NH2 | 0.31 | 37 (LXW22) | cGRGDd-D(NMe)Val-c-NH2 | 0.63 |
| 18 (LXW12) | R1-cGRGDdvc-NH2 | 1.12 | 38 (LXW63) | cGRGDd-DIng-c-NH2 | 0.32 |
| 19 (LXW76) | R2-cGRGDdvc-NH2 | 0.71 | 39 (LXW48) | cGRGDd-DBta-c-NH2 | 0.25 |
| 20 (LXW77) | R3-cGRGDdvc-NH2 | 2.7 | 40 (LXW64) | cGRGDd-DNal1-c-NH2 | 0.07 |
| 21 (LXW78) | R4-cGRGDdvc-NH2 | 2.9 | 41 (LXW65) | cGRGDd-DNal2-c-NH2 | 0.13 |
Substitution of D-Cys1 and/or D-Cys8
The cyclic OBOC peptide libraries from which LXW7 was discovered utilized N- and C-terminal D-Cys to form the disulfide bridge (21). To determine the importance of the chirality at these Cys residues, we replaced each or both D-Cys at N- and/or C-termini with L-Cys. The replacement of one D-Cys at N- or C-termini (peptide 2 and peptide 3) caused 10- and 16-fold decrease in potency, respectively, than the parent compound. Substitution of D-Cys with L-Cys on both termini (peptide 4) resulted in a complete loss of binding affinity (Table 1).
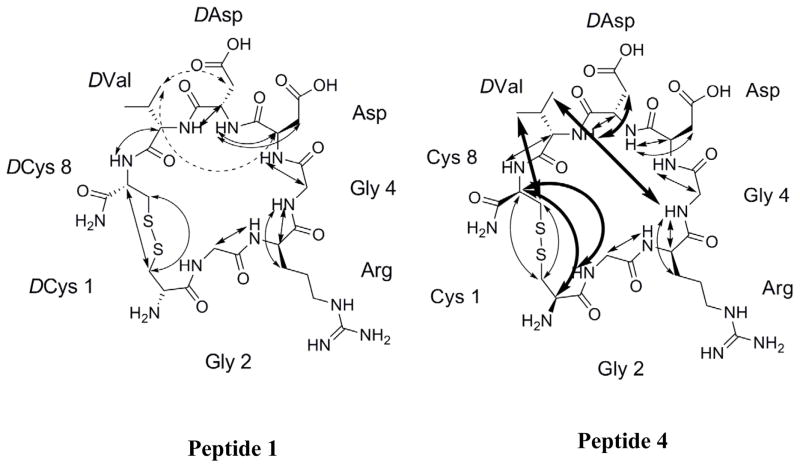
We have conducted NMR study of LXW7 and peptide 4 (LXW11) to examine the conformational changes after replacement of the flanking D-Cys with the opposite enantiomers. The complete chemical shift assignment of proton and carbon resonances for LXW7 and peptide 4 are presented in supplementary data Table S5 and S6, using one- (1H NMR and 13C NMR) and two- dimensional NMR experiments (HMQC, HMBC, TOCSY, and NOESY). Conformations of LXW7 and peptide 4 have been established on the basis of NOESY data (Figure 1, supplementary data Figure S1). In comparison of NOEs of LXW7 and peptide 4, proton correlations in LXW7 between γ CH of D-Val and α CH of Asp, and the β CH of D-Asp were absent in peptide 4. More inter-residue NOEs were observed in peptide 4. New proton correlations in peptide 4 were indicated between the γ CH of D-Val and β CH of Cys8, and the NH of Gly4, which suggested that they are located on the same side of the ring. New correlation was observed between β CH of D-Asp and NH of D-Val, exhibiting that the side chain of D-Asp was more close to the peptide backbone. Other new correlations in peptide 4 were shown between α CH of Cys8 and α CH of Cys1, NH of Gly2. The NOE intensity between two α CH of Cys1 and Cys8 in peptide 4 is much stronger than the intensity between D-Cys1 and D-Cys8 in LXW7, which should indicate that conformation of peptide 4 is more constrained than that of LXW7.
