Abstract
Free full text

EWS/FLI-1 Silencing and Gene Profiling of Ewing Cells Reveal Downstream Oncogenic Pathways and a Crucial Role for Repression of Insulin-Like Growth Factor Binding Protein 3†
Abstract
Ewing tumors are characterized by abnormal transcription factors resulting from the oncogenic fusion of EWS with members of the ETS family, most commonly FLI-1. RNA interference targeted to the junction between EWS and FLI-1 sequences was used to inactivate the EWS/FLI-1 fusion gene in Ewing cells and to explore the resulting phenotype and alteration of the gene expression profile. Loss of expression of EWS/FLI-1 resulted in the complete arrest of growth and was associated with a dramatic increase in the number of apoptotic cells. Gene profiling of Ewing cells in which the EWS/FLI-1 fusion gene had been inactivated identified downstream targets which could be grouped in two major functional clusters related to extracellular matrix structure or remodeling and regulation of signal transduction pathways. Among these targets, the insulin-like growth factor binding protein 3 gene (IGFBP-3), a major regulator of insulin-like growth factor 1 (IGF-1) proliferation and survival signaling, was strongly induced upon treating Ewing cells with EWS/FLI-1-specific small interfering RNAs. We show that EWS/FLI-1 can bind the IGFBP-3 promoter in vitro and in vivo and can repress its activity. Moreover, IGFBP-3 silencing can partially rescue the apoptotic phenotype caused by EWS/FLI-1 inactivation. Finally, IGFBP-3-induced Ewing cell apoptosis relies on both IGF-1-dependent and -independent pathways. These findings therefore identify the repression of IGFBP-3 as a key event in the development of Ewing's sarcoma.
Ewing tumors, the second most frequent bone tumors in adolescents and young adults, are characterized by the presence of specific gene fusions which most frequently involve the EWS gene on chromosome 22 and the FLI-1 gene on chromosome 11 (8). Less frequently, EWS is fused with other members of the ETS family, including ERG, ETV1, E1A-F, or FEV (1). As a result of these gene fusions, tumor cells express a chimeric protein that contains the amino-terminal part of EWS and the DNA binding domain of the ETS transcription factor. EWS belongs to the TET family of proteins, which also includes TAFII68 and TLS/FUS (4). All three proteins from this family have been shown to be involved in cancer-specific translocations, following a general scheme, which fuses the N-terminal domain of the TET partner to a variety of DNA binding domains, each fusion being highly specific for a tumor type. Numerous reports have addressed the mechanisms of tumoral transformation induced by TET fusion proteins and particularly EWS/FLI-1, the most frequent representative.
Considerable attention has focused on the search for specific downstream target genes that may mediate EWS/FLI-1 transforming properties. Although the specificity of the EWS/FLI-1 fusion gene for Ewing tumors suggests that the cell context is critical for EWS/FLI-1-induced oncogenesis, the parental Ewing cell of origin is currently unknown. This issue precludes the development of homologous cell systems that may be particularly helpful to design strategies aimed at the identification of specific target genes. Given this concern, all approaches to seek EWS/FLI-1 target genes have been based on differential screenings using ectopic overexpression of EWS/FLI-1 in heterologous cell systems. A number of attractive targets have been identified, but most of these targets remain to be validated in the Ewing context. Differential screenings performed in Ewing cells with EWS/FLI-1 being turned on or off could constitute an alternate strategy to heterologous models. Several studies have reported the use of RNA antisense for silencing EWS/ETS fusion (12, 13, 18, 31), but no systematic screening for changes in the expression profile has been reported using this approach. Recently, the advent of RNA interference and the ability to specifically silent gene expression using small interfering RNAs (siRNAs) have allowed us to consider new means to inhibit EWS/FLI-1 in Ewing cells (9).
In this study, we have combined the siRNA strategy with the DNA microarray technique in order to identify genes that are regulated by EWS/FLI-1. Among these genes, we particularly focused our attention on the insulin-like growth factor binding protein 3 gene (IGFBP-3), a major regulator of cell proliferation and apoptosis.
MATERIALS AND METHODS
Antibodies and oligonucleotides.
Antibodies against AKT, phospho-AKT (Ser 473), p44/42 mitogen-activated protein kinase (MAPK), phospho-p44/42 MAPK (Thr 202 and Tyr 204), and RAF-1 were obtained from Cell Signaling (Beverly, Mass.). IGFBP-3 antibody was purchased from Upstate Cell Signaling Solutions (Charlottesville, Va.). Hemagglutinin (HA), histidine (His), and Flag antibodies were acquired from Roche Diagnostic (Meylan, France), Santa Cruz Biotechnology (Santa Cruz, Calif.), and Sigma-Aldrich (Saint-Quentin Fallavier, France), respectively. The cyanin-3-conjugated anti-rabbit antibodywas from Jackson ImmunoResearch Laboratories (West Grove, Penn.), and fluorescein isothiocyanate (FITC)-conjugated anti-mouse antibody was from DakoCytomation (Trappes, France). Anti-FLI-1 antibody (7.3) was described previously (16).
