Abstract
Free full text

JARID1D is a suppressor and prognostic marker of prostate cancer invasion and metastasis
Abstract
Entire or partial deletions of the male-specific Y chromosome are associated with tumorigenesis, but whether any male-specific genes located on this chromosome play a tumor-suppressive role is unknown. Here, we report that the histone H3 lysine 4 (H3K4) demethylase JARID1D (also called KDM5D and SMCY), a male-specific protein, represses gene expression programs associated with cell invasiveness and suppresses the invasion of prostate cancer cells in vitro and in vivo. We found that JARID1D specifically repressed the invasion-associated genes MMP1, MMP2, MMP3, MMP7, and Slug by demethylating trimethyl H3K4, a gene-activating mark, at their promoters. Our additional results demonstrated that JARID1D levels were highly downregulated in metastatic prostate tumors compared to normal prostate tissues and primary prostate tumors. Furthermore, the JARID1D gene was frequently deleted in metastatic prostate tumors, and low JARID1D levels were associated with poor prognosis in prostate cancer patients. Taken together, these findings provide the first evidence that an epigenetic modifier expressed on the Y chromosome functions as an anti-invasion factor to suppress the progression of prostate cancer. Our results also highlight a preclinical rationale for using JARID1D as a prognostic marker in advanced prostate cancer.
Introduction
Histone lysine methylation plays a pivotal role in epigenetically regulating gene expression at the genome-wide level (1, 2). This modification is associated with either gene activation or silencing, depending on the site of lysine residues. For instance, methylated histone H3 lysine 4 (H3K4) is related to active or poised gene states, whereas methylated H3K27 is linked to gene repression. Lysine residues can be methylated in three different states: mono-, di- and trimethylation (me1, me2, and me3). Distribution patterns and functions of the three methylation states in the mammalian genome are often distinct but overlap substantially. For example, trimethyl H3K4 (H3K4me3) is generally located at the transcription initiation sites (3). Its levels are positively linked to the transcription frequency of active genes but can be found at poised promoters, which also contain the repressive mark H3K27me3. Most of dimethyl H3K4 (H3K4me2) is co-localized with H3K4me3 at the 5′-ends of active genes, and its subset is localized at the enhancers and bodies of active genes.
Histone lysine methylation is reversibly modulated by specific lysine methyltransferases and demethylases (4, 5). H3K4 methylation is established by several methyltransferases, including mixed lineage leukemia (MLL) 1–5, SET1, SET7 (alias SET9), SMYD1, and SMYD2 (6). It can be reversed by several H3K4 demethylases, including LSD1, LSD2, and JARID1A–D (7-13).
The entire or partial regions of the male-specific Y chromosome are deleted in up to 52% of prostate tumors, although the entire loss of the Y chromosome may be rare in these male-specific tumors (14). Of interest, alterations of the Y chromosome also occur in other types of cancer, including testicular tumors, male-predominant esophageal tumors (about 33%), male bladder tumors (10-40%), and male pancreatic tumors (15-19). Vijayakumar et al. showed that the re-introduction of a Y chromosome in the Y chromosome–deficient human prostate cancer cell line PC-3 impeded tumor formation in vivo (20). These studies indicate that the Y chromosome has a tumor-suppressive function in addition to its roles in sex determination and male fertility. This notion is further supported by a recent report showing that the loss of the Y chromosome in peripheral blood is correlated with a high risk of cancer (21). However, whether a male–specific protein expressed on the Y chromosome may function as a tumor or metastasis suppressor remains unknown.
We previously showed that the histone demethylase JARID1D (also known as KDM5D and SMCY) downregulates gene expression by demethylating H3K4me2 and H3K4me3 (7). Of H3K4 methyltransferases and demethylases, only JARID1D is male-specific, because only JARID1D is localized on the Y chromosome. The JARID1D gene is often deleted in prostate tumor samples (15). Therefore, we sought to determine the role of JARID1D in prostate cancer. We found that JARID1D suppresses the invasion, but not proliferation or migration, of prostate cancer cells and represses invasiveness-associated gene programs in these cells. We demonstrated that JARID1D's demethylase activity is necessary for the repression of multiple invasiveness genes, including MMP1, MMP2, MMP3, MM7P, and Slug (also known as SNAI2). In metastatic prostate tumors, JARID1D mRNA and protein levels were commonly decreased. Low JARID1D mRNA and protein levels were correlated with poor survival in prostate cancer patients. Our results indicate that JARID1D is an anti-invasiveness factor and a prognostic marker for prostate cancer.
Materials and Methods
Cell lines, antibodies, and expression plasmids
DU145 and PC3 prostate cell lines and HEK293T cells were purchased from American Type Culture Collection (ATCC), which confirms cell lines using short tandem repeat analysis, and were cultured within 15 times of passages. Cell culture reagents were obtained from Invitrogen/Gibco. Anti-JARID1D antibodies were from Bethyl (A310-624A) and Santa Cruz (sc-83944). Anti-JARID1A (A300-897A), anti-JARID1B (A301-813A), and anti-JARID1C (A301-034A) antibodies were from Bethyl. Anti-H3, anti-H3K4me3, and anti-H3K27me3 antibodies were from Millipore. Normal Rabbit IgG was from santa cruz (sc-2027). Anti-FLAG (M2, F1804) and anti-β–Actin (A5441) antibodies were from Sigma Aldrich. The anti-p84 antibody was from GeneTex. JARID1D and its catalytic mutant (HE/AA) were cloned into the pFlag-CMV2 vector (Sigma). We previously reported that JARID1D's catalytic mutant mJARID1D (HE/AA) in a baculoviral recombinant form is enzymatically inactive (7).
Knockdown and ectopic expression experiments
Three independent small hairpin (sh) RNAs against JARID1D were used for knockdown of JARID1D. shJARID1D-N1 (TRCN0000234751), shJARID1D-N4 (TRCN0000234754), and shLuciferase (shLuc; control shRNA) in the puromycin-resistant PLKO.1 vector were purchased from Sigma Aldrich. shJARID1D-4 (GCCAACCATGTGCAATGTAAC) was cloned into the PLKO.1 vector. Lentiviruses were generated by co-transfecting HEK293T cells with a shJARID1D (or shLuciferase)-expressing plasmid, a packing plasmid (deltaR8), and an envelope plasmid (VSV-G). Fourteen hours later, the medium was replaced with Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Media with virus particles were used to infect the DU145 prostate cancer cells. Infected cells were selected by puromycin for 72 hours. Knockdown efficiency was determined by quantitative reverse transcription polymerase chain reaction (RT–PCR) and Western blot analysis.
To ectopically express JARID1D and its catalytic mutant mJARID1D, we transfected the PC3 prostate cancer cells with the JARID1Dor mJARID1D cDNA construct using Lipofectamine 3000 (Life Technologies) or the Continuum transfection reagent (GEMINI) according to the manufacturer's instructions. Following 24-48 hours of incubation, the cells were harvested, and the total RNAs were isolated for further analysis.
