Abstract
Background
The RTS,S/AS01 vaccine targets the circumsporozoite protein of Plasmodium falciparum and has partial protective efficacy against clinical and severe malaria disease in infants and children. We investigated whether the vaccine efficacy was specific to certain parasite genotypes at the circumsporozoite protein locus.Methods
We used polymerase chain reaction-based next-generation sequencing of DNA extracted from samples from 4985 participants to survey circumsporozoite protein polymorphisms. We evaluated the effect that polymorphic positions and haplotypic regions within the circumsporozoite protein had on vaccine efficacy against first episodes of clinical malaria within 1 year after vaccination.Results
In the per-protocol group of 4577 RTS,S/AS01-vaccinated participants and 2335 control-vaccinated participants who were 5 to 17 months of age, the 1-year cumulative vaccine efficacy was 50.3% (95% confidence interval [CI], 34.6 to 62.3) against clinical malaria in which parasites matched the vaccine in the entire circumsporozoite protein C-terminal (139 infections), as compared with 33.4% (95% CI, 29.3 to 37.2) against mismatched malaria (1951 infections) (P=0.04 for differential vaccine efficacy). The vaccine efficacy based on the hazard ratio was 62.7% (95% CI, 51.6 to 71.3) against matched infections versus 54.2% (95% CI, 49.9 to 58.1) against mismatched infections (P=0.06). In the group of infants 6 to 12 weeks of age, there was no evidence of differential allele-specific vaccine efficacy.Conclusions
These results suggest that among children 5 to 17 months of age, the RTS,S vaccine has greater activity against malaria parasites with the matched circumsporozoite protein allele than against mismatched malaria. The overall vaccine efficacy in this age category will depend on the proportion of matched alleles in the local parasite population; in this trial, less than 10% of parasites had matched alleles. (Funded by the National Institutes of Health and others.).Free full text

Genetic Diversity and Protective Efficacy of the RTS,S/AS01 Malaria Vaccine
Associated Data
Abstract
BACKGROUND
The RTS,S/AS01 vaccine targets the circumsporozoite protein of Plasmodium falciparum and has partial protective efficacy against clinical and severe malaria disease in infants and children. We investigated whether the vaccine efficacy was specific to certain parasite genotypes at the circumsporozoite protein locus.
METHODS
We used polymerase chain reaction–based next-generation sequencing of DNA extracted from samples from 4985 participants to survey circumsporozoite protein polymorphisms. We evaluated the effect that polymorphic positions and haplotypic regions within the circumsporozoite protein had on vaccine efficacy against first episodes of clinical malaria within 1 year after vaccination.
RESULTS
In the per-protocol group of 4577 RTS,S/AS01-vaccinated participants and 2335 control-vaccinated participants who were 5 to 17 months of age, the 1-year cumulative vaccine efficacy was 50.3% (95% confidence interval [CI], 34.6 to 62.3) against clinical malaria in which parasites matched the vaccine in the entire circumsporozoite protein C-terminal (139 infections), as compared with 33.4% (95% CI, 29.3 to 37.2) against mismatched malaria (1951 infections) (P = 0.04 for differential vaccine efficacy). The vaccine efficacy based on the hazard ratio was 62.7% (95% CI, 51.6 to 71.3) against matched infections versus 54.2% (95% CI, 49.9 to 58.1) against mismatched infections (P = 0.06). In the group of infants 6 to 12 weeks of age, there was no evidence of differential allele-specific vaccine efficacy.
CONCLUSIONS
These results suggest that among children 5 to 17 months of age, the RTS,S vaccine has greater activity against malaria parasites with the matched circumsporozoite protein allele than against mismatched malaria. The overall vaccine efficacy in this age category will depend on the proportion of matched alleles in the local parasite population; in this trial, less than 10% of parasites had matched alleles. (Funded by the National Institutes of Health and others.)
Malaria induces substantial morbidity and mortality worldwide1 and has proved to be a challenge for vaccine-development efforts. The recently renewed effort to control, eliminate, and hopefully eradicate malaria will have a greater likelihood of success if a vaccine can be combined with other intervention methods, such as drug-administration campaigns and insect-vector control.2,3 The most advanced candidate vaccine for protection against Plasmodium falciparum malaria infection, RTS,S/AS01, is a monovalent recombinant protein vaccine that targets a fragment of the circumsporozoite protein parasite antigen. RTS,S/AS01 was evaluated in a large randomized, controlled, phase 3 trial, conducted at 11 study sites in Africa between 2009 and 2013, in which the efficacy, safety, and immunogenicity of the vaccine was assessed in more than 15,000 children. The vaccine confers moderate protective efficacy against clinical disease and severe malaria that wanes over time,4-7 a finding concordant with results of multiple phase 2 trials.8-13 Higher protection was observed among young children (5 to 17 months of age at first vaccination) than among infants (6 to 12 weeks of age at first vaccination).4-7
The mechanism by which the vaccine confers protection is incompletely understood, and differing hypotheses exist regarding the relative importance of B-cell–mediated versus T-cell–mediated immunity to circumsporozoite protein and other antigens.14-17 The circumsporozoite protein is expressed on the surface of the parasite during the infective sporozoite stage and contains a conserved NANP–NVDP tandem repeat with a length polymorphism ranging from 37 to 44 repeat units,18 which is thought to represent the dominant B-cell epitope.19 There are numerous polymorphisms within the C-terminal region of circumsporozoite protein,18,20-22 some of which reside within described T-cell epitopes (Th2R and Th3R)23 that may also function as B-cell epitopes.24 Although the immune response provoked by RTS,S/AS01 may be distinct from the natural response to circumsporozoite protein,25,26 the partial protective efficacy of the vaccine may be due in part to allelic specificity. Evidence of naturally acquired allele-specific immunologic protection has been observed for the merozoite surface protein 1 antigen27 (but not circumsporozoite protein28) in a prospective cohort study, and allele-specific protection has been reported in a field trial of a vaccine based on apical membrane antigen 1.29
Previous genetic analyses of parasite samples from three different RTS,S phase 2 studies did not detect an association between protective efficacy and the genetic similarity of the parasite to 3D7 (the vaccine construct parasite line)30-32; however, our study has both a larger sample and improved sequencing technology. Next-generation sequencing (Illumina MiSeq, PacBio) of polymerase-chain-reaction (PCR) amplicons obtained from samples from malaria-infected participants makes a more sensitive genetic investigation of allele-specific protection possible and allows for more immunologically relevant analyses of multivariant haplotypes. Using sieve-analysis methods (Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org) that have previously been applied to the detection of allele-specific vaccine efficacy against human immunodeficiency virus type 1,33,34 we analyzed the relationship between vaccine efficacy, in two age categories with regard to two defined trial end points, and the parasite circumsporozoite protein at three levels: the entire C-terminal amplicon haplotype (95 amino acids), defined haplotypic regions of the C-terminal (10 to 17 amino acids), and individual polymorphic positions. We also investigated the relationship between vaccine efficacy and NANP–NVDP repeat count, and we included the serine repeat antigen 2 (SERA-2) locus as a control; SERA-2 was not in the vaccine, and therefore no differential efficacy was expected with regard to the parasite genotype at this locus.