The siRNAs for EWS/FLI1 (5′ GGC AGC AGA ACC CUU CUU A-dCdG)and control (5′ GGC AGC AGA GUU CAC UGC U-dCdG) were purchased from Proligo (Paris, France), and siRNA for IGFBP-3 was purchased from Dharmacon and MWG Biotech (Ebersberg, Germany). The oligonucleotides used for reverse transcription-PCR (RT-PCR) experiments were as follows: for the EWS/FLI-1 fusion gene, GCACCTCCATCCTACCCTCCT (forward) and TGGCAGTGGGTGGGTCTTCAT (reverse); for the DKK1 gene, GCATGCGTCACGCTATGTGC (forward) and TGAAGACAAGGTGGTTCTTCTGG (reverse); for the DKK3 gene, GATGTTCCGCGAGGTTGAGG (forward) and CCAACCTTCGTGTCTGTGTTGG (reverse); for the IGFBP-5 gene, CCGCGAGCAAGTCAAGATCG (forward) and AGCCTTCAGCTCGGAGATGC (reverse); for the LOX gene, GGATACGGCACTGGCTACTTCC (forward) and GCGCATCTCAGGTTGTACATGG (reverse); for the CHI3L1 gene, GAATTCCAGGCCGGTTCACC (forward) and GCTGGCCGAGGATTCTATGG (reverse); for the CYP1B1 gene, GGCCACTATCACTGACATCTTCG (forward) and TCCAATTCTGCCTGCACTCG (reverse); for the CD44 gene, TTGCATTGCAGTCAACAGTCG (forward) and CCTCTCCGTTGAGTCCACTTGG (reverse); for the glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH), CTTCAACAGCGACACCCACT (forward) and GTGGTCCAGGGGTCTTACTC (reverse); and for the IGFBP-3 gene, GCACTGCTACAGCATGCAGAGC (forward) and CAGCTGCTGGTCATGTCCTTGG (reverse). IGFBP-3 oligonucleotides for the electrophoretic mobility shift assay (EMSA) were (sense strand only) AGTATTTAAGGAACGGATGTAAACCTGGG (position −1829) and ATGGCACGGTTTCCTGTAAACAAG (position −1686). IGFBP-3 oligonucleotides for chromatin immunoprecipitation (ChIP) were AGTATTTAAGGAACGGATGT (forward) and CAGCACCGTCATTTTCTTAT (reverse).
siRNA transfection experiments.
A total of 4 × 105 A673 type 1 Ewing cells were plated and propagated in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (Abcys SA, Paris, France). The cells were transfected 24 h later with 200 pmol of each siRNA duplex using OligofectAMINE (Invitrogen, Cergy Pontoise, France). For transfection on six-well plates, 10 μl of a 20 μM oligonucleotide stock solution was diluted in 200 μl of Opti-MEM (Invitrogen) and 10 μl of OligofectAMINE reagent was mixed into 50 μl of Opti-MEM. After 10-min incubation, both solutions were gently mixed and incubated at room temperature for 20 min and then added to cells freshly incubated with 1.5 ml of antibiotic- and serum-free medium. Twenty-four hours after transfection, cells were washed and grown for another 48 h. Cells were either harvested or subjected to a second round of transfection.
FACS analysis.
For cell cycle analysis, cells fixed in 70% ethanol for 3 h at 4°C were stained with propidium iodide (50 μg/ml). To evaluate apoptosis, cells were labeled using annexin V-FITC apoptosis detection kit (BD Biosciences, San Diego, Calif.) following the manufacturer's recommendations. A total of 10,000 cells in each sample were subjected to fluorescence-activated cell sorting (FACS) analysis (FACScalibur; BD Biosciences), and the data were processed using CELL Quest software (BD Biosciences).
DNA microarray analysis.
Experimental procedures for GeneChip microarray were performed according to the manufacturer's instructions (Affymetrix GeneChip expression analysis technical manual, Affymetrix, Santa Clara, Calif.) using the HG-U133A array. The expression data were analyzed using the Affymetrix MicroArray Suite version 5.0, dChip, and Rosetta Resolver software programs.
Quantitative RT-PCR.
Total RNA samples (2 μg) were reverse transcribed with oligonucleotide-random hexamers using the GeneAmp RNA PCR core kit (Applied Biosystems, Courtaboeuf, France). Quantitative RT-PCR using the fluorescent SYBR green method (Applied Biosystems) was performed according to the manufacturer's instructions.
ChIP.
Briefly, cross-linking was performed with 20 × 106 cells for 7 min. Cells were then lysed in 1.5 ml of radioimmunoprecipitation assay buffer and sonicated for 10 min at 20% duty cycles. Cell lysates precleared with protein A-Sepharose were incubated for 2 h with 2 μl of anti-HA, anti-His, or anti-Flag antibodies or 0.2 μl of anti-FLI-1 (7.3) antibodies. After sequential washes and elution of immunoprecipitates, cross-links were reversed overnight at 65°C. Proteins were then digested, and DNA was ethanol precipitated and resuspended in 15 μl of Tris-EDTA buffer. One microliter of purified DNA was used for 25 cycles of PCR amplification (Applied Biosystems) using IGFBP-3 promoter-specific primers.
EMSA.
Duplex primers were purified on nondenaturing 6% acrylamide gel before [γ32P]ATP PNK end labeling. EMSA experiments were conducted as described previously (16). Reaction mixtures were incubated with 2 μl (or the indicated amount) of in vitro-translated EWS/FLI-1 or FLI-1 with or without 1 μl of a 1/10 dilution of antibody 7.3 ascitic fluid.
Luciferase assay.