Cell proliferation, migration, and invasion assays
For the cell proliferation assays, cells infected with viruses containing shLuc or shJARID1D were seeded in 24 well plates at a density of 15,000 cells per well in triplicate, and cells were counted 1, 2, 3, and 4 days after plating. Migration assays were performed as previously described (22-24) with some modification. Briefly, cells (1.0 × 105) in 500 μl of serum free DMEM were seeded on the porous membrane of each insert, and 500 μl of DMEM media containing 10% fetal bovine serum was added to each well. Twenty-four hours later, the cells that had migrated to the other side of the membrane were fixed with 4% paraformaldehyde and stained with crystal violet. For the cell invasion assay, DU145 cells (1.0 × 105) and PC3 cells (1.0 × 105) were suspended in DMEM with 0.1% bovine serum albumin and seeded on the Matrigel-coated membrane in each insert. Twenty-four hours later, the cells that had invaded the Matrigel and moved to the other side of the membrane were fixed and stained as in the migration assays.
Mouse xenograft studies
DU145 cells were transfected with the pGL4.51 [luc2/CMV/Neo] plasmid (Promega) and selected with 500 μg/ml G418. Two weeks later, single-cell colonies that stably expressed bioluminescence signals were selected using the IVIS 100 Imaging System (Xenogen). A clone (DU145-Luc2, #9) was used in the mouse xenograft studies. DU145-Luc2 cells were infected with viruses containing shScramble or shJARID1D-4. Cells were selected for at least three days using puromycin.
The cells were injected into the tail veins of 6- to 8-week-old male athymic nu/nu mice (1 × 106 cells per mouse) obtained from The University of Texas MD Anderson Cancer Center. After cell implantation, metastasized loci and tumor growth were monitored using the IVIS 100 Imaging System. Eight to ten weeks after implantation, the mice were humanely euthanized, and their lung tissues were collected for standard histological examination. MD Anderson's Institutional Animal Care and Use Committee approved the care and use of the mice used in these experiments.
RNA isolation, cDNA synthesis, and quantitative RT–PCR
Total RNAs were extracted using RNeasy kits (Qiagen). cDNAs were synthesized using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. Quantitative RT–PCR was carried out in triplicate using SYBR-green. Gene expression levels were measured quantitatively using CFX Manager software (Bio-Rad) and were normalized to GAPDH. The relative fold is the fold change compared to the control. The RT-PCR primer sequences are listed in Supplementary Table S1.
cDNA microarray
DU145 cells were infected with shLuciferase- or shJARID1D-containing lentiviruses three times and selected with puromycin for 72 hours. Total RNAs were prepared using an RNeasy Kit (Qiagen), and knockdown efficiency was assessed by quantitative RT-PCR. Affymetrix U133P GeneChips were used for the microarray experiments.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were carried out as previously described (25). Rabbit immunoglobulin (IgG) was used as a control for the ChIP assay, and the PCR values of ChIP DNA were normalized to input. The ChIP PCR primer sequences are listed in Supplementary Table S2.
ChIP-Seq
For H3K4me3 ChIP-Seq, DU145 prostate cells infected with shLuc- or shJARID1D-containing viruses were cross-linked with 1% formaldehyde at room temperature for 10 min and then sonicated to fragment genomic DNA sizes to 200–500 bp. ChIP was performed using anti-H3K4me3 antibody. After library preparation and quantification, DNA was subjected to HiSeq 2500 sequencing platform (UCI Genomics HTF). Reads were aligned to the human genome (UCSC hg19) using the Bowtie aligner. If a read maps to more than one genomic locations, the read alignment was suppressed (26). Then the unmapped reads were removed using Sam tool. MACS software was used for peak calling. In MACS, the parameter with no model and a shift size of 100bp was chosen, and input data were used as a control for normalization (27). Bigwig track were generated to visualize ChIP-seq data in the UCSC genome browser. Integrative analysis of gene profile around TSS and heatmap generation was performed using Cistrome platform (28)
Immunohistochemistry (IHC)
The avidin-biotin immunoperoxidase method was used to visualize proteins in tissue microarrays (Biomax PR955 and PR8011). Tissues in microarrays were de-paraffinized, treated with citrate buffer and heated with a microwave for 20 min. Then, microarrays were incubated with the primary antibodies (e.g., anti-JARID1D) for 1 h at room temperature. Staining intensity was assessed by Chromavision Automated Cellular Imaging System (ACIS-III) from Dako.
Analysis of expression levels in tumor samples and of patient survival
For the Oncomine database analysis, the mRNA levels were normalized to the median value of the dataset, and expression levels were log2-transformed (www.oncomine.com). Kaplan-Meier overall survival curves were generated for patients for whom follow-up data were available. The log-rank (Mantel-Cox) test was used to analyze survival differences between groups.
Statistical analysis
Data are presented as the means ± SEMs (error bars). A Student's t-test was used to assess whether differences between two different groups were statistically significant. P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) indicate statistically significant changes.
Results
JARID1D suppresses the invasive but not the proliferative or migratory abilities of prostate cancer cells
As an initial step towards understanding the role of JARID1D in prostate cancer events, we determined whether JARID1D regulates the proliferation, migration, and invasion of prostate cancer cells. In particular, we examined the effect of JARID1D knockdown on these cellular characteristics using the widely used and weakly metastatic prostate cancer cell line DU145 (29, 30). All three shJARID1Ds used in this study specifically reduced JARID1D mRNA and proteins levels (Fig. 1A, Supplementary Fig. S1), but JARID1D depletion did not have a significant effect on the proliferative and migratory abilities of DU145 cells (Fig. 1B, C). The cell migration assay, because it lacks an extracellular matrix barrier, scores the simple migratory abilities of cancer cells. In contrast, the invasion assay measures the abilities of cancer cells to degrade and invade a Matrigel matrix that mimics the in vivo matrix barrier. Therefore, we also examined the effect of JARID1D knockdown on cell invasion. In sharp contrast to cell migration, cellular invasiveness was highly increased by JARID1D depletion (Fig. 1D). These results indicate that JARID1D may act as an epigenetic repressor of the invasion, but not proliferation or migration, of prostate cancer cells.