METHODS
STUDY DESIGN AND SEQUENCE-DATA GENERATION
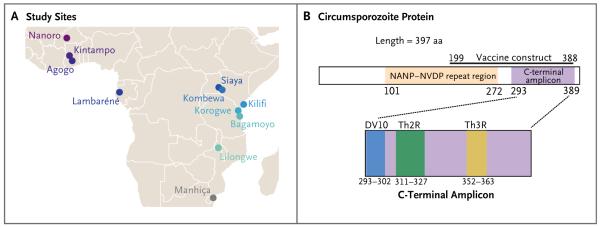
The RTS,S phase 3 trial design has been described previously (for details, see ClinicalTrials.gov number NCT00866619).4-7 Samples were analyzed from vaccine recipients at all 11 trial sites, spanning seven countries in Africa (Fig. 1A). Samples representing two protocol-specified end points were sequenced in the per-protocol cohort (i.e., participants who received all three vaccinations at months 0, 1, and 2): primary clinical malaria (first or only episode of clinical malaria with a parasite density >5000 per cubic millimeter), occurring between 14 and 385 days after the third vaccination, and parasite positivity (parasite density >0 per cubic millimeter, regardless of whether symptoms were present), occurring at 18 months after vaccination.3
Samples from participants were received as dried blood spots on Whatman FTA sample cards. The methods we used for DNA extraction, PCR amplification, and sequencing are described in the Supplementary Appendix. The circumsporozoite protein C-terminal and SERA-2 amplicons were sequenced on an Illumina MiSeq platform. The NANP–NVDP amplicon was sequenced on a PacBio platform because of its greater length. Fig. 1A shows the respective locations of the NANP–NVDP repeat region and C-terminal amplicon within the circumsporozoite protein. The analyses included MiSeq data for 4421 circumsporozoite protein C-terminal samples and 4499 SERA-2 samples and PacBio data for 3137 circumsporozoite protein NANP–NVDP samples (Tables S1 through S4 and the statistical analysis plan in the Supplementary Appendix). All MiSeq and PacBio amplicon sequence data were submitted to the NCBI Sequence Read Archive35 (BioProject PRJNA235895).
STUDY OVERSIGHT
The trial was sponsored by GlaxoSmithKline Biologicals, the developer and manufacturer of the vaccine, and was funded by both GlaxoSmithKline Biologicals and the PATH Malaria Vaccine Initiative, which received a grant from the Bill and Melinda Gates Foundation. Several of the academic authors, in collaboration with the four authors who are employed by GlaxoSmithKline Biologicals, designed the study and the analysis plan. Several of the academic authors collected the data, wrote the manuscript, and vouch for the accuracy and veracity of the reported data, and the first and last authors made the decision to submit the manuscript for publication. GlaxoSmithKline Biologicals reviewed the manuscript before it was submitted for publication. Additional details regarding author contributions are provided in the Supplementary Appendix. The trial protocol was approved by all relevant ethics review boards, and written informed consent, indicated with either a signature or a thumb-print, was obtained from the children’s parents or guardians.4-7
STATISTICAL ANALYSIS
The analyses that were prespecified for this study are described in the statistical analysis plan (see the Supplementary Appendix). In brief, we performed sieve analyses of translated amino acid sequences to assess differences in vaccine efficacy with respect to the end points of first or only episodes of clinical malaria and parasite positivity, according to whether the parasite was a match or a mismatch with the 3D7 vaccine strain at the circumsporozoite protein C-terminal and according to the number of NANP–NVDP repeats present. We refer to differential vaccine efficacy according to a given parasite feature as a “sieve effect.” For the primary analysis of clinical malaria, two haplotype-specific measures of vaccine efficacy were assessed: cumulative vaccine efficacy, which is defined as one minus the ratio (RTS,S/AS01 vs. control) of cumulative incidences of the first or only episode of clinical malaria with a specific haplotype by a given number of days (t) beyond 14 days after the third vaccination; and hazard-ratio vaccine efficacy, which is defined as one minus the ratio (RTS,S/AS01 vs. control) of instantaneous incidences of the end point under the assumption that incidences (RTS,S/AS01 vs. control) are proportional over time. Aalen–Johansen nonparametric maximum-likelihood estimation with stratification according to study site was used to estimate the cumulative vaccine efficacy against 3D7-matched malaria and 3D7-mismatched malaria, with Wald tests used to identify nonzero vaccine efficacy and a sieve effect of differential vaccine efficacy. Targeted maximum-likelihood estimation36 was used to address the same objectives with adjustment for all relevant baseline participant covariates (listed in Table S5 in the Supplementary Appendix). The hazard-ratio vaccine efficacy was estimated with cause-specific Cox models stratified according to study site, with the use of score tests to identify nonzero vaccine efficacy and Wald tests for sieve effects.37 The analysis methods were selected to be in close alignment with those used for assessment of overall vaccine efficacy in the original published analyses.4-7 For the parasite-positivity end point, we used analysis methods similar to those used for the cumulative vaccine efficacy analysis of the primary clinical malaria end point.