A fragment of the IGFBP-3 promoter from positions −2584 to +63 was amplified by PCR (forward oligonucleotide, CGTAGCGGCCGCCTCCTGACCTCTATTCGGATGGTTATTCG; reverse oligonucleotide, CGTAAAGCTTGAAGCTGTGGAATCCAGGCAGGAAGCGG) from MCF7 genomicDNA and introduced between NotI and HindIII sites of the pREP4-luc vector (generous gift of Keji Zhao). A total of 4 × 105 HeLa cells were seeded into six-well plates in duplicate and grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Cells were transfected by Effecten transfection reagents (QIAGEN, Courtaboeuf, France) 24 h later with 400 ng of the reporter plasmid, 4 ng of pRL-SV40 Renilla luciferase control, and 250 ng of ΔEB-78/EWS/FLI-1, ΔEB-78/EWS/FLI-1-derived mutants, or ΔEB-78/FLI-1 (2). Forty-eight hours posttransfection, cells were lysed and assayed for luciferase activity using the dual luciferase reporter assay system (Promega, Charbonnières-les-bain, France) according to the manufacturer's instructions.
Immunodetection.
For immunofluorescence experiments, 103 HeLa cells grown in eight-well plates (Nunc GmbH, Wiesbaden, Germany) were transfected by the Effectene procedure (QIAGEN) with 6.5 ng of ΔEB-78/EWS/FLI-1, fixed, permeabilized, and stained as previously described (16). Mitochondrial RAF-1 was detected as previously described (19).
RESULTS
In order to abolish EWS/FLI-1 expression in Ewing cell lines, we designed a siRNA duplex specifically directed against the fusion point of EWS/FLI-1 type 1 transcript (Fig. (Fig.1A).1A). This EWS/FLI-1-specific siRNA (siEF1) and a control siRNA (siCT) were used to transfect A673 Ewing cells expressing EWS/FLI-1 type 1 transcripts. A single round of transfection of siEF1 led to a specific inhibition of the expression of the EWS/FLI-1 fusion gene that was almost complete after two successive rounds of transfections (Fig. 1B and C). In agreement with the specificity of siEF1 toward the EWS/FLI-1 fusion gene, the levels of expression of EWS and FLI-1 were not modified (Fig. (Fig.1B).1B). Therefore, all siRNA experiments described hereafter were performed using this double-transfection strategy following the scheme shown in Fig. Fig.1D1D.
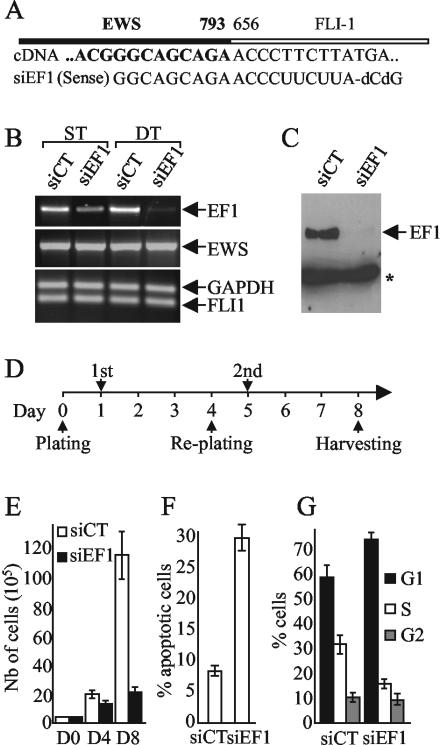
Induction of apoptosis and cell cycle arrest by inhibition of expression of the EWS/FLI-1 fusion gene using specific siRNA. (A) Scheme of type 1 EWS/FLI-1 junction and sequence of the cDNA and siEF1. (B) Inhibition of the expression of EWS/FLI-1 transcript by siEF1. RNA from Ewing cells transfected with either siEF1 or siCT were subjected to competitive RT-PCR experiments. GAPDH was used as an internal control. ST and DT, single and double transfections, respectively. (C) Immunoblot with the 7.3 monoclonal antibody. The position of a cross-reactive band used for standardization (16) is indicated by an asterisk. (D) Temporal scheme of the double-transfection strategy. The first (1st) and second (2nd) siRNA transfections are indicated. (E to G) Growth inhibition, apoptosis, and cell cycle arrest induced by the silencing of EWS/FLI-1 in the A673 Ewing cell line. Seventy-two hours after the second transfection, the cells were counted (E), analyzed by FACS for apoptosis using an annexin V detection kit assay (F), and analyzed for cell cycle progression using propidium iodide staining (G). The means ± standard deviations (error bars) of two independent experiments are shown. D0, D4, and D8, days 0, 4, and 8 after the initial plating, respectively.
We then investigated the phenotype of EWS/FLI-1 knockdowned Ewing cells. Analysis of growth curves demonstrated a marked growth inhibition in siEF1-transfected cells compared to siCT-treated cells (Fig. (Fig.1E).1E). The first transfection led to a slight decrease in the number of cells at day 4, and the second transfection resulted in a complete growth arrest. The timing of the growth arrest paralleled the decrease in the amount of the EWS/FLI-1 protein (Fig. (Fig.1C).1C). FACS analysis with annexin V and propidium iodide stains demonstrated that this dramatic inhibition of cell growth could be related to both a strong increase in the number of apoptotic cells (Fig. (Fig.1F)1F) and a significant reduction of the number of cells in the S phase with parallel accumulation of cells in the G1 phase of the cell cycle (Fig. (Fig.1G1G).