The NMR-derived structures of LXW7 and peptide 4 were superimposed and final ensembles of 15 acceptable structures for each peptide are shown in supplementary data Figure S2 and Table S7. The energy minimized average structures of LXW7 and peptide 4 are illustrated in Figure 2. The two peptides adopt entirely different structures. LXW7 forms a bowl-like conformation, in which side chains of Arg, Asp, D-Asp and D-Val point to outside of the peptide ring. This might enhance the interaction of LXW7 with the αvβ3 integrin compared to peptide 4. The structure of peptide 4 was more compact than that of LXW7 (Figure 2). The bowl-like structure of LXW7 was twisted in peptide 4, which resulted in a drastic change of amino acids side chains orientation. The side chain of D-Val and Asp in peptide 4 was buried inside. The side chain of D-Asp approached to the peptide ring from the other side of the ring in comparison to LXW7. Therefore amino acids side chains cannot interact with αvβ3 integrin efficiently. These explained the complete loss of binding affinity of peptide 4 to K562/αvβ3+ cells.
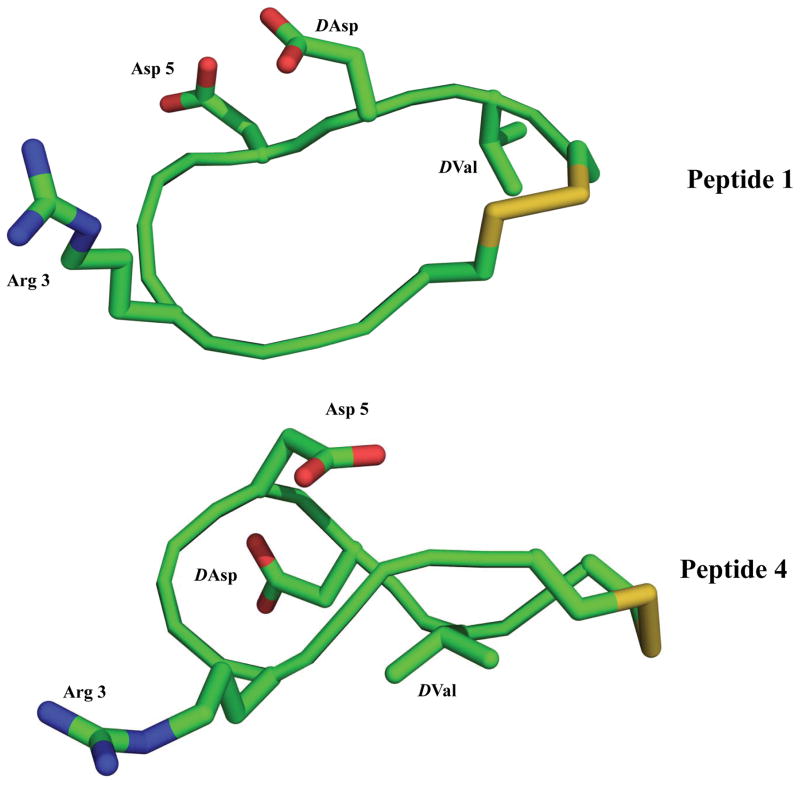
Ribbon representation of the energy minimized average NMR structures of peptide 1 and peptide 4. Side-chain atoms of key residues are shown as sticks. Color code: green, carbon; blue, nitrogen; red, oxygen; yellow, sulfur.
Introducing bulky penicillamine (Pen or β, β-dimethylcysteine, the structure in Table 1C) can often increase peptide proteolytic stability. It is also often used in investigation of disulfide bridge role in peptide biological activity (22–24). D-Pen was adopted to replace D-Cys1 and D-Cys8 singularly or simultaneously (peptide 5, peptide 6 and peptide 7, Table 1). However the replacements caused 4 to 10 times loss of binding affinity. This may be attributed to the constraint exerted by the bulky β dimethyl groups of the two D-Pen side chains. The above results indicated that disulfide bond formed between two D-Cys was required for maintaining the integrin-binding 3D conformation of LXW7.