Many participants had complex malaria infections generated through multiple parasite founder genotypes. Consequently, sieve analyses were performed on data sets composed of one founder haplotype randomly selected from each participant, with “multiple outputation”38 used to aggregate results (details are provided in the statistical analysis plan). We also performed sieve analyses on data sets in which participants infected with parasites of one or more founder haplotypes were classified as having “any match” to 3D7 or “no match” to 3D7.
In addition to investigating the previously described Th2R and Th3R epitopes,23 we analyzed haplotype frequencies in a previously undefined genomic region that we designate as DV10 (representing 10 amino acid positions, 293 through 302, bounded by amino acids aspartate [D] and valine [V]) and a linkage disequilibrium (LD) haplotype based on the 6 positions (314, 317, 352, 354, 356, and 357) that we found to exhibit LD (which we assessed for sites with a minor-allele frequency of at least 3% and an r2 of at least 0.1) with two or more other positions at the five largest study sites (Fig. S9 in the Supplementary Appendix).
All analyses were performed separately for each age category. Multiplicity adjustment of sieve-effect P values across the epitope haplotypes and positions was applied separately to the two age categories, the two studied proteins (circumsporozoite protein and SERA-2), and the two measures of vaccine efficacy (cumulative efficacy and hazard-ratio efficacy). Adjustment for family-wise error rate (Holm–Bonferroni39) and false discovery rate (Q values; Benjamini–Hochberg40) was applied. Results with Q values of 0.20 or less for all multiply compared loci or unadjusted P values of 0.05 or less for the full circumsporozoite protein C-terminal amplicon were considered to indicate statistical significance; a P value that controlled for the family-wise error rate and was 0.05 or less indicated more stringent significance. All P values and Q values are two-sided.
RESULTS
PARTICIPANTS AND SAMPLES
Fig. 2, and Fig. S2 in the Supplementary Appendix, summarize sample size and follow-up information for the per-protocol group of participants who were 5 to 17 months of age; in this cohort, P. falciparum genetic data were analyzed from 1181 RTS,S/AS01-vaccine recipients and 909 control-vaccine recipients in whom the primary clinical malaria end point was confirmed, as well as from 284 RTS,S/AS01-vaccine recipients and 208 control-vaccine recipients in whom the parasite-positivity end point was confirmed. Fig. S3 and S4 in the Supplementary Appendix show this information for the per-protocol group of infants who were 6 to 12 weeks of age, and Fig. S5 through S8 in the Supplementary Appendix provide information on the samples studied for both end points and age categories for the NANP–NVDP repeat amplicon.
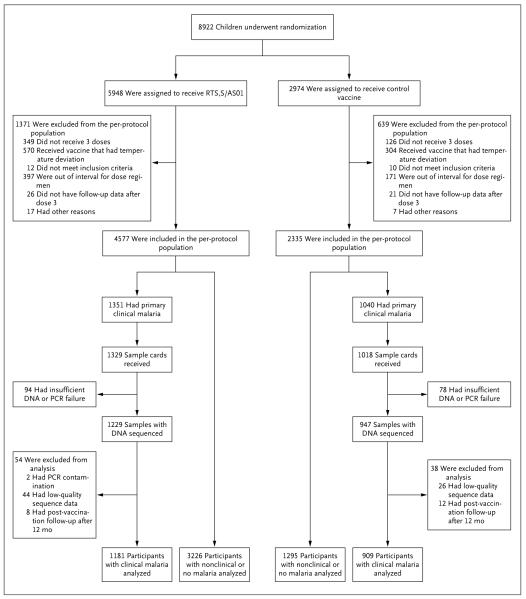
Vaccine recipients were considered to be out of interval for the dose regimen if they did not receive booster vaccinations according to the schedule specified by the trial protocol. The sample cards used were Whatman FTA cards. PCR denotes polymerase chain reaction.
COMPLEXITY OF INFECTION
The majority of samples from participants with primary clinical malaria in both age categories (68% of samples from infants 6 to 12 weeks of age and 65% of samples from children 5 to 17 months of age) had complex infections, defined as being founded by two or more distinct parasite lineages. In the older age category, the distribution of complexity of infection was shifted toward fewer parasite lineages in RTS,S/AS01-vaccine recipients than in control-vaccine recipients (complex infections in 61% vs. 71% of samples, P<0.001 by Wald test) (Fig. 3A), whereas in the younger age category there was no evidence of a different distribution between the RTS,S/AS01 and control groups (67% and 70%, respectively; P = 0.43) (Fig. S10 in the Supplementary Appendix). This observation for the older age category is concordant with findings in two phase 2 trials of the related RTS,S/AS02 vaccine,31,32 and there were fewer 3D7-matching full-amplicon haplotypes in infections of high complexity in the RTS,S/AS01-vaccinated group than in the control group (Fig. S11 in the Supplementary Appendix).