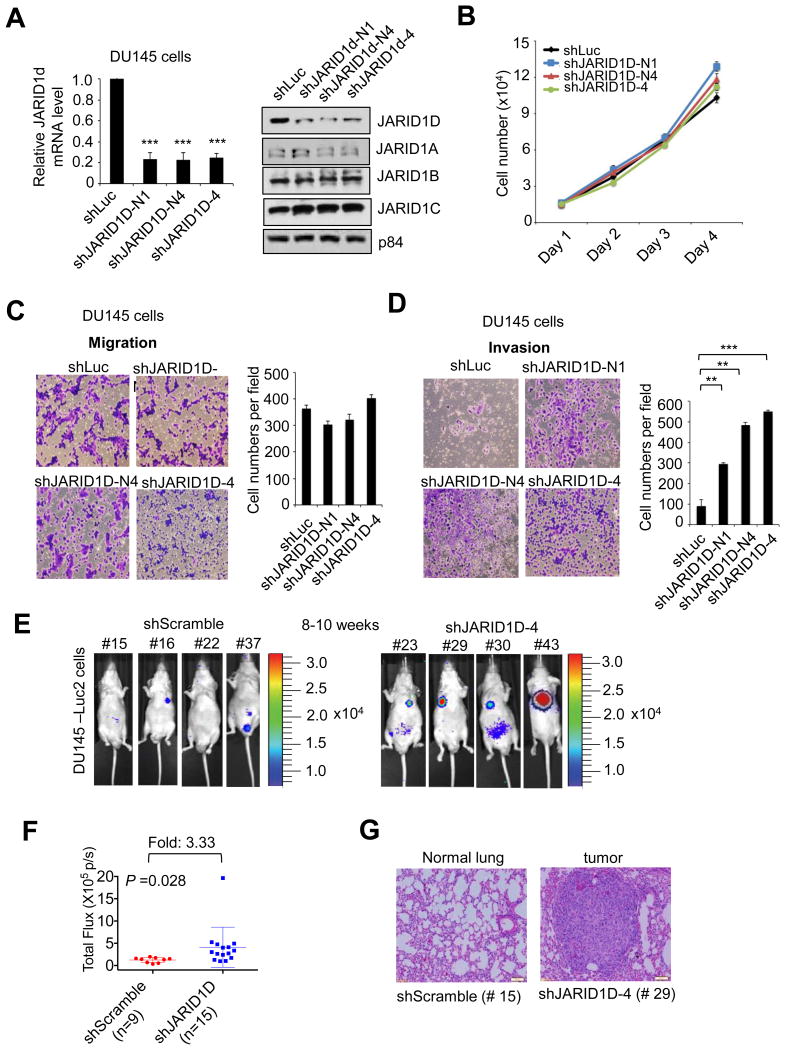
(A) Analysis of JARID1D mRNA and JARID1A–D protein levels following shRNA treatment. shRNAs were used to deplete JARID1D in DU145 prostate cancer cells. Total RNAs were analyzed using quantitative RT-PCR. The relative fold represents the expression levels in individual samples over those in control shLuciferase (shLuc) samples. For Western blot analysis, nuclear proteins were extracted and blotted with anti-JARID1D and anti-p84 antibodies. (B–D) Effects of JARID1D knockdown on the proliferation (B), migration (C) and invasion (D) of DU145 cells. Cells that had migrated and invaded were counted in at least five fields. (E & F) Effects of JARID1D knockdown on the metastatic abilities of DU145 cells in an intravenous mouse xenograft model. Metastasized loci were monitored using a bioluminescence imaging system. Representative bioluminescent images of mice (8-10th weeks after tail vein injections) are shown (dorsal view for No. 43 and ventral view for all the others) (E). Quantified bioluminescent signals were individually plotted for the shScramble and shJARID1D groups (shScramble, n = 9; shJARID1D, n = 15) (F). (G) Representative images of hematoxylin and eosin–stained lung tissues in shScramble and shJARID1D mice.
JARID1D inhibits the invasiveness of prostate cancer cells in vivo
To assess the effect of JARID1D on the invasiveness of prostate cancer cells in vivo, we used an intravenous mouse xenograft model. For this experiment, we generated a DU145 cell line expressing a luciferase (DU145-Luc2 cells) that enabled us to monitor tumor growth in vivo. Following the treatment of DU145-Luc2 cells with shJARID1D or control shRNA, cells were injected into tail veins of mice. In this xenograft model, cancer cells invade into a distant organ during extravasation (departure from the blood system) and grow as a tumor at the metastatic site (primarily, the lung). This mouse xenograft experiment demonstrated that 8th–10th weeks after cell injection, luciferase signals in the shJARID1D mice (n = 15) were significantly higher than those in the control shRNA mice (n = 9) (Fig. 1E and F). Tumor formation in shJARID1D group mice was confirmed by hematoxylin and eosin staining (Fig. 1G). These results suggest that JARID1D suppresses the invasive capacities of DU145 prostate cancer cells in vivo.
JARID1D represses the expression of invasiveness-associated genes
To determine how JARID1D suppresses the invasive ability of prostate cancer cells, we compared the whole genome mRNA expression profile of JARID1D-depleted DU145 cells to that of control (shLuciferase-treated) DU145 cells using Affymetrix microarrays. Consistent with JARID1D's inhibition of cellular invasiveness, gene ontology analysis showed that JARID1D knockdown up-regulated gene expression programs associated with cellular invasiveness, including cell adhesion, cytoskeleton organization, and extracellular matrix organization (Fig. 2A and B). The number of invasiveness-associated genes upregulated by JARID1D knockdown was higher than the number of those genes downregulated by JARID1D knockdown (Fig. 2A vs. Supplementary Fig. S2). Interestingly, expression levels of several invasion-associated genes, including the matrix metalloproteinase 1 (MMP1) and the transcriptional repressor gene Slug, were increased by JARID1D knockdown (Fig. 2B). Our quantitative RT-PCR analysis confirmed that JARID1D knockdown upregulated expression levels of MMP1 and Slug (Fig. 2C).