Substitution of Gly2
The RGD containing peptide sequence, GRGDSP was initially reported by Pierschbacher et.al., which had better inhibition effect of cell attachment to fibronectin than the peptide RGDS, indicating the flanking sequence of RGD also affected peptide activity (25). Cyclo(GRGDSPA) was subsequently reported to be more effective at inhibiting cell attachment to vitronection than to fibronectin (26). For the design of cyclic RGD containing peptides, Gly or Ala was added as spacer flanking the RGD sequence. Our previous OBOC library screening effort for αvβ3 integrin ligands with 20% down-substituted library (cX2RGDX6X7c) showed 100% preference for Gly at the X2 position (21). In this study, replacement of Gly2 of LXW7 with D-Ala yielded peptide 8 (D amino acids were used in this position for our previous library design) and led to a 7.8 time loss of binding affinity to K562/αvβ3+ cell (Table 1). Gly lacks side chain and it provides conformational flexibility to the peptide backbone and affords the appropriate spatial orientation, resulting in binding affinity enhancement (27, 28). Introducing D-Ala limits backbone flexibility and results in interruption of the proton correlations between α CH of Gly and NH of Arg (Figure 1). Interestingly, replacement of Gly2 with β-alanine (peptide 9) also caused 11 times reduction of binding affinity to αvβ3-K562 cells (Table 1). The additional -CH2- of β-Alanine changed the peptide ring size and might bring too much flexibility to the peptide backbone. Together, the above results indicate that Gly in the N-terminal side of the RGD motif is crucial in LXW7 for cell binding. Even slight change in this position is not acceptable.
Substitution of the RGD triad
The previous docking stimulation of LXW7 suggested that the orientation of the Arg side-chain with Asp-218 (in α-subunit of integrin αvβ3) might not be optimal (21). In order to clarify this point, and also to increase the peptide stability toward trypsin digestion at Arg side chain, we replaced Arg with three arginine homologues, HoArg, Agb, and Agp (structures in Table 2), to afford peptides 10–12 (Table 1). However, replacement of Arg with any of these three different arginine homologues resulted in a drastic loss of binding affinity. The optimization of Arg side chain interaction with integrin αvβ3 was not achieved using singular replacement of Arg. Once again, it indicates the dominant effect of RGD motif in LXW7 binding affinity to αvβ3 integrin. Therefore we did not make any further modification on Gly4 and Asp5.
Table 2
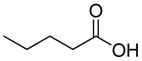
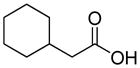
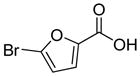
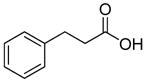
Structures of building blocks used at SAR study of peptide 1 (LXW7).
 |
 |
 |
 |
 |
 |
 |
 |
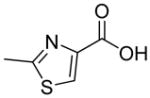
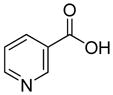
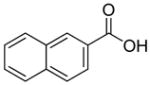
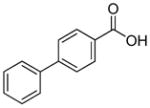
| R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 |
 |
 |
 |
 |
 |
 |
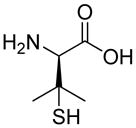 |
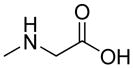 |
| R9 | R10 | R11 | R12 | R13 | R14 | D-Pen | Sar |
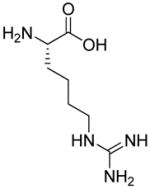 |
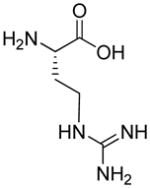 |
 |
 |
 |
 |
 |
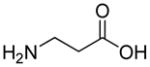 |
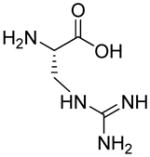
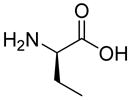
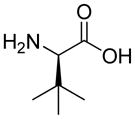
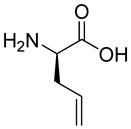
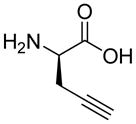
| HoArg | Agb | Agp | DAbu | DBug | DAgl | DPra | β-Alanine |
Substitution of D-Asp6
The previous OBOC screening study (21) showed that D-Asp and D-Glu appeared with higher frequency (63%) at X6 position in the library, indicating that acidic residues are favorable at this position. The prior docking simulation study on LXW7 pointed out that D-Asp6 was able to form a salt bridge with Arg-214 in the β-subunit of integrin αvβ3. This additional interaction probably contributes to the specific binding of LXW7. Therefore we did not do further SAR study at this position. However in the highly focused OBOC library design for further optimization of LXW7, twenty-three acidic and polar amino acids were chosen as building blocks at the X6 position (supplementary data Table S2).