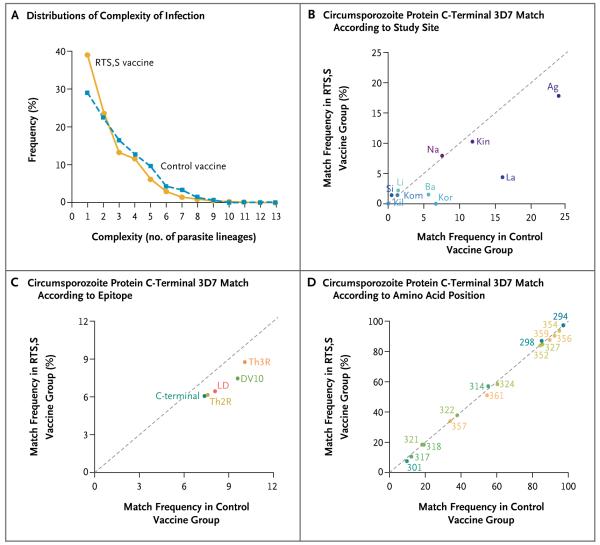
The denominator for the frequencies in Panels B through D is the number of samples representing the primary clinical malaria end point that had sequence data available. Ag denotes Agogo, Ba Bagamoyo, Kil Kilifi, Kin Kintampo, Kom Kombewa, Kor Korogwe, La Lambaréné, Li Lilongwe, Na Nanoro, and Si Siaya.
POPULATION VARIATION PROFILE
We searched for a sieve effect based on perfect 3D7 match–mismatch in the C-terminal of the circumsporozoite protein at three scales: the full-amplicon haplotype (95 amino acids), four described epitopes or polymorphism cluster haplotypes (10 to 17 amino acids apiece), and 25 individual polymorphic positions. In the category of children 5 to 17 months of age, the frequency of haplotypes with an exact match to the 3D7 vaccine strain across all polymorphic positions varied considerably among study sites (Fig. 3B). In addition, there was a lower frequency of 3D7-matching haplotypes among RTS,S/AS01-vaccine recipients than among control vaccine recipients, especially at geographic sites with at least a 5% frequency of 3D7-matching haplotypes in the control-vaccine group. Similar differences were evident with respect to the epitope haplotype frequencies (Fig. 3C). The frequency of alleles matching the 3D7 vaccine strain at individual polymorphic positions was variable (Fig. 3D). In the category of infants 6 to 12 weeks of age, the frequencies of 3D7 matching at all three scales in the RTS,S/AS01-vaccinated group were similar to those in the control-vaccinated group (Fig. S10 in the Supplementary Appendix).
C-TERMINAL REGION SIEVE EFFECTS
Through 1 year after vaccination, we detected 139 clinical malaria cases with a perfect full-amplicon 3D7 match (Fig. 4A) and 1951 cases that were mismatched (Fig. 4B). During this period, the cumulative vaccine efficacy against clinical malaria with a perfect full-amplicon 3D7 match was 50.3% (95% confidence interval [CI], 34.6 to 62.3), and that against mismatched clinical malaria was 33.4% (95% CI, 29.3 to 37.2), with vaccine efficacy significantly higher against matched malaria (P = 0.04 by Wald test for the sieve effect) (Fig. 4D and Fig. 5A). The covariate-adjusted analysis gave almost identical results (Table S5 in the Supplementary Appendix). The cumulative vaccine efficacy was higher against matched malaria than against mismatched malaria throughout the follow-up period; for example, during the period through month 6, vaccine efficacy against matched malaria was 70.2% (95% CI, 56.1 to 79.8), and that against mismatched malaria was 56.3% (95% CI, 51.1 to 60.9) (P = 0.05 for the sieve effect) (Fig. 4C). Cumulative vaccine efficacy and sieve effects for the circumsporozoite protein C-terminal also varied in magnitude among study sites when the sites were analyzed individually in the older age category (Table S6 in the Supplementary Appendix).
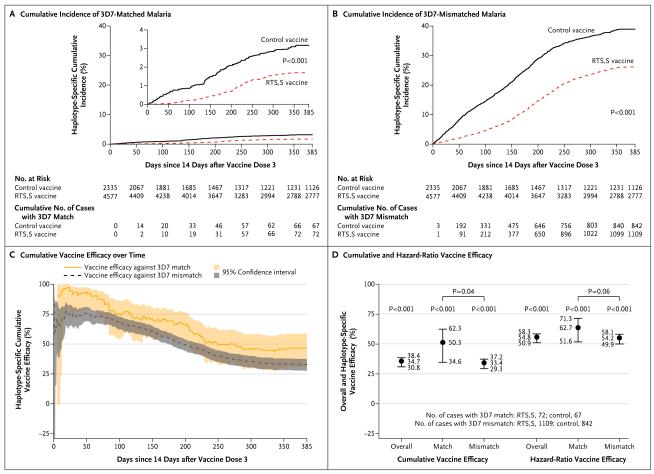
Shown is the cumulative incidence of 3D7-matched malaria (Panel A) and 3D7-mismatched malaria (Panel B) among RTS,S/AS01-vaccine recipients and control-vaccine recipients during 12 months of post-vaccination follow-up, the cumulative vaccine efficacy (one minus the ratio [RTS,S/AS01 vs. control] of cumulative incidences of the first or only episode of clinical malaria with a specific haplotype) against 3D7-matched and 3D7-mismatched malaria over the entire post-vaccination follow-up period (Panel C), and the cumulative vaccine efficacy and hazard-ratio vaccine efficacy (one minus the ratio [RTS,S/AS01 vs. control] of instantaneous incidences of the end point under the assumption that incidences are proportional over time) against 3D7-matched and 3D7-mismatched malaria at 12 months after vaccination, with tests for differential haplotype-specific vaccine efficacy (Panel D). The I bars in Panel D indicate 95% confidence intervals.