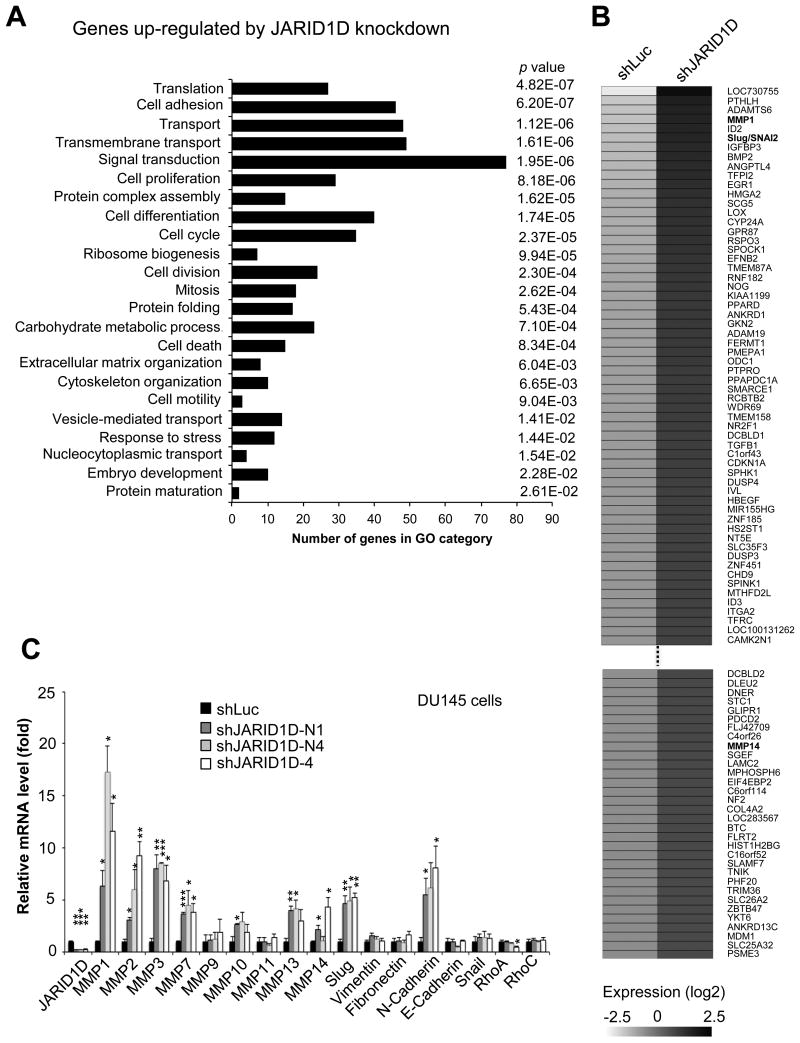
(A) Gene ontology analysis for genes up-regulated by JARID1D knockdown in DU145 cells. The mRNA levels were measured using the Affymetrix U133P gene expression microarray. Gene ontology analysis was performed for 1416 genes whose expression levels in shJARID1D group were 1.5-fold higher than those in shLuciferase group (i.e., shJARID1D/shLuciferase: >1.5). (B) Heatmap of genes upregulated by JARID1D knockdown in DU145 cells as measured by microarray. (C) Effects of JARID1D knockdown on the expression of invasiveness-associated genes. Quantitative RT-PCR analysis of genes showed that JARID1D knockdown increased MMP1, MMP2, MMP3, MMP7, MMP10, MMP13, MMP14, N-Cadherin, and Slug mRNA levels in DU145 cells.
MMPs are known to facilitate cell invasion and metastasis by enzymatically degrading extracellular matrix components (31). The mesenchymal cell-cell adhesion molecule N-cadherin and Slug promote epithelial-to-mesenchymal transition (EMT), a reversible, temporary but complex process in which immotile epithelial cells convert to motile and invasive mesenchymal cells. In contrast, the epithelial cell-cell adhesion molecule E-cadherin inhibits this process (32, 33). However, the expression of multiple well-known invasion-associated genes, including several MMPs, E-Cadherin, and N-cadherin, was not detected by the Affymetrix microarrays (Fig. 2B). Therefore, we manually analyzed the effect of JARID1D knockdown on these genes using quantitative RT-PCR. Our results demonstrated that in addition to the upregulation of MMP1 and Slug, JARID1D knockdown also up-regulated the MMP2, MMP3, MMP7, MMP10, MMP13, MMP14, and N-cadherin genes in DU145 cells (Fig. 2C). In agreement with JARID1D's lack of effect on cell migration, JARID1D depletion did not affect the expression of several genes associated with cell migration, such as RhoA and RhoC. These results suggest that JARID1D suppresses cell invasion by transcriptionally repressing multiple invasion-associated genes.
JARID1D's enzymatic activity is required for JARID1D's anti-invasive function
A histone demethylase can regulate cellular functions dependent or independent of its catalytic activity. For example, we have shown that the demethylase activity of the H3K27 demethylase UTX (also called KDM6A) is indispensable for the regulation of the proliferative and invasive abilities of breast cancer cells (24). In contrast, the demethylase activity of UTX is dispensible for several other biological processes, such as T-box family member–dependent gene expression (34), mesoderm differentiaion of mouse embryonic stem cells (35), and mouse embryonic development (36).
To assess whether the enzymatic activity of JARID1D is necessary for cellular invasiveness and to complement shRNA-mediated JARID1D knockdown experiments, we determined the effect of JARID1D's ectopic expression in the prostate cancer cell line PC3, which lacks endogenous JARID1D because of the absence of the Y chromosome. As shown in Fig. 3A and B, wild type JARID1D and its catalytic mutant mJARID1D were similarly expressed, and their overexpression had no obvious effect on total cellular H3K4me3 levels. Our results also showed that JARID1D but not mJARID1D significantly decreased the invasive abilities of PC3 cells in vitro (Fig. 3C). Consistent with this, the ectopic expression of JARID1D but not that of mJARID1D significantly reduced the expression levels of the MMP1, MMP2, MMP3, MMP7, and Slug genes in PC3 cells (Fig. 3D–I). These results indicate that JARID1D has an anti-invasiveness function that depends largely on JARID1D's catalytic activity.
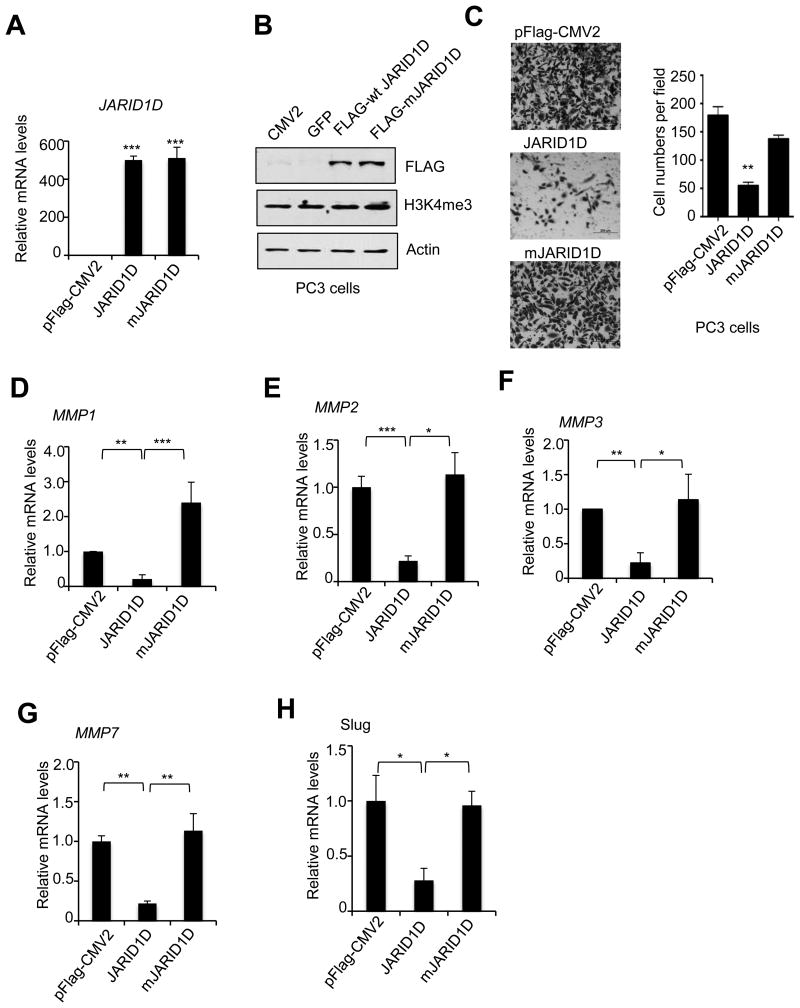
(A and B) Ectopic expression of wild type JARID1D and its catalytic mutants (mJARID1D) in PC3 prostate cancer cells. PC3 cells were transfected with control plasmids (pFLAG-CMV2 and GFP), pFLAG-CMV2-JARID1D, or pFLAG-CMV2-mJARID1D. The mRNA levels of JARID1D and mJARID1D were measured using quantitative RT-PCR (A). JARID1D protein and H3K4me3 levels were assessed using Western blot analysis (B). (C) Effects of ectopic expression of JARID1D and mJARID1D on the invasiveness of PC3 cells. Representative images are shown. Cells that had invaded were counted in at least five fields. (D–H) Effects of JARID1D and mJARID1D on MMP1 (D), MMP2 (E), MMP3 (F), MMP7 (G), and Slug (H) expression in PC3 cells.