Substitution of D-Val7
Various amino acids (hydrophobic amino acids, polar amino acids and negative charged amino acids) were found at D-Val position from the previous OBOC library screening effort (21). These results may be explained in two ways: first, D-Val position was not fully optimized and further optimization may increase binding affinity; second, D-Val position was not important for the peptide binding to cells, therefore diverse properties of amino acids appeared in this position. However our prior docking stimulation study of LXW7 indicated that side-chain of D-Val bound in an almost identical position as the D-Phe side-chain in cyclo(RGDfV). D-Val did contribute to the binding affinity of LXW7 to the integrin. Therefore we predicted that X7 position is a potential site for LXW7 optimization. Based on D-Val structure, we introduced several D-form hydrophobic amino acids into this position, giving peptide 13–17 (Table 1). Replacement with D-Abu (LXW25, D-Abu structure in Table 2) has similar binding affinity with LXW7 (Table 1). The replacement with D-Bug (LXW18, D-Bug structure in Table 2) led to a slightly increased binding affinity (Table 1). Other three compounds (LXW16, 17 and 26, special amino acids structures in Table 2) have 4–5 times decrease in binding affinity (Table 1). The slight changes of amino acid structure at X7 position caused variable binding affinity results. The above results indicated that it is a potential position for further optimization. Therefore, in the design of two highly focused OBOC libraries, we introduced seventy-one different natural and unnatural amino acids to this position (supplementary data Table S3 and S4).
Modification of the N-terminus
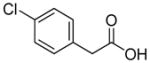
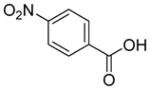
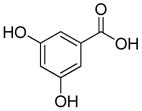
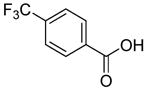
In previous studies, LXW7 binding to αvβ3 integrin was based on the structural feature that LXW7 had free N-terminus. The electrostatic interaction between integrin and basic N-terminus of peptide may play an important role in binding affinity. Since peptides with uncharged N-termini have increased cell permeability and more stable against aminopeptidase digestion, we explored the effect of N-capping on binding affinity. Fourteen analogues of LXW7 (peptide 18–31, structures of R1 to R14 in Table 2) were designed and synthesized (Table 1). Linear LXW7 was first constructed on Rink amide resin; then the resin was aliquot and reacted with various aliphatic and aromatic carboxylic acids. Finally cyclization was done in solution after peptide cleavage from resin with TFA cocktail. Capping of the N-terminus did not result in significant changes in binding affinity. Only 2 substitutions (LXW 77 and 78, Table 1) triggered a modest loss of potency. Six analogues (LXW80, 83, 84, 85, 86 and 89, Table1) have similar binding affinity as LXW7. The above results indicated that N-terminus doesn’t have any significant impact on the binding affinity. One major application of the integrin-binding ligand is to use it as vehicle for drug delivery. The well-known head-to-tail cyclic peptide cyclo(RGDfV), although has high binding affinity and specificity against αvβ3 integrin, lacks a handle for attachment of drug payload. Introducing a handle via replacement of D-Phe or Val by Lys or Glu, resulted in a 2–8 fold decrease in binding affinity (21). The LXW peptides such as LXW7 and LXW64, were discovered through OBOC combinatorial library screening utilizing an on-bead whole-cell binding assay. They all have a built-in C-terminus handle because these ligands were discovered while tethered to the bead. The result of the current N-capping experiment suggests that the N-terminus, if not capped, can also be used as a handle for attachment of drug payload. Therefore, unlike cyclo(RGDfV) which has no handle for tethering, the LXW peptides have two handles for functionalization with drugs and/or imaging agents.