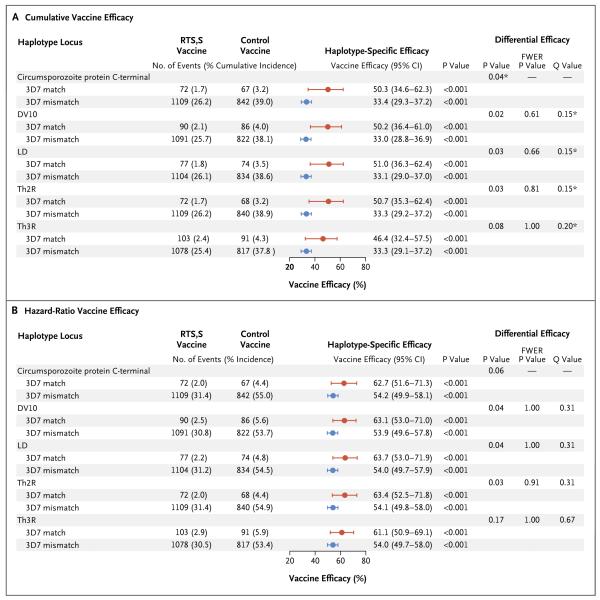
Shown are the cumulative vaccine efficacy (Panel A) and hazard-ratio vaccine efficacy (Panel B) for the prevention of clinical malaria in which parasites were matches or mismatches with the RTS,S/AS01 vaccine strain (3D7) at each haplotype locus. Estimates were stratified according to study site. For each haplotype locus, the calculation of haplotype-matched vaccine efficacy included only clinical malaria end-point events with samples in which parasites matched 3D7 at the given locus; the calculation of haplotype-mismatched vaccine efficacy included only clinical malaria end-point events with samples in which parasites mismatched 3D7 at the given locus. Asterisks indicate that the difference in efficacy was significant (Q value ≤0.2 for all 28 multiply compared haplotype loci and unadjusted P≤0.05 for the full circumsporozoite protein C-terminal amplicon). FWER denotes family-wise error rate.
The hazard-ratio vaccine efficacy over the 12 months of post-vaccination follow-up was also higher against 3D7-matched malaria than against 3D7-mismatched malaria (62.7% [95% CI, 51.6 to 71.3] vs. 54.2% [95% CI, 49.9 to 58.1]; P = 0.06 for the sieve effect) (Fig. 5B, and Table S7 in the Supplementary Appendix). The overall vaccine efficacy was similar to the efficacy against mismatched malaria, because more than 90% of the infections were mismatched (Fig. 4D). Post hoc analysis defining the haplotypes of a malaria case as “any match” or “no match” to 3D7 also identified a sieve effect for the circumsporozoite protein C-terminal (P<0.001 for the cumulative vaccine efficacy sieve effect; P = 0.002 for the hazard-ratio vaccine efficacy sieve effect) (Tables S8 and S9 in the Supplementary Appendix). In contrast, the cumulative and hazard-ratio vaccine efficacies were similar against full-amplicon–matched malaria and full-amplicon–mismatched malaria in the category of infants 6 to 12 weeks of age (P = 0.58 for the sieve effect) (Tables S10 and S11 and Fig. S12 in the Supplementary Appendix), as well as against SERA-2– matched malaria and SERA-2–mismatched malaria in both age categories (P values for the sieve effect >0.30) (Tables S12 through S15 and Fig. S13 and S14 in the Supplementary Appendix).
Among the participants who were 5 to 17 months of age, there were also significant cumulative vaccine efficacy sieve effects (Q value ≤0.2) for the Th2R and Th3R epitopes, the Th2R–Th3R LD haplotype, and the DV10 region (Fig. 5A), as well as for the individual amino acid positions 299, 301, 317, 354, 356, 359, and 361 (Table 1). The cumulative vaccine efficacy tended to decrease with the number of mismatches with 3D7 at these seven amino acid positions (Fig. S15 in the Supplementary Appendix). Hazard-ratio analyses of epitopes and regions (Fig. 5B) and individual amino acid positions (Table S7 in the Supplementary Appendix) yielded differential vaccine efficacy results that were consistent with those from the cumulative vaccine efficacy analysis, at reduced levels of significance. In the younger age category, vaccine efficacy against malaria that matched 3D7 at individual circumsporozoite protein C-terminal amino acid positions was similar to that against malaria with mismatches (all Q values >0.20) (Tables S10 and S11 in the Supplementary Appendix). No evidence of sieve effects was found for individual positions in either age category for the SERA-2 locus (Table S12 through S15 in the Supplementary Appendix).