Total RNAs from Ewing cells transfected with siEF1 or siCT were isolated and used to perform duplicate hybridization experiments with Affymetrix HG-U133A arrays that contain 22,000 probe sets. Results were analyzed using the Affymetrix Microarray Suite version 5, Resolver (Rosetta Inpharmatics), and dChip software programs. Statistical comparison by analysis of variance using the Rosetta software program leads to the identification of 114 probe sets, corresponding to 86 genes, the expression of which differed significantly between siEF1- and siCT-treated cells (Table (Table1).1). Interestingly enough, the number of genes up-regulated by siEF1, and therefore expected to be down-regulated by EWS/FLI-1, was much higher than the number of genes with the reverse pattern, since only six genes were significantly down-regulated by siEF1. Analysis of the differentially expressed genes using the dChip Gene Ontology classifier identified two highly significant functional clusters (Table (Table1).1). One group includes a variety of genes involved in signal transduction, particularly receptor binding. More specifically, this group contains secreted regulators of the Wnt, insulin-like growth factor 1 (IGF-1), and epidermal growth factor pathways and intracellular inhibitors of the MAPK and STAT pathways. The second functional cluster includes a number of molecules involved in the formation of, remodeling of, and adhesion to the extracellular matrix (ECM). In order to confirm these DNA array results, the levels of expression of eight genes up-regulated in the presence of siEF1 (IGFBP-3, IGFBP-5, DKK1, DKK3, LOX, CHI3L1, CYP1B1, and CD44) were further analyzed by quantitative RT-PCR experiments. As shown in Table Table1,1, quantitative RT-PCR experiments unambiguously confirmed the microarray results, with changes higher than those found with DNA chips.
TABLE 1.
Clusters of genes regulated by EWS/FLI-1 in Ewing cellse
| Functional gene cluster and accession no. | Genea | Fold change
| ||
|---|---|---|---|---|
| Probe setb | QPCRc | |||
| Extracellular genes | ||||
    NM_012242.1 NM_012242.1 | DKK1d | 31.7 | 94 | |
    L27560.1 L27560.1 | IGFBP-5d | 27.8 | 166 | |
    NM_004369.1 NM_004369.1 | COL6A3 | 24.8 | ||
    M31159.1 M31159.1 | IGFBP-3d | 17.5 | 555 | |
    NM_001432.1 NM_001432.1 | EREGd | 11.4 | ||
    M80927.1 M80927.1 | CHI3L1 | 8.7 | 39 | |
    NM_002317.1 NM_002317.1 | LOX | 7.4 | 70 | |
    AU148057 AU148057 | DKK3d | 5.9 | 27 | |
    NM_002309.2 NM_002309.2 | LIFd | 5.3 | ||
    NM_000358.1 NM_000358.1 | TGFBI | 5.0 | ||
    M57731.1 M57731.1 | CXCL2d | 4.5 | ||
    J04177 J04177 | COL11A1 | 4.5 | ||
    M11734.1 M11734.1 | CSF2d | 4.3 | ||
    NM_001511.1 NM_001511.1 | CXCL1d | 4.2 | ||
    NM_004591.1 NM_004591.1 | CCL20d | 4.0 | ||
    S69738.1 S69738.1 | CCL2d | 3.9 | ||
    NM_000064.1 NM_000064.1 | C3d | 3.7 | ||
    NM_002852.1 NM_002852.1 | PTX3 | 3.5 | ||
    NM_000204.1 NM_000204.1 | IF | 3.5 | ||
    U16307.1 U16307.1 | GLIPR1 | 3.4 | ||
    AL574096 AL574096 | TFP12 | 3.4 | ||
    NM_002658.1 NM_002658.1 | PLAUd | 3.4 | ||
    M15330 M15330 | IL1bd | 3.3 | ||
    NM_001124.1 NM_001124.1 | ADMd | 3.3 | ||
    AY029208.1 AY029208.1 | COL6A2 | 3.1 | ||
    NM_000610.1 NM_000610.1 | CD44 | 2.9 | 12 | |
    BC003355.1 BC003355.1 | LAMA5 | 2.9 | ||
    NM_002318.1 NM_002318.1 | LOXL2 | 2.9 | ||
    AU144167 AU144167 | COL3A1 | 2.8 | ||
    NM_003358.1 NM_003358.1 | UGCG | 2.7 | ||
    NM_002966.1 NM_002966.1 | S100A10d | 2.7 | ||
    NM_002993.1 NM_002993.1 | CXCL6d | 2.7 | ||
    AA292373 AA292373 | COL6A1 | 2.7 | ||
    NM_002084.2 NM_002084.2 | GPX3 | 2.6 | ||
    NM_002178.1 NM_002178.1 | IGFBP-6d | 2.5 | ||
    AY029180.1 AY029180.1 | PLAURd | 2.3 | ||
    AI264196 AI264196 | FBN1 | 2.2 | ||
| Signal transduction genes | ||||
    AB005043.1 AB005043.1 | SOCS1 | 4.8 | ||
    U58111.1 U58111.1 | VEGF-C | 4.3 | ||
    NM_002045.1 NM_002045.1 | GAP43 | 3.6 | ||