JARID1D occupies multiple invasiveness-associated genes and demethylates H3K4me3 at their promoters
JARID1D represses gene expression by demethylating H3K4me3 at the gene promoter regions (7). Therefore, genes upregulated by JARID1D knockdown may be targeted by JARID1D. We tested this possibility by performing ChIP experiments. In particular, we examined different promoter regions of the MMP1, MMP2, MMP3, MMP7, and Slug genes. Our ChIP results showed that JARID1D occupied specific promoter regions in the MMP1, MMP2, MMP3, MMP7, and Slug genes (Fig. 4A-E).

(A-E) Quantitative ChIP analysis of JARID1D levels at the MMP1 (A), MMP2 (B), MMP3 (C), MMP7 (D), and Slug (E) genes in DU145 cells. The relative occupancy represents the fold changes in PCR values from specific antibody over those from the IgG. Schematic representation of the promoter regions of the MMP1, MMP2, MMP3, MMP7, and Slug genes are shown. Two primer sets were used to amplify the proximal promoter regions in individual genes. The lines and primer set numbers below indicate the PCR-amplified regions. TSS, transcriptional start site.
Because we previously showed that JARID1D demethylates H3K4me3 to repress gene expression (7), we sought to determine whether JARID1D demethylates H3K4me3 at the promoters of JARID1D-repressed genes in DU145 cells. Specifically, we assessed the effect of JARID1D knockdown on H3K4me3 levels at gene promoters using a ChIP-Seq approach. JARID1D knockdown slightly increased the average of H3K4me3 ChIP-Seq peaks of all the gene-regulatory regions spanning −6kb to +6kb (Fig. 5A, Supplementary Fig. S3) and also augmented the averaged ChIP-Seq peak of H3K4me3 signals of the top 150 genes upregulated by shJARID1D (Fig. 5B; Supplementary Table S3; See also Fig. 2B). The averaged H3K4me3 ChIP-Seq peak of the 150 genes was slightly higher than that of all the gene-regulatory regions (Fig. 5A), suggesting that JARID1D preferentially represses genes whose H3K4me3 levels are higher than the average H3K4me3 level. Modest increases of H3K4me3 peaks at the 150 genes upon JARID1D knockdown were correlated with substantial increases in their gene expression (Fig. 2C vs. Fig. 5B).
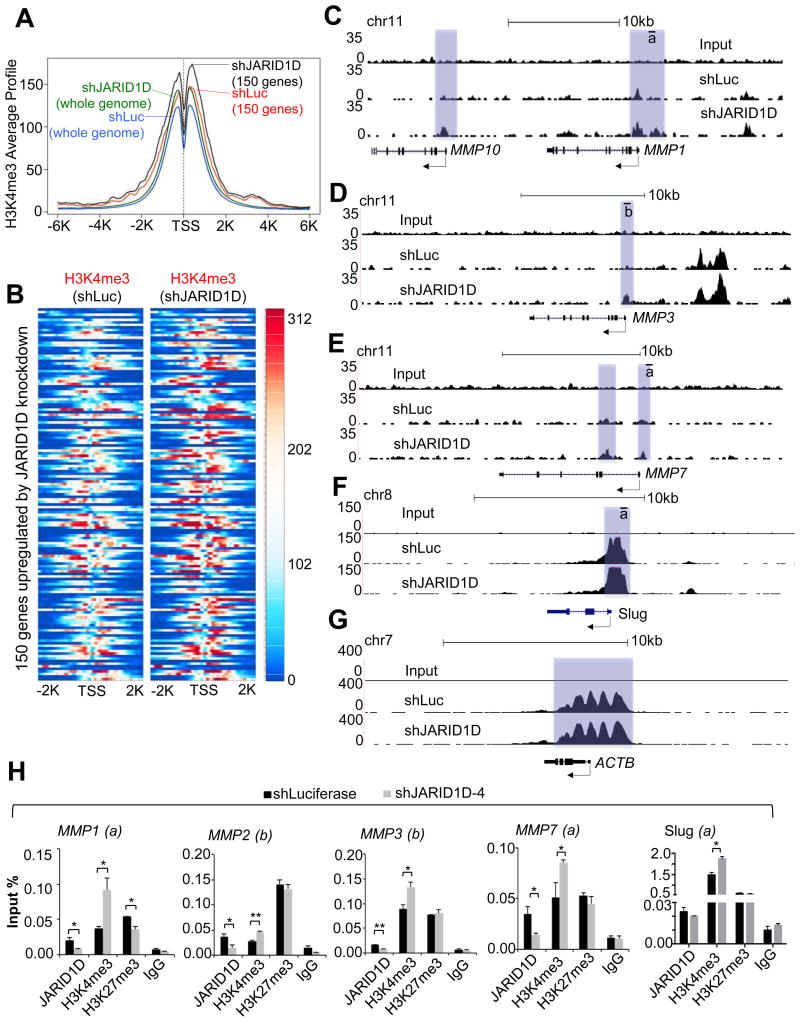
(A) Comparison of averaged H3K4me3 ChIP-seq profiles for whole genome or 150 genes between shLuc- and shJARID1D-treated DU145 cells. The top 150 genes up-regulated by JARID1D knockdown were analyzed. The gene list is summarized in Supplementary Table S3. The profiling resolution is 50 bp. (B) Heatmap for H3K4me3 ChIP-seq signals in 150 genes. (C–G) The genome browser view of H3K4me3 signals at the MMP1 (C) and MMP10 (C), MMP3 (D), MMP7 (E), Slug (F), and ACTB (β-Actin) (G) genes in shLuc and shJARID1D-treated DU145 cells. Arrows indicate transcriptional direction for individual genes, and the proximal promoters with H3K4me3 signals are highlighted. (H) Effects of JARID1D knockdown on H3K4me3 levels at the MMP1, MMP2, MMP3, MMP7, and Slug promoters. The chromatin levels of JARID1D, H3K4me3, H3K27me3 and IgG at these promoters were compared between shLuc- and shJARID1D-4–treated DU145 cells using a quantitative ChIP assay. The lines and letters below indicate the PCR-amplified regions (see also Fig. 4).