Table 1
Cumulative Vaccine Efficacy against Primary Clinical Malaria.*
| Locus and Amino Acid Position† | 3D7-Matched Malaria‡ | 3D7-Mismatched Malaria‡ | Differential Efficacy | ||||
|---|---|---|---|---|---|---|---|
| Vaccine Efficacy (95% CI) | P Value | Vaccine Efficacy (95% CI) | P Value | P Value | FWER P Value§ | Q Value | |
| percent | percent | ||||||
| DV10 | |||||||
| 294 | 34.8 (30.8 to 38.6) | <0.001 | 31.3 (−6.4 to 55.6) | 0.09 | 0.83 | 1.00 | 0.89 |
| 295 | 34.9 (31.0 to 38.6) | <0.001 | 12.6 (−78.8 to 57.2) | 0.71 | 0.44 | 1.00 | 0.71 |
| 296 | 34.8 (30.9 to 38.5) | <0.001 | 7.1 (−310.9 to 79.0) | 0.92 | 0.67 | 1.00 | 0.89 |
| 298 | 33.5 (29.1 to 37.7) | <0.001 | 41.7 (30.1 to 51.4) | <0.001 | 0.20 | 1.00 | 0.41 |
| 299¶ | 35.4 (31.5 to 39.1) | <0.001 | −62.8 (−186.0 to 7.3) | 0.09 | 0.003 | 0.08 | 0.08 |
| 301¶ | 49.2 (35.3 to 60.1) | <0.001 | 33.1 (28.9 to 37.0) | <0.001 | 0.03 | 0.81 | 0.15 |
| 302 | 34.8 (30.9 to 38.5) | <0.001 | 20.0 (−108.2 to 69.3) | 0.64 | 0.69 | 1.00 | 0.89 |
| 303 | 34.6 (30.7 to 38.3) | <0.001 | 68.4 (−100.9 to 95.0) | 0.22 | 0.45 | 1.00 | 0.71 |
| Th2R | |||||||
314 | 32.7 (26.2 to 38.2) | <0.001 | 37.3 (30.9 to 43.0) | <0.001 | 0.33 | 1.00 | 0.61 |
317¶ | 45.9 (33.3 to 56.2) | <0.001 | 33.1 (28.9 to 37.1) | <0.001 | 0.06 | 1.00 | 0.19 |
| 318 | 36.9 (25.8 to 46.4) | <0.001 | 34.2 (29.8 to 38.4) | <0.001 | 0.65 | 1.00 | 0.89 |
| 320 | 34.4 (30.4 to 38.1) | <0.001 | 64.0 (21.0 to 83.6) | 0.01 | 0.14 | 1.00 | 0.32 |
| 321 | 35.7 (24.3 to 45.3) | <0.001 | 34.5 (30.1 to 38.7) | <0.001 | 0.85 | 1.00 | 0.89 |
| 322 | 34.8 (27.2 to 41.5) | <0.001 | 34.7 (29.4 to 39.6) | <0.001 | 0.99 | 1.00 | 0.99 |
| 324 | 37.3 (32.0 to 42.2) | <0.001 | 30.7 (23.3 to 37.4) | <0.001 | 0.16 | 1.00 | 0.35 |
| 327 | 35.5 (31.2 to 39.5) | <0.001 | 30.1 (16.5 to 41.4) | <0.001 | 0.42 | 1.00 | 0.71 |
| Th3R | |||||||
| 349 | 34.8 (30.9 to 38.5) | <0.001 | 25.4 (−84.1 to 69.8) | 0.53 | 0.81 | 1.00 | 0.89 |
352 | 35.1 (30.7 to 39.2) | <0.001 | 32.8 (20.0 to 43.5) | <0.001 | 0.73 | 1.00 | 0.89 |
354¶ | 36.0 (32.0 to 39.8) | <0.001 | 10.8 (−22.6 to 35.1) | 0.48 | 0.05 | 1.00 | 0.17 |
| 355 | 34.7 (30.8 to 38.4) | <0.001 | 54.1 (−286.1 to 94.5) | 0.47 | 0.71 | 1.00 | 0.89 |
356¶ | 36.2 (32.2 to 40.1) | <0.001 | 15.8 (−7.8 to 34.3) | 0.17 | 0.04 | 0.85 | 0.15 |
357 | 35.3 (27.3 to 42.5) | <0.001 | 34.4 (29.3 to 39.2) | <0.001 | 0.86 | 1.00 | 0.89 |
| 359¶ | 36.1 (32.0 to 40.1) | <0.001 | 22.2 (5.2 to 36.2) | 0.01 | 0.07 | 1.00 | 0.20 |
| 361¶ | 39.3 (33.6 to 44.4) | <0.001 | 29.3 (22.4 to 35.5) | <0.001 | 0.03 | 0.81 | 0.15 |
 The linkage disequilibrium (LD) haplotype includes this amino acid position.
The linkage disequilibrium (LD) haplotype includes this amino acid position.For the parasite-positivity end point at 18 months after dose 3, in the older age category, the estimates of vaccine efficacy tended to be higher for circumsporozoite protein C-terminal 3D7-matched malaria than for 3D7-mismatched malaria (e.g., vaccine efficacy, 53% vs. 30%; P = 0.19 for the full amplicon), although none of the differences were significant (Table S16 in the Supplementary Appendix). In contrast, there was no evidence at all of a sieve effect for the circumsporozoite protein C-terminal in the younger age category with regard to this end point (Table S17 in the Supplementary Appendix), and there was no evidence of a sieve effect for SERA-2 in either age category (Tables S18 and S19 in the Supplementary Appendix).
NANP–NVDP REPEAT REGION
In 3137 samples representing the clinical malaria end point with sequence data that could be evaluated from the B-cell epitope repeat region, the NANP–NVDP repeat count ranged from 37 to 44, with a mode of 40 repeats. There was a nonsignificant trend toward declining cumulative vaccine efficacy with increasing NANP–NVDP repeat count in the older age category (P = 0.07) (Fig. S16 in the Supplementary Appendix) and no significant differential vaccine efficacy according to repeat count in the younger age category (P = 0.89) (Fig. S17 in the Supplementary Appendix). We did not assess the dependence of vaccine efficacy on NANP–NVDP repeat amino acid sequences because the vaccine construct contains a truncated repeat region (18.5 NANP–NVDP repeats).
DISCUSSION
The discovery that RTS,S/AS01 vaccine efficacy is higher against clinical malaria with infections matching the 3D7 vaccine construct at epitope haplotypes and amino acid positions than it is against infections not matching 3D7 is not entirely unexpected, given the polymorphism at the C-terminal of the circumsporozoite protein antigenic locus and previous observations of allele-specific immune responses to other parasite proteins.27,29 This differential cumulative vaccine efficacy result could be a false positive result, given that the result for the full circumsporozoite protein C-terminal was of borderline significance (P = 0.04) and that the analysis of four epitopes and 24 amino acid sites gave no Holm–Bonferroni-adjusted P values below 0.05. However, 11 of the 28 tests yielded Q values less than or equal to 0.20; consequently, we expect at least 80% of these 11 results to be true positives.