    BE620457 BE620457 | NRP1 | 2.6 | ||
    BC005047.1 BC005047.1 | DUSP6 | 2.5 | ||
    BC001422.1 BC001422.1 | PGF | 2.4 | ||
    NM_004414.2 NM_004414.2 | DSCR1 | 2.3 | ||
    BF575514 BF575514 | PBEF | 2.1 | ||
    BC003096.1 BC003096.1 | RIL | 2.1 | ||
    AW242315 AW242315 | PTGER3 | −2.0 | ||
    NM_006472.1 NM_006472.1 | TXNIP | −2.8 | ||
| Other genes | ||||
    NM_000050.1 NM_000050.1 | ASS | 21.1 | ||
    NM_005651.1 NM_005651.1 | TDO2 | 7.4 | ||
    AL050025.1 AL050025.1 | AP1G1 | 6.1 | ||
    BF752277 BF752277 | FLJ20151 | 5.6 | ||
    NM_002526.1 NM_002526.1 | NT5E | 5.1 | ||
    NM_000963.1 NM_000963.1 | PTGS2 | 5.0 | ||
    AI346835 AI346835 | TM4SF1 | 4.9 | ||
    AI653981 AI653981 | LICAM | 4.1 | ||
    NM_017458.1 NM_017458.1 | MVP | 3.9 | ||
    NM_005824.1 NM_005824.1 | LRRC17 | 3.9 | ||
    NM_015577.1 NM_015577.1 | RAI14 | 3.8 | ||
    NM_000104.2 NM_000104.2 | CYP1B1 | 3.2 | 15 | |
    L03203.1 L03203.1 | PMP22 | 3.2 | ||
    NM_005978.2 NM_005978.2 | SI00A2 | 3.2 | ||
    NM_016352.1 NM_016352.1 | CPA4 | 3.2 | ||
    NM_005382.1 NM_005382.1 | NEF3 | 3.1 | ||
    NM_014600.1 NM_014600.1 | EHD3 | 3.1 | ||
    NM_005195.1 NM_005195.1 | CEBPD | 3.1 | ||
    NM_013451.1 NM_013451.1 | FERIL3 | 3.0 | ||
    NM_005532.1 NM_005532.1 | IFI27 | 2.9 | ||
    AA576961 AA576961 | PHLDA1 | 2.9 | ||
    AK026420.1 AK026420.1 | DMN | 2.9 | ||
    M19267.1 M19267.1 | TPM1 | 2.8 | ||
    NM_006158.1 NM_006158.1 | NEFL | 2.8 | ||
    AF043337.1 AF043337.1 | IL8 | 2.4 | ||
    NM_003186.2 NM_003186.2 | TAGLN | 2.3 | ||
    D13889.1 D13889.1 | Id1 | 2.3 | ||
    NM_018444.1 NM_018444.1 | PPM2C | 2.2 | ||
    NM_001423.1 NM_001423.1 | EMP1 | 2.2 | ||
    AW149417 AW149417 | OAZ | 2.2 | ||
    NM_014320.1 NM_014320.1 | HEBP2 | 2.2 | ||
    M18767.1 M18767.1 | CIS | 2.1 | ||
    NM_007286.1 NM_007286.1 | SYNPO | 2.1 | ||
    NM_012288.1 NM_012288.1 | TRAM2 | 2.0 | ||
    NM_000859.1 NM_000859.1 | HMGCR | −2.0 | ||
    AC005378 AC005378 | CNTNAP2 | −2.1 | ||
    NM_014778.1 NM_014778.1 | NUPL1 | −2.2 | ||
    NM_006741.1 NM_006741.1 | PPPIR1A | −3.0 | ||
In the top ranking genes that are modulated in siEF1-treated cells, various genes encode soluble factors involved in cell survival. These genes include DKK1 and DKK3, which are inhibitors of the Wnt/β-catenin pathway, and IGFBP-3 and IGFBP-5, two major regulators of the IGF-1 pathway. Our interest focused on IGFBP-3, which encodes the most predominant form of IGFBPs. IGFBP-3 prevents the interaction of IGF-1 with its receptor (IGF-1R), inhibits cell proliferation, and induces apoptosis in a number of human cancer cells through IGF-1-dependent and -independent mechanisms of action (6, 20, 21, 29). IGFBP-3 is present in two different probe sets in the HG-U133A array, and both were found equally modulated (mean changes of 17.51- and 16.62-fold). This induction was confirmed by quantitative RT-PCR that detected a ratio of 555 ± 128 between siEF1- and siCT-transfected cells. Quantitative RT-PCR was used to investigate the level of expression of IGFBP-3 after EWS/FLI-1 knockdown in another Ewing cell line. Following a two-transfection procedure similar to that used for A673 cells, siEF1 was also able to silence EWS/FLI-1 expression in the EW24 cell line, which expresses a type 1 EWS/FLI-1 fusion gene (Fig. (Fig.2A).2A). This also resulted in a strong induction of IGFBP-3 (Fig. (Fig.2B).2B). The expression of IGFBP-3 in 12 Ewing cell lines harboring different fusion types was investigated. All cell lines, whatever the fusion type, expressed low levels of IGFBP-3.
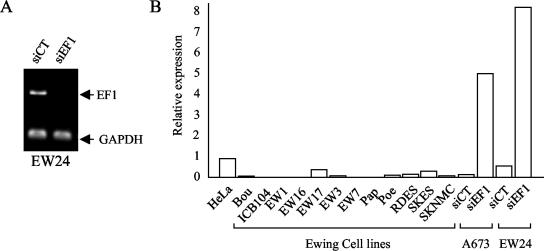
Analysis of IGFBP-3 mRNA expression levels in Ewing cell lines by quantitative RT-PCR. (A) Inhibition of EWS/FLI-1 expression by siEF1 in the EW24 cell line. (B) Levels of expression of IGFBP-3 in Ewing cell lines and A673 or EW24 cells treated with siEF1 or siCT are indicated relative to the level of expression of IGFBP-3 in HeLa cells.