N-methylation scan of LXW7
Incorporation of N-methylated amino acids into peptides changes its backbone conformation and increases peptide membrane permeability and proteolytic stability. It has been often used to affect peptide potency and selectivity to receptor subtypes (29, 30). N-methylation scan strategy was applied to LXW7, from X2 position to X7 position, resulting peptides 32–37 (Table 1). Their structures and physicochemical characteristics are given in Table 1 and supplementary data Table S1. Among N-methylated amino acids, D form of N-methylated aspartic acid was synthesized according to published method (31). N-methylated Arg and D-Val led to slight loss of binding affinity to αvβ3 integrin. The N-methylation of Gly2 triggered a 4 folds loss of potency. This confirmed the importance of Gly2 in LXW7, which has a big impact on the peptide potency. N-methylation at other position (peptide 34–36, table 1) caused drastic loss of binding affinity. The N-methylation scan did not yield any additional potent peptides in comparison to LXW7. Despite this, we included some N-methylated amino acids as building blocks in the focused OBOC libraries design in order to detect their combination effect with amino acids replacements at other positions.
Design, Synthesis, and Screening of Two Highly Focused OBOC Peptide Libraries
Based on above SAR studies, we concluded that: (1) D-Cysteine disulfide bond bridge is necessary for maintaining the integrin-binding conformation of the peptide; (2) Even slight change on structure of Gly2 is not acceptable for the peptide binding to K562/αvβ3+ cells; (3) RGD motif has dominant effect on peptide binding to K562/αvβ3+; (4) Position 6 and 7 of LXW7 are potential places for optimization. Based on these and our previous study results, two highly focused OBOC cyclic peptide libraries with the general sequence of cyclo (cGRGDX6X7c) were designed and synthesized (Figure 3A and B, Supplementary Data “Synthesis of focused libraries”). The focused libraries were synthesized on TentaGel resin utilizing the standard “split-mix” synthetic method (16). Topologically segregated bilayer beads were generated using a biphasic solvent approach (32). The library compounds were displayed on the 4% of the outer layer, and the coding peptide tags were synthesized on the bead interior. Our previous OBOC libraries (even with 20% down-substitution on the bead surface) had many positive beads presenting very strong binding with K562/αvβ3+ cells. Therefore it could be challenging to identify and isolated the strongest binding beads. To overcome this problem, we down-substituted the free amine groups on the outer layer of the beads to 4% prior to library synthesis, so that the screening stringency is higher. The synthetic route is shown in Figure 3B. The X6 position was diversified with 23 negatively charged or polar amino acids (supplementary data Table S2). The X7 position was varied with 71 amino acids, including hydrophobic, polar and negative charged