We also examined the effect of JARID1D knockdown on H3K4me3 ChIP-Seq peaks at individual genes using our ChIP-Seq data. JARID1D knockdown modestly increased H3K4me3 levels at MMP1, MMP3, MMP7, and Slug promoters whereas it had no obvious effect on H3K4me3 levels at the ACTB promoter (Fig. 5C–G). However, some genes, including MMP2, did not show a detectable H3K4me3 ChIP-Seq signal upon JARID1D knockdown, although their expression levels were upregulated by JARID1D knockdown (data not shown). In the analysis of chromatin occupancy of histone marks and chromatin-binding proteins, the sensitivity of quantitative ChIP is often higher than that of ChIP-Seq. Therefore, we performed quantitative ChIP to manually detect H3K4me3 levels at the MMP2 promoter. Our quantitative ChIP results showed that JARID1D knockdown increased H3K4me3 levels at the MMP2 promoter in addition to the MMP1, MMP3, MMP7, and Slug promoters (Fig. 5H). These results indicate that the JARID1D-catalyzed demethylation of H3K4me3 is linked to the JARID1D-mediated repression of several invasion-associated genes.
JARID1D levels are low in metastatic prostate tumors, and the JARID1D gene is frequently deleted in metastatic prostate cancer but not primary tumors
To determine whether JARID1D expression is dysregulated in primary and metastatic prostate tumors, we compared JARID1D levels among normal tissues, primary prostate tumors, and metastatic prostate tumors using immunohistochemical (IHC) analysis. Compared with those in normal tissues (10%, 2/21), JARID1D protein levels in metastatic prostate tumors were drastically down-regulated (100%, 6/6), and those in primary prostate tumors were substantially decreased (41%, 28/68) (Fig. 6A and B). We also determined whether JARID1D mRNA levels are decreased in prostate tumor samples using the publicly available database Oncomine. The JARID1D mRNA levels in metastatic prostate tumors were significantly lower than those in primary prostate tumors and normal tissues (Fig. 6C and Supplementary Fig. S4A and B).
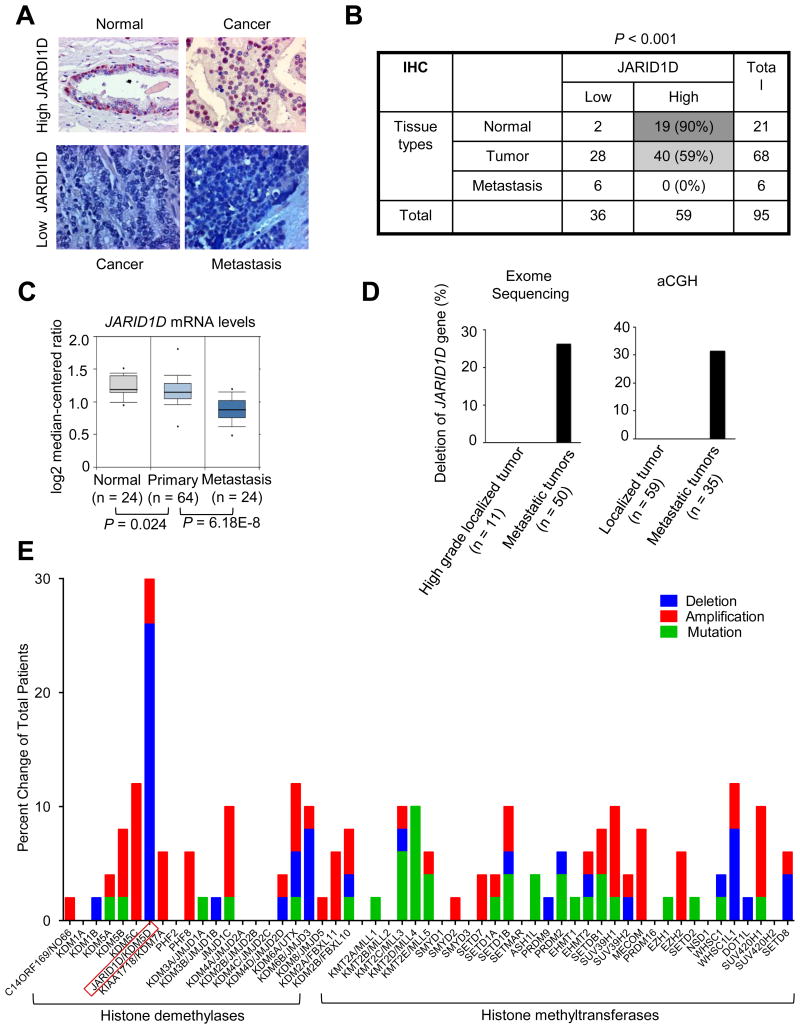
(A) Representative images of immunohistochemical staining for JARID1D expression in human normal prostate tissues, primary prostate tumors, and metastatic prostate tumors. (B) Immunohistochemical analysis of JARID1D protein levels in normal prostate tissues (n = 21), primary prostate tumors (n = 68) and metastatic prostate tumors (n = 6). (C) Analysis of JARID1D mRNA levels using a dataset called “Yu prostate cancer” from the publicly available database Oncomine. (D) Database analysis for the frequency of JARID1D gene deletion. Using exome sequencing data from the Grasso prostate cancer dataset from the Oncomine database, we analyzed the JARID1D gene's deletion in localized prostate tumors (n=11) and metastatic prostate tumors (n=50) (Left panel). Tumors with -2 (a high-level copy loss) were considered to have gene deletion (37). Using array comparative genomic hybridization (aCGH) data of the Grasso prostate cancer dataset from the Oncomine database, we analyzed the JARID1D gene's deletion (Right panel). Less than -1 in log 2 (tumor/normal tissue ratio) in gene copy numbers was considered to have gene deletion (37). (E) Analysis of the mutation and copy number alteration of multiple histone lysine methyltransferases and demethylases in 50 metastatic prostate tumors using exome sequencing data from the Grasso prostate cancer dataset (cBio Portal). Tumors with -2 (a high-level copy loss) were considered to have gene deletion. Data in the bar graph in Fig. 6D are re-used here for direct comparison.