To more precisely document the link between EWS/FLI-1 and IGFBP-3 expression, we first assessed whether ectopic EWS/FLI-1 was able to down-regulate the expression of the endogenous IGFBP-3 gene. Immunofluorescence experiments showed that whereas IGFBP-3 staining was strong in nontransfected cells, it was not detected in HeLa cells expressing EWS/FLI-1 (Fig. (Fig.3A).3A). This result strongly supported the hypothesis of EWS/FLI-1 being responsible for IGFBP-3 repression.
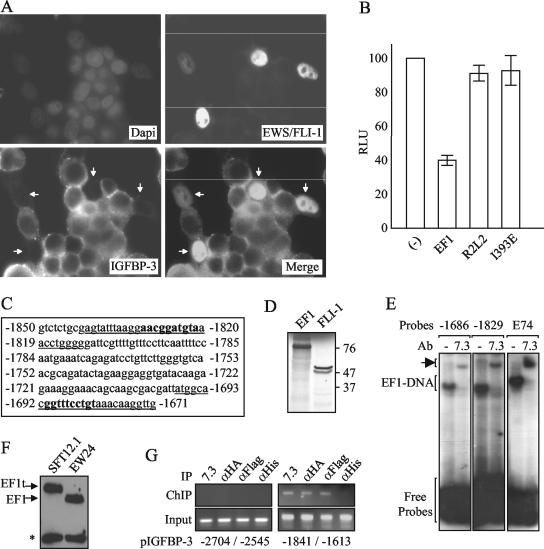
Down-regulation of IGFBP-3 expression by EWS/FLI-1 and direct binding of EWS/FLI-1 to the promoter region. (A) Inhibition of endogenous IGFBP-3 expression by ectopic EWS/FLI-1. HeLa cells transfected by EWS/FLI-1 were stained by the monoclonal 7.3 (FITC) and polyclonal anti-IGFBP-3 (cyanin-3) antibodies. Dapi, 4′,6′-diamidino-2-phenylindole. Merge, combined EWS/FLI-1 and IGFBP-3 stainings. Arrows, EWS/FLI-1-transfected cells. (B) Inhibition of the IGFBP-3 promoter by EWS/FLI-1. The IGFBP-3 promoter from positions −2584 to +63 was cloned upstream of the luciferase gene of pRep4 plasmid. Light intensity (in relative light units [RLU]) is shown on the y axis. The means ± standard deviations (error bars) from two independent duplicate experiments are shown. R2L2 and I393E are DNA binding-deficient mutants of EWS/FLI-1. Empty expression vector (−) was used as a control. (C) Potential EWS/FLI-1 binding sites within the IGFBP-3 promoter. EWS/FLI-1 consensus binding sites are indicated by bold type, and oligonucleotides used for EMSA experiments are underlined. (D) In vitro-translated EWS/FLI-1 and FLI-1. The fastest migrating FLI-1 band results from internal initiation at a downstream methionine residue (17). The positions of molecular size markers (in kilodaltons) are shown to the right of the gel. (E) EWS/FLI-1 binding to two potential sites located around positions −1686 and −1829 upstream of the transcription start site (7) in the presence (7.3) and absence (−) of the 7.3 monoclonal antibody (Ab). The arrow indicates the supershift of the EF1-DNA complex by the 7.3 antibody. E74 is the control oligonucleotide that contains a specific EWS/FLI-1 binding site (2). (F) Expression of a triple-tagged version of EWS/FLI-1 (EF1t) from the EW24-derived SFT12.1 cells. *, cross-reactive band. (G) Binding of EWS/FLI-1 to the IGFBP-3 promoter in vivo. ChIP experiments from SFT12.1 cell extracts using three different specific antibodies (anti-EWS/FLI-1 [αEWS/FLI-1] [7.3], anti-HA [αHA], or anti-Flag [αFlag]) or a control (anti-His [αHis]) antibody. The −1841/−1613 IGFBP-3 promoter fragment, according to the position of the transcription start site, is immunoprecipitated with the specific antibodies, whereas the −2704/−2545 fragment is not. The control PCR on extracts before immunoprecipitation (input) is shown.
To further explore this hypothesis, the IGFBP-3 promoter was cloned upstream of the luciferase reporter gene. The luciferase activity was then measured after transfection of HeLa cells with expression vectors encoding EWS/FLI-1 or two DNA binding-deficient mutants of EWS/FLI-1 (R2L2 and I393E) (2, 10). Results showed that EWS/FLI-1 decreased luciferase activity compared to empty vector (Fig. (Fig.3B).3B). This down-regulation of the IGFBP-3 promoter was not observed with DNA binding-deficient mutants of EWS/FLI-1, strongly suggesting that this phenomenon was dependent on DNA binding. The sequence of the IGFBP-3 promoter contains several potential ETS protein binding sites. Among these sites, band shift experiments showed that in vitro-translated EWS/FLI-1 could efficiently bind two sites located at positions −1686 and −1829 (Fig. (Fig.3C,3C, D, and E). The specificity of binding to these two sites was documented by competition experiments (data not shown) and by incubation with the anti-FLI-1 7.3 antibody that could completely shift the EWS/FLI-1/DNA complex (Fig. (Fig.3E3E).