amino acids (Supplementary Data Table S3 and S4). To overcome sequencing ambiguity due to similar retention times of amino acids, the building blocks for position X7 were divided into two groups after X6 coupling (Supplementary Data Table S3 and S4) to form two sub-libraries. After coupling of D-Cys1, on-resin deprotection of amino acids side chain protecting groups was carried out with TFA cocktail cleavage for two hours. On-resin cyclization was allowed to proceed over two days until Ellman test was negative. The two highly focused OBOC cyclic peptide libraries (total diversity of 1633) were screened against K562/αvβ3+ cells in parallel using our standard whole-cell binding assay (8, 33). Ten strong binding beads were identified and physically isolated for microsequencing. Four of them (LXW48, 63–65) were re-synthesized on Rink amide resin and purified by RP-HPLC after cleavage (Supporting Information Table S1). The sequences of re-synthesized peptides are shown in Table 1. At position X6, D-Asp was highly favored, which confirmed our previous study result. The D-Asp was able to form a salt bridge with Arg-214 in the β-subunit of αvβ3. This additional interaction probably contributed to the specific binding of LXW7 to αvβ3 integrin (21). At position 7, hydrophobic amino acids with bulky side chain, were preferred. The IC50 measurements of the peptides inhibiting biotinylated LXW7 binding with K562/αvβ3+ cells were determined. The results indicated that peptide 40 (LXW64) (structure in Figure 3C) had the highest binding affinity to αvβ3 integrin with 6.6 times higher binding affinity than LXW7. LXW64 was biotinylated at the C-terminus via a long hydrophilic linker (structure in Figure 3C), mixed with streptavidin-PE and used as probe for flow cytometry analysis of its binding to αIIbβ3, αvβ5 and αvβ3 transfected K562 and parent K562 cells (α5β1) (Figure 4A). The results demonstrated that LXW64 had similar selectivity with LXW7 towards different integrins. It bound strongly to αvβ3–K562 cells, weakly to αvβ5-K562 cells and αIIbβ3-K562 cells, and has no binding to K562 cells (Figure 4A).
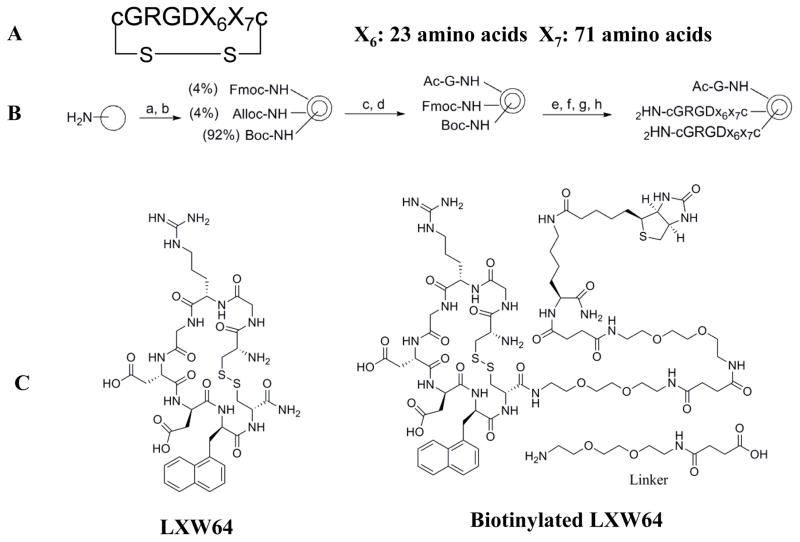
A. Focused OBOC libraries structure; B. Focused library synthesis. a) 1. Swelling in water for 24 h; 2. Fmoc-OSu (0.04 equiv), Alloc-OSu (0.04 equiv.) and DIPEA, DCM/diethyl ether (55: 45), vigorously shaking 30min; b) Boc2O (3 equiv), DIPEA (6 equiv) in DMF, 2 h c): Pd(PPh3)4 (0.2 equiv), PhSiH3 (20 equiv) in DCM, twice (30 min each); d) Ac-Gly-OH (3 equiv.), DIC (3equiv.) and HOBt (3 equiv.), 2h; e) 1. 20% piperidine in DMF, 2 x 10 min.; 2. 50% TFA in DCM (2x30min); f) Split-mix combinatorial synthetic approach using Fmoc-L-amino acids; Four grams resin was split into two focused libraries after coupling of Fmoc-D-Cys(Trt)-OH; g) TFA: phenol: water: thioanisole: Tis (10:0.5:0.5:0.5:0.25, v/w/v/v/v), 3 h; h) 500mL mixture of water, acetic acid and DMSO (75:5:20, pH=6), shaking for 2 days. C. Chemical structure of LXW64 and Biotinylated LXW64.