Because the Y chromosome, on which the JARID1D gene is localized, frequently undergoes complete or partial deletion in prostate tumors (15), we assessed the status of the JARID1D gene in primary and metastatic prostate tumors and compared it with those of other histone methylation modifiers in prostate tumors. Our analysis of publicly available database showed that the JARID1D gene was more frequently lost in metastatic prostate cancer than in primary prostate tumors (Fig. 6D) (37). Of interest, the JARID1D gene was deleted more frequently than were genes encoding other histone lysine methyltransferases and demethylases in metastatic tumors (Fig. 6E).
Low JARID1D levels are associated with high MMP1 and MMP3 mRNA levels and with poor prognosis in prostate cancer patients
To determine whether JARID1D levels are associated with the expression of certain JARID1D target genes in prostate tumor samples, we compared the expression levels of JARID1D to those of its target genes using the cBioPortal (NCBI GEO Database #GSE21032). Our database analysis showed that JARID1D mRNA levels were inversely correlated with the mRNA levels of MMP1 and MMP3 but not those of other JARID1D target genes, including MMP2, MMP7, MMP10, and Slug (Fig. 7A and B; Supplementary Fig. S5).
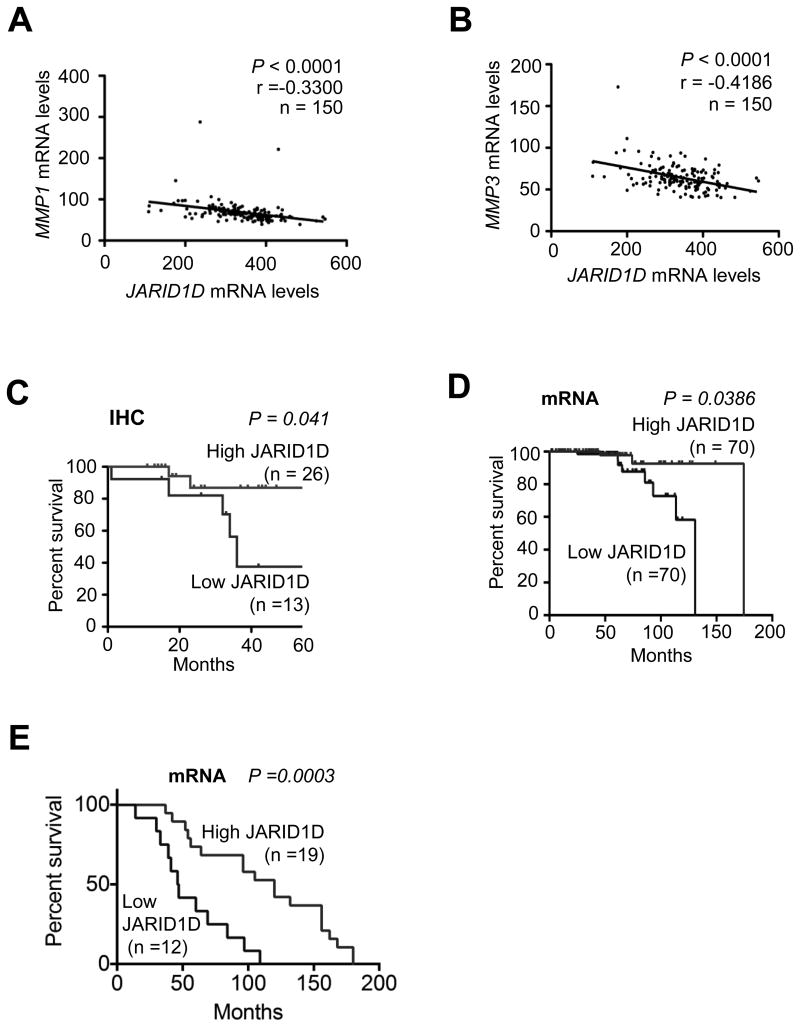
(A and B) Correlation analysis of JARID1D mRNA levels with either MMP1 (A) or MMP3 (B) mRNA levels in a dataset from the publicly available database cBioPortal (NCBI GEO Database #GSE21032). (C) Kaplan-Meier survival rate analysis of a set of prostate cancer patients grouped by IHC–assessed JARID1D levels. Tumor samples in tissue microarrays were analyzed by IHC. (D) Kaplan-Meier survival rate analysis of a set of prostate cancer patients grouped by JARID1D mRNA levels. A prostate cancer patient dataset with follow-up history (NCBI GEO Database #: GSE21032) from cBioPortal was used; the mean JARID1D level for the entire group was used as the cutoff value. (E) Survival analysis of castration-resistant prostate cancer patients grouped by JARID1D mRNA levels. Data with follow-up history in the Grasso prostate cancer dataset from the Oncomine database were used for this analysis.
To determine whether JARID1D levels are linked to the clinical outcomes of prostate cancer patients, we performed a Kaplan-Meier survival analysis of 39 prostate cancer patients grouped by immunochemistry-assessed JARID1D protein levels. This analysis revealed that low JARID1D protein levels were significantly associated with poor overall survival in prostate cancer patients (Fig. 7C). Moreover, our analysis of the publicly available databases showed that low JARID1D mRNA levels also signified poor overall survival in prostate cancer patients in two different cohorts of prostate cancer patients (Fig. 7D and E). These results suggest that JARID1D is a novel prognostic marker of prostate cancer.
Discussion
Our results indicate that the male-specific histone demethylase JARID1D suppresses the invasive ability of prostate cancer cells in vitro and in vivo by downregulating invasion-associated gene programs. Compared with normal prostate tissues, primary prostate tumors had low JARID1D levels, and metastatic prostate tumors had even lower JARID1D levels than primary prostate tumors did. These findings are strengthened by our public database analysis, which showed that the JARID1D gene was frequently deleted in metastatic prostate tumors but not primary tumors. Furthermore, our findings demonstrated that low JARID1D mRNA and protein levels were linked to poor survival in prostate cancer patients, indicating that JARID1D is a prognostic marker for prostate cancer.
The overall function of the Y chromosome appears to be associated with tumor suppression (20, 21). However, multiple Y-chromosome-specific genes, including Sry and TSPY, have been reported to have oncogenic function (38, 39). The current study, which indicates that JARID1D has a metastasis-suppressive role in prostate cancer progression, provides new molecular insight into how the partial or entire loss of the Y chromosome may reduce or nullify its overall tumor-suppressive function. To our knowledge, JARID1D is the first Y chromosome–specifically expressed protein to be identified as an anti-invasiveness factor. Walport et al. recently reported that the Y chromosome contains an additional histone methylation modifier called UTY. (40) UTY has a weak H3K27 demethylase activity, but its function in prostate tumorigenesis is not known.