The current study had greater power to detect allele-specific protection than did previous evaluations of RTS,S in phase 2 trials, because of three factors: a larger sample, the inclusion of study sites harboring a higher frequency of 3D7-matching haplotypes, and the use of PCR-based next-generation sequencing to resolve the haplotypes that make up mixed infections. Our main result of significant sieve effects for the primary clinical malaria end point in the age category of 5 to 17 months was based on a large sample, with measured genetic data from 2145 total clinical malaria cases. In contrast, the sieve analysis of the parasite-positivity end point in participants 5 to 17 months of age, which yielded nonsignificant results, had many fewer cases (genetic data from 507 total cases). However, in terms of estimates of vaccine efficacy, the sieve effects were slightly stronger for the parasite-positivity end point, which suggests that the lack of significance could be due to lower statistical power rather than to a true lack of differential protection. Post hoc power calculations showed only 30% power to detect the 23-percentage-point difference between vaccine efficacies (53% against malaria matching the 3D7 circumsporozoite protein C-terminal vs. 30% against mismatched malaria) with regard to the parasite-positivity end point, as compared with 51% power to detect such a difference with regard to the clinical malaria end point. Therefore, the selective vaccine protection may have operated for both clinical malaria and parasite positivity. In contrast, there was no evidence for allele-specific vaccine efficacy in the age category of 6 to 12 weeks, despite the significant over all protection in this group. This result implies a qualitative difference in the vaccine response, in addition to the previously reported quantitative difference in anti-circumsporozoite antibody titers in the younger age category.4 The biologic mechanisms underlying the differences between the age categories remain to be elucidated, but they could include a role of maternal antibody, interactions with other vaccine responses, or differences in immune-response capacity between infants and children. Further immunologic analyses could clarify the mechanisms of selective versus nonselective vaccine-induced immunity.
Genetic surveillance of circumsporozoite protein sequences in parasite populations could inform the development of future vaccine candidates targeting polymorphic malaria parasite proteins. The genotype-specific vaccine efficacy results we report here complement previous estimates of RTS,S/AS01 efficacy in this phase 3 trial; the previously reported 12-month hazard-ratio vaccine efficacy of 55.8% against clinical malaria among children 5 to 17 months of age4 can now be interpreted as a multiplicative weighted mean of hazard-ratio vaccine efficacy values of 62.7% against matched parasites (139 infections) and 54.2% against mismatched parasites (1951 infections) (Fig. 5B). The observed variation among study sites in infections with a perfect vaccine match in the circumsporozoite protein C-terminal (Fig. 3B, and Fig. S18 and S19 in the Supplementary Appendix) may help to explain previously reported variation in overall vaccine efficacy among study sites, although the magnitude of this contribution is expected to be low because of the overall rarity of the 3D7 haplotype.6 Broader deployment of the vaccine could result in increased selection on the 3D7 haplotype or its component epitopes and amino acid alleles. Sieve analysis of next-generation sequencing data constitutes an approach for understanding partial vaccine efficacy.
Acknowledgments
Supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract no. HHSN272200900018C); the Bill and Melinda Gates Foundation; the PATH Malaria Vaccine Initiative; GlaxoSmithKline Biologicals; and the Harvard Malaria Initiative.
We thank the members of the RTS,S Clinical Trials Partnership Committee for their support and assistance, the staff of the Broad Institute Genomics Platform for data generation, James Bochicchio for project management, and Ted Holzman (Fred Hutchinson Cancer Research Center) for assistance with computational analysis.
APPENDIX
The authors’ full names and academic degrees are as follows: Daniel E. Neafsey, Ph.D., Michal Juraska, Ph.D., Trevor Bedford, Ph.D., David Benkeser, M.P.H., Clarissa Valim, M.D., Sc.D., Allison Griggs, M.Sc., Marc Lievens, M.Sc., Salim Abdulla, M.D., Ph.D., Samuel Adjei, M.B., Ch.B., D.T.M., Tsiri Agbenyega, M.B., Ch.B., Ph.D., Selidji T. Agnandji, M.D., Pedro Aide, M.D., Ph.D., Scott Anderson, B.S., Daniel Ansong, M.B., Ch.B., John J. Aponte, M.D., Ph.D., Kwaku Poku Asante, M.D., Ph.D., Philip Bejon, Ph.D., Ashley J. Birkett, Ph.D., Myriam Bruls, M.Sc., Kristen M. Connolly, B.S., Umberto D’Alessandro, M.D., Ph.D., Carlota Dobaño, Ph.D., Samwel Gesase, M.D., Brian Greenwood, M.D., Jonna Grimsby, Ph.D., Halidou Tinto, Ph.D., Mary J. Hamel, M.D., D.T.M.&H., Irving Hoffman, P.A., M.P.H., Portia Kamthunzi, M.D., Simon Kariuki, Ph.D., Peter G. Kremsner, M.D., Amanda Leach, M.R.C.P.C.H., Bertrand Lell, M.D., Niall J. Lennon, Ph.D., John Lusingu, M.D., Ph.D., Kevin Marsh, M.D., Francis Martinson, M.D., Ph.D., Jackson T. Molel, M.Sc., Eli L. Moss, B.S., Patricia Njuguna, M.B., Ch.B., M.Med., Christian F. Ockenhouse, M.D., Ph.D., Bernhards Ragama Ogutu, Ph.D, Walter Otieno, M.B., Ch.B., M.Med., Lucas Otieno, M.B., Ch.B., M.P.H., Kephas Otieno, M.P.H., Seth Owusu-Agyei, Ph.D., Daniel J. Park, Ph.D., Karell Pellé, Ph.D., Dana Robbins, B.S., Carsten Russ, Ph.D., Elizabeth M. Ryan, B.S., Jahit Sacarlal, M.D., Ph.D., Brian Sogoloff, B.S., Hermann Sorgho, Ph.D., Marcel Tanner, Ph.D., Thor Theander, M.D., D.Sc., Innocent Valea, Pharm.D., Ph.D., Sarah K. Volkman, Sc.D., Qing Yu, M.Sc., Didier Lapierre, M.D., Bruce W. Birren, Ph.D., Peter B. Gilbert, Ph.D., and Dyann F. Wirth, Ph.D.