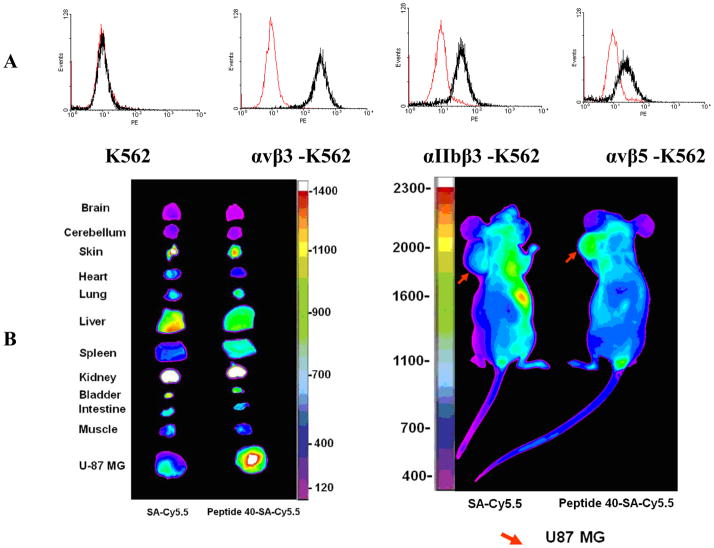
A. Series of integrin transfected K562 cells were stained with LXW64. Black curves: cells were treated with 1μM biotinylated LXW64 and Streptavidin-PE successively, and measured with Flow Cytometry. Red curves represent cells without treatment of biotinylated LXW64, as negative controls. LXW64 showed significant positive binding with αvβ3, weak cross-reaction with αvβ5 and αIIbβ3, and no binding with α5β1expressed on K562 cells. B. In Vivo and Ex Vivo near-infrared fluorescence images.
To demonstrate the utility of LXW64 as useful vehicle for in vivo tumor targeting, biotinylated LXW64 was mixed with streptavidin-cy5.5 and injected via tail vein into nude mice bearing αvβ3 integrin expressing glioblastoma U-87MG xenograft. Near infra-red imaging of excised organs and tumors confirmed preferential tumor uptake of peptide/strep-cy5.5 conjugate but not strep-cy5.5 alone (Figure 4B, supplementary data, “In vivo and ex vivo optical imaging”). Kidney uptake in both cases is due to the known affinity of streptavidin to renal tissues. (34, 35) The targeting specificity and efficiency of LXW64 to αvβ3 integrin on glioblastoma and several other epithelial tumors indicates that it is an excellent candidate vehicle for delivery of radionuclides, cytotoxic agents or drug-loaded nanoparticles for cancer imaging and therapy.
Conclusions
Systematic SAR studies had been conducted on LXW7 (a cyclic RGD containing peptide) from C- to N-terminus, providing 36 analogues. Their binding affinities to αvβ3-K562 cells were evaluated using a competitive binding assay. Based on the results of the SAR studies, two highly focused, 4% surface down-substituted cyclic OBOC peptide libraries were designed, synthesized and screened against αvβ3-K562 cells. The four top peptides were resynthesized and their binding affinity was measured. LXW64 demonstrated a 6.6-fold increase of binding affinity compared to LXW7. Cell binding studies to K562 cells with different integrin expression indicated that LXW64 had similar selectivity with LXW7 towards different integrins. In vivo and ex vivo optical imaging studies further demonstrated that LXW64 efficiently targeted αvβ3 integrin in tumor xenograft model.