Global H3K4me3 levels are significantly increased in many advanced prostate tumors, including prostate cancer metastases (41). Our data indicated that low JARID1D levels were not correlated with high global H3K4me3 levels in prostate tumor samples (data not shown). In addition, our results showed that JARID1D overexpression had no detectable effect on global H3K4me3 levels while JARID1D knockdown only weakly increased global H3K4me3 levels (Fig. 5A). These results are not surprising, because global H3K4me3 levels may be co-regulated by several H3K4 methyltransferases and demethylases. In support of this, previous studies have shown that total cellular levels of the H3K4me3 demethylase JARID1B and the H3K4me2 demethylase LSD1 are increased in prostate tumors, although these studies included small numbers of tumors (42, 43). Together, these findings suggest that low JARID1D levels enhance cellular invasion by increasing H3K4me3 levels at specific invasiveness-associated genes (rather than at the whole genomic level) and activating such genes.
Metastasis is a complicated process in which tumor cells undergo the detachment from the primary site, intravasation into blood vessels, extravasation to a different body location, and colonization at the secondary site (44). Mounting evidence suggests that EMT is required for metastasis. In the present study, JARID1D knockdown increased expression levels of key EMT regulators, such as N-Cadherin and Slug. In contrast, JARID1D did not affect the expression of E-Cadherin (Fig. 2C), suggesting that E-Cadherin may not be an important factor for the JARID1D-mediated regulation of cell invasion. In line with this, Nieman et al. reported that overexpression of N-Cadherin enhances the mobility and invasion of cancer cells regardless of E-Cadherin levels (45). We also found that JARID1D represses the expression of multiple MMPs, such as MMP1, MMP2, MMP3, MMP7, MMP10, MMP13, and MMP14 (Fig. 2C). Interestingly, MMPs have been known to induce EMT. For example, the induced expression of MMP3 drives the molecular and phenotypic conversion of normal epithelial cells to mesenchymal cells (46). Reciprocally, EMT induction results in increased levels of MMPs. For instance, the expression of Snail1 in cancer cells triggers an MMP-14- and MMP-15-dependent invasion program (47). Therefore, it is likely that JARID1D represses certain EMT pathways in addition to several MMP genes to inhibit the invasiveness of prostate cancer cells.
Increased invasive abilities of cancer cells are essential for metastasis, and these abilities are in principle under epigenetic control. In fact, histone methylation modifiers, which epigenetically regulate gene expression, are positively or negatively associated with metastasis processes. There are multiple examples for metastasis-promoting histone methylation modifiers. Min et al. found that the H3K27 methyltransferase EZH2 may induce prostate tumorigenesis and metastasis by repressing expression of the tumor-suppressor protein DAB2IP (48). Ren et al. reported that EZH2 might enhance cancer cell invasion by downregulating expression of the metastasis suppressor RKIP (49). Cao et al. showed that JARID1A (also known as RBP2 and KDM5A) promotes breast cancer progression and metastasis (50). Similarly, Luo et al. demonstrated that the histone demethylase KDM4C might enhance breast tumor growth and metastasis (51). SMYD3 is upregulated in many cancer types, including pancreatic and lung tumors, and may promote cancer cell invasion by increasing expression of the matrix metalloproteinase 9 (52). In addition to overexpression of aforementioned metastasis-promoting histone modifiers, the losses of metastasis-inhibiting histone methylation modifiers may enhance cancer cell metastasis. As described in the present study, JARID1D is lost and downregulated in a substantial portion of metastatic prostate tumors. The H3K9 methyltransferase SETDB1, which appears to inhibit metastasis of lung cancer cells, is strongly downregulated in lung cancer cells (53).
In summary, we identify JARID1D as an anti-invasiveness histone methylation modifier that acts against prostate cancer cell metastasis by repressing metastatic gene programs. Our work uncovers a previously unknown invasion-suppressive mechanism by which a Y chromosome–specifically expressed histone demethylase epigenetically inhibits prostate cancer metastasis.
Supplementary Material
1
2
3
4
Acknowledgments
We thank Dr. Bingnan Gu, Dr. Amrish Sharma, Dr. Zhenbo Han, and Mr. Su Zhang for their technical assistance and Joan Ritho, Dr. Chunru Lin, and Dr. Liuqing Yang for supplying reagents. We are also thankful to Joseph Munch for manuscript editing. This study was supported by grants to M.G.L. from the NIH (CA157919 and GM095659), the Cancer Prevention and Research Institute of Texas (CPRIT RP110183), and the Center for Cancer Epigenetics at MD Anderson (including Solexa allowance) and by a postdoctoral fellowship to N.L. from the Center for Cancer Epigenetics at MD Anderson.
References
Full text links
Read article at publisher's site: https://doi.org/10.1158/0008-5472.can-15-0906
Read article for free, from open access legal sources, via Unpaywall:
https://cancerres.aacrjournals.org/content/canres/76/4/831.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The impact of mosaic loss of the Y chromosome (mLOY) in men of advanced age.
Biogerontology, 25(6):943-955, 02 Sep 2024
Cited by: 0 articles | PMID: 39223433
Review
Curcumin blunts epithelial-mesenchymal transition to alleviate invasion and metastasis of prostate cancer through the JARID1D demethylation.
Cancer Cell Int, 24(1):303, 01 Sep 2024
Cited by: 0 articles | PMID: 39218854 | PMCID: PMC11366129
Comparing the roles of sex chromosome-encoded protein homologs in gene regulation.
Genes Dev, 38(13-14):585-596, 20 Aug 2024
Cited by: 0 articles | PMID: 39048311
Review
Sex chromosome-encoded protein homologs: current progress and open questions.
Nat Struct Mol Biol, 31(8):1156-1166, 09 Aug 2024
Cited by: 2 articles | PMID: 39123067
Review
Y Chromosome Loss and Implications for Oncology.
Mol Cancer Res, 22(7):603-612, 01 Jul 2024
Cited by: 0 articles | PMID: 38647375 | PMCID: PMC11217729
Review Free full text in Europe PMC
Go to all (77) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (3 citations) GEO - GSE21032
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes.
Mol Cell, 63(3):470-484, 28 Jul 2016
Cited by: 85 articles | PMID: 27477906 | PMCID: PMC4975651
Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression.
Proc Natl Acad Sci U S A, 113(22):6259-6264, 16 May 2016
Cited by: 81 articles | PMID: 27185910 | PMCID: PMC4896695
Curcumin blunts epithelial-mesenchymal transition to alleviate invasion and metastasis of prostate cancer through the JARID1D demethylation.
Cancer Cell Int, 24(1):303, 01 Sep 2024
Cited by: 0 articles | PMID: 39218854 | PMCID: PMC11366129
Involvement of the multiple tumor suppressor genes and 12-lipoxygenase in human prostate cancer. Therapeutic implications.
Adv Exp Med Biol, 407:41-53, 01 Jan 1997
Cited by: 35 articles | PMID: 9321930
Review
Funding
Funders who supported this work.
NCI NIH HHS (1)
Grant ID: R01 CA157919
NIGMS NIH HHS (1)
Grant ID: R01 GM095659