The authors’ affiliations are as follows: the Broad Institute of Massachusetts Institute of Technology and Harvard (D.E.N., C.V., A.G., S. Anderson, K.M.C., J.G., N.J.L., E.L.M., D.J.P., K.P., D.R., C.R., E.M.R., B.S., S.K.V., Q.Y., B.W.B.), Cambridge, and Harvard T.H. Chan School of Public Health (C.V., K.P., S.K.V., D.F.W.) and Simmons College School of Nursing and Health Sciences (S.K.V.), Boston — all in Massachusetts; Fred Hutchinson Cancer Research Center (M.J., T.B., D.B., C.V., P.B.G.) and University of Washington (D.B., P.B.G.), Seattle; GlaxoSmithKline Vaccines, Rixensart, Belgium (M.L., M.B., A.L., D.L.); Ifakara Health Institute, Bagamoyo (S. Abdulla, J.T.M.), and National Institute for Medical Research, Korogwe (S.G., J.L.) — both in Tanzania; School of Medical Sciences, Kwame Nkrumah University of Science and Technology, Kumasi (S. Adjei, T.A.), and Kintampo Health Research Centre, Kintampo (K.P.A., B.G., S.O.-A.) — both in Ghana; Albert Schweitzer Hospital, Lambaréné, Gabon (S.T.A., D.A., P.G.K., B.L.); Institute of Tropical Medicine, University of Tübingen, Tübingen, Germany (S.T.A., P.G.K., B.L.); Centro de Investigaçåo em Saùde de Manhiça, Manhiça, Mozambique (P.A., J.J.A., C.D., J.S.); ISGlobal, Barcelona Center for International Health Research, Hospital Clinic–Universitat de Barcelona, Barcelona (P.A., J.J.A., C.D.); PATH Malaria Vaccine Initiative, Washington, DC (A.J.B., C.F.O.); Medical Research Council Unit, Banjul, Gambia (U.D.); London School of Hygiene and Tropical Medicine, London (B.G.); Institut de Recherche en Sciences de la Santé, Nanoro, Burkina Faso (H.T., H.S., I.V.); Centers for Disease Control and Prevention (CDC), Atlanta (M.J.H.); University of North Carolina (UNC), Chapel Hill (I.H.); UNC Project, Lilongwe, Malawi (P.K., F.M.); Kenya Medical Research Institute (KEMRI)–Wellcome Trust Research Program, Kilifi (P.B., K.M., P.N.), KEMRI–CDC Research and Public Health Collaboration, Kisumu (S.K., K.O.), and KEMRI–Walter Reed Project, Kombewa (B.R.O., W.O., L.O., K.P.) — all in Kenya; Swiss Tropical and Public Health Institute, Basel, Switzerland (M.T.); and the University of Copenhagen, Copenhagen (T.T.).
Footnotes
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1056/nejmoa1505819
Read article for free, from open access legal sources, via Unpaywall:
https://www.nejm.org/doi/pdf/10.1056/NEJMoa1505819?articleTools=true
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1056/nejmoa1505819
Article citations
Plasmodium vivax antigen candidate prediction improves with the addition of Plasmodium falciparum data.
NPJ Syst Biol Appl, 10(1):133, 13 Nov 2024
Cited by: 0 articles | PMID: 39537634 | PMCID: PMC11561111
Vaccine-induced human monoclonal antibodies to PfRH5 show broadly neutralizing activity against P. falciparum clinical isolates.
NPJ Vaccines, 9(1):198, 24 Oct 2024
Cited by: 0 articles | PMID: 39448626 | PMCID: PMC11502735
Whole-sporozoite malaria vaccines: where we are, where we are going.
EMBO Mol Med, 16(10):2279-2289, 16 Sep 2024
Cited by: 0 articles | PMID: 39284948 | PMCID: PMC11473726
Review Free full text in Europe PMC
Monoclonal antibodies to the circumsporozoite proteins as an emerging tool for malaria prevention.
Nat Immunol, 25(9):1530-1545, 28 Aug 2024
Cited by: 0 articles | PMID: 39198635
Review
Genetic structure of apical membrane antigen-1 in Plasmodium falciparum isolates from Pakistan.
Parasites Hosts Dis, 62(3):302-312, 26 Aug 2024
Cited by: 0 articles | PMID: 39218629 | PMCID: PMC11366544
Go to all (223) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
BioProject
- (1 citation) BioProject - PRJNA235895
Clinical Trials
- (1 citation) ClinicalTrials.gov - NCT00866619
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Safety and efficacy of the RTS,S/AS01E candidate malaria vaccine given with expanded-programme-on-immunisation vaccines: 19 month follow-up of a randomised, open-label, phase 2 trial.
Lancet Infect Dis, 11(10):741-749, 22 Jul 2011
Cited by: 85 articles | PMID: 21782519
Immunogenicity of the RTS,S/AS01 malaria vaccine and implications for duration of vaccine efficacy: secondary analysis of data from a phase 3 randomised controlled trial.
Lancet Infect Dis, 15(12):1450-1458, 02 Sep 2015
Cited by: 194 articles | PMID: 26342424 | PMCID: PMC4655306
First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children.
N Engl J Med, 365(20):1863-1875, 18 Oct 2011
Cited by: 532 articles | PMID: 22007715
The malaria vaccine--status quo 2013.
Travel Med Infect Dis, 11(1):2-7, 01 Jan 2013
Cited by: 13 articles | PMID: 23454205
Review
Funding
Funders who supported this work.
NCRR NIH HHS (1)
Grant ID: UL1 RR025758
NIAID NIH HHS (3)
Grant ID: HHSN272200900018C
Grant ID: R37 AI054165
Grant ID: U19 AI110818
PHS HHS (1)
Grant ID: HHSN272200900018C
Wellcome Trust (1)
Oxford part of Kenya MOP renewal 2010 - 2015
Professor Kevin Marsh, University of Oxford
Grant ID: 092654