Our prior research revealed that Gly2, RGD motif and D-Asp were critical for K562/αvβ3+ cell binding (21). For the optimal binding affinity, our continuous experiments further demonstrates that the peptide portion of the targeting ligands of αvβ3-K562 cells must contain (1) the disulfide linkage of D-Cystine, (2) Gly at position 2, (3) RGD motif, (4) acidic amino acid (D-Aspartic acid) at position 6, and (5) bulky hydrophobic amino acid at position 7. These features were required for our peptide ligand binding with αvβ3-K562 cell. Furthermore, both the N-terminus and C-terminus of LXW64 are free to serve as handles for ligation to toxin, nucleic acid, drugs, or drug-loaded nanoparticles, making it an excellent vehicle for efficient delivery of imaging and therapeutic agents against glioblastoma and other cancers.
Acknowledgments
Financial information: This work was supported by NIH Grants awarded to K.S. Lam: R01CA115483 and R01EB012569.
We thank Dr. Diana Lac for editorial assistance.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/1535-7163.mct-15-0544
Read article for free, from open access legal sources, via Unpaywall:
https://mct.aacrjournals.org/content/molcanther/15/2/232.full.pdf
Citations & impact
Impact metrics
Article citations
EGFR- and Integrin αVβ3-Targeting Peptides as Potential Radiometal-Labeled Radiopharmaceuticals for Cancer Theranostics.
Int J Mol Sci, 25(15):8553, 05 Aug 2024
Cited by: 0 articles | PMID: 39126121 | PMCID: PMC11313252
Review Free full text in Europe PMC
Systemic Dendrimer-Peptide Therapies for Wet Age-Related Macular Degeneration.
Pharmaceutics, 15(10):2428, 05 Oct 2023
Cited by: 0 articles | PMID: 37896188 | PMCID: PMC10609940
Orthogonal Combinatorial Raman Codes Enable Rapid High-Throughput-Out Library Screening of Cell-Targeting Ligands.
Research (Wash D C), 6:0136, 17 May 2023
Cited by: 1 article | PMID: 37214198 | PMCID: PMC10198463
Cross-talk between cancer stem cells and immune cells: potential therapeutic targets in the tumor immune microenvironment.
Mol Cancer, 22(1):38, 21 Feb 2023
Cited by: 41 articles | PMID: 36810098 | PMCID: PMC9942413
Review Free full text in Europe PMC
Targeting Epigenetic Modifiers of Tumor Plasticity and Cancer Stem Cell Behavior.
Cells, 11(9):1403, 21 Apr 2022
Cited by: 18 articles | PMID: 35563709 | PMCID: PMC9102449
Review Free full text in Europe PMC
Go to all (23) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The use of one-bead one-compound combinatorial library technology to discover high-affinity αvβ3 integrin and cancer targeting arginine-glycine-aspartic acid ligands with a built-in handle.
Mol Cancer Ther, 9(10):2714-2723, 21 Sep 2010
Cited by: 44 articles | PMID: 20858725 | PMCID: PMC3571112
Pegylated Arg-Gly-Asp peptide: 64Cu labeling and PET imaging of brain tumor alphavbeta3-integrin expression.
J Nucl Med, 45(10):1776-1783, 01 Oct 2004
Cited by: 148 articles | PMID: 15471848
αVβ3 Integrin-Targeted Radionuclide Therapy with 64Cu-cyclam-RAFT-c(-RGDfK-)4.
Mol Cancer Ther, 15(9):2076-2085, 15 Jul 2016
Cited by: 23 articles | PMID: 27422811
Structure-Activity Relationship of RGD-Containing Cyclic Octapeptide and αvβ3 Integrin Allows for Rapid Identification of a New Peptide Antagonist.
Int J Mol Sci, 21(9):E3076, 27 Apr 2020
Cited by: 1 article | PMID: 32349271 | PMCID: PMC7246635
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: R01 CA115483
Grant ID: R01CA115483
NEI NIH HHS (2)
Grant ID: R01 EY012347
Grant ID: R01EY012347
NIBIB NIH HHS (2)
Grant ID: R01 EB012569
Grant ID: R01EB012569
NIH (3)
Grant ID: R01EB012569
Grant ID: R01CA115483
Grant ID: R01EY012347