Finally, ChIP experiments were performed using the SFT12.1 Ewing cell line. In this cell line derived from the EW24 cell line, the endogenous type 1 EWS/FLI-1 gene has been replaced by homologous recombination with a triple-tagged (Flag, vesicular stomatitis virus, and HA) version of this gene (Fig. (Fig.3F).3F). Using this cell line, we were able to immunoprecipitate a fragment containing the sites of the IGFBP-3 promoter at positions −1686 and −1829 with either the anti-EWS/FLI-1 (7.3), anti-HA, or anti-Flag antibodies but not with the anti-His control antibody (Fig. (Fig.3G).3G). In contrast, specific ChIP was not observed with an upstream fragment (Fig. (Fig.3G).3G). This result suggested that EWS/FLI-1 was present nearby the IGFBP-3 promoter in vivo. Taken together, our results showed that EWS/FLI-1 was able to bind the IGFBP-3 promoter in vitro and in vivo, possibly allowing the direct repression of its activity.
In contrast to EWS/FLI-1, FLI-1 induced a moderate activation of this promoter (Fig. (Fig.4A).4A). EWS/FLI-1 was able to counteract this FLI-1-induced activation in a manner dependent on DNA binding, since none of the DNA binding mutants could challenge the effect of FLI-1. Band shift experiments confirmed that FLI-1 could bind the sites at positions −1829 and −1686 and could compete for EWS/FLI-1 binding (Fig. (Fig.4B4B and data not shown). In agreement with the hypothesis that FLI-1 and EWS/FLI-1 have opposite effects on the regulation of IGFBP-3 transcription, we could show that ectopic expression of FLI-1 in Ewing cells led to a dose-dependent increase in the level of IGFBP-3 expressed, indicating that changes in the balance of FLI-1 and EWS/FLI-1 alter IGFBP-3 expression in Ewing cells (Fig. (Fig.4C4C).
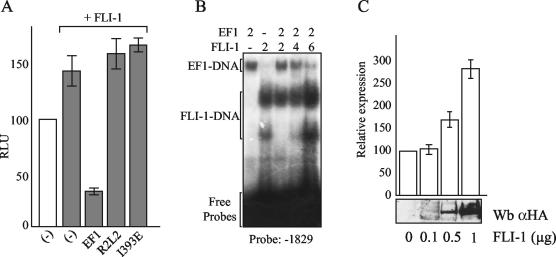
Role of the balance of FLI-1 and EWS/FLI-1 expression in IGFBP-3 regulation. (A) Inhibition of the moderate FLI-1-induced activation of the IGFBP-3 promoter by EWS/FLI-1 but not by DNA binding-deficient mutants. Light intensity (in relative light units [RLU]) is shown on the y axis. Luciferase construct and expression vectors are identical to those used in Fig. Fig.3B.3B. Empty expression vector (−) was used as a control. The means ± standard deviations (error bars) of duplicate experiments are shown. (B) FLI-1 and EWS/FLI-1 binding on the −1829 probe. The reaction mixtures were incubated with no (−) or 2, 4, and 6 microliters of siEF1 or FLI-1. EMSA experiments show that FLI-1 can compete EWS/FLI-1 binding in a dose-dependent manner. Both proteins were produced by in vitro translation as shown in Fig. Fig.3D.3D. The two complexes observed with FLI-1 correspond to the two bands of FLI-1. (C) Increased endogenous IGFBP-3 expression induced by FLI-1 transfection in Ewing cells. IGFBP-3 levels were analyzed by quantitative RT-PCR after transfection of A673 cells with increasing amounts of the FLI-1 expression vector. Relative expression of IGFBP-3 compared to that in untransfected A673 cells is shown. The means ± standard deviations (error bars) of duplicate experiments are shown. The expression level of FLI-1 evaluated by immunoblotting is shown beneath the diagram. Wb αHA, Western blotting with anti-HA antibody.
Next we addressed the role of IGFBP-3 induction in the increased apoptotic death of siEF1-treated cells. Figure Figure5A5A shows that an IGFBP-3-specific siRNA could very efficiently inhibit the IGFBP-3 expression induced by siEF1. The simultaneous transfection of IGFBP-3-specific siRNA and siEF1 led to strong decreases in the amount of apoptotic cells compared to cells transfected with siEF1 alone, hence indicating that IGFBP-3 induction is a key step for siEF1-induced apoptosis.
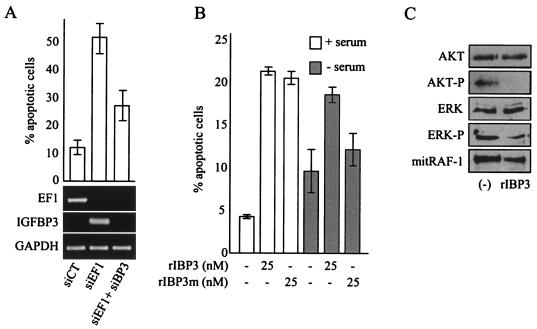
IGFBP-3-dependent apoptosis induced by siEF1 in Ewing cells. (A) Inhibition of siEF1-induced apoptosis by silencing of IGFBP-3. The high apoptotic rate induced by siEF1 can be reduced dramatically by siRNA-based blockade of IGFBP-3 induction. The levels of expression of EWS/FLI-1 and IGFBP-3, measured by competitive RT-PCR, are shown. (B) IGF-1-dependent and -independent pathways of IGFBP-3-induced apoptosis in Ewing cells. Recombinant IGFBP-3 (rIBP3) or a version of rIBP3 with a mutated NLS (rIBP3m) were incubated in the presence (+) or absence (−) of 10% serum. The means ± standard deviations (error bars) of duplicate experiments are shown in panels A and B.(C) Regulation of the IGF-1 downstream pathways by 25 nM IGFBP-3 treatment of Ewing cells. Untreated cells (−) were used as controls. AKT-P, phosphorylated AKT; ERK-P, phosphorylated ERK; mitRAF-1, mitochondrial RAF-1.
This was confirmed by the observation of a strong induction of apoptosis following the addition of recombinant IGFBP-3 in the culture medium of A673 cells (Fig. (Fig.5B).5B). In contrast, conditioned medium from siEF1-treated A673 cells had no effect on untreated A673 cells, suggesting that the concentration of IGFBP-3 in this conditioned medium was not sufficient to induce apoptosis (data not shown). Down-regulation of EWS/FLI-1 may induce a high local concentration of IGFBP-3 that may in turn induce apoptosis of secreting and neighboring cells. Induction of apoptosis was also observed with a recombinant IGFBP-3 mutated in the nuclear localization signal (NLS) (Fig. (Fig.5B).5B). This mutant of IGFBP-3 has been shown to bind IGF-I and IGF-II similarly to wild-type IGFBP-3 and to inhibit IGF-I interaction with the IGF receptor; however, this mutant is unable to enter cells and mediate the IGF-independent, intracellular actions of IGFBP-3 (15). This indicated that inhibition of the IGF-1 pathway was sufficient to induce apoptosis in Ewing cells. Nevertheless, IGFBP-3 could also strongly induce apoptosis in serum-free medium and hence in the absence of IGF-1, indicating that IGFBP-3 may also act through an IGF-1-independent mechanism. The observation that the IGFBP-3 with mutated NLS was much less efficient in inducing apoptosis in serum-free medium suggested that this IGF-1-independent pathway requires cellular entry and nuclear translocation of IGFBP-3 (Fig. (Fig.5B5B).
Finally, we investigated the activated state of the pathways downstream of IGF-1 in Ewing cells. Figure Figure5C5C shows that the levels of expression of AKT and ERK were not modified by IGFBP-3 treatment. However, IGFBP-3 treatment resulted in a dramatic decreases in the amounts of the phosphorylated forms of these proteins. In addition, IGFBP-3 could decrease the amount of mitochondrial RAF-1. Together, these results strongly suggest that the IGF-1 pathway is constitutively active in A673 cells and that IGFBP-3-mediated apoptosis could rely on the negative modulation of AKT, ERK, and mitochondrial RAF-1 pathways.
DISCUSSION
Ewing tumor cells express specific gene fusions, most frequently between EWS and FLI-1, whose oncogenic properties have been documented by a number of reports (1). In agreement with the role of this fusion gene in the growth of Ewing cells, we show that the silencing of EWS/FLI-1 has profound antiproliferative effects correlated to a strong induction of apoptosis. This gene fusion constitutes a simple genetic event that results in the synthesis of an abnormal transcription factor. This is expected to lead to pleiotropic cellular effects, collectively contributing to the full transformation of the Ewing parental cell. Understanding the oncogenic properties of this aberrant transcription factor therefore requires identification of the downstream targets.
To address this issue, we have analyzed 22,000 Affymetrix DNA chips from cells with EWS/FLI-1 expression silenced by specific siRNA compared to mock-treated cells and identified 86 genes that exhibit a twofold change in duplicate experiments. The products of these genes could be classified in two major functional clusters. The first functional cluster points to components of the ECM. This cluster includes collagens and lysyl-oxidase enzymes involved in the structure and reticulation of the ECM (11), as well as plasminogen activator urokinase together with its receptor, which participate in the remodeling of ECM (28). This cluster also contains a number of proteins involved in membrane plasticity, cell-cell contacts, and cell-ECM interactions. As Ewing tumors are highly aggressive neoplasms with a strong propensity to spread into neighboring tissues, it will be of considerable interest to document the roles of these EWS/FLI-1-induced modifications of components of the ECM on the invasive potential of Ewing cells.
The second functional cluster contains genes involved in the regulation of a variety of signal transduction pathways, including negative regulators of the Wnt, IGF-1, STAT, and MAPK pathways, which are critical for the control of the apoptosis or survival-proliferation balance and directly involved in a variety of human cancers.
In this study, we focused our attention on the strong induction of the IGFBP-3 gene observed after the silencing of EWS/FLI-1. IGFBP-3 is the main circulating carrier protein for IGFs, and one of its major roles is to inhibit IGF-1 action by altering receptor-ligand interaction (36). Evidence that supports the critical role of the IGF-1 pathway in cancer development has been accumulating. Epidemiological studies indicate that high IGF-1 and low IGFBP-3 levels in serum are associated with increased risk of cancer (5). Furthermore, the IGF-1R is overexpressed in many tumors, and in vitro experiments have indicated that activation of IGF-1R is involved in the development of a number of cancers (34, 35). The role of this pathway in oncogenesis is further supported by the observation that cells in which the IGF-1R has been inactivated require the presence of this receptor for full transformation by a variety of oncogenic agents (3, 26, 30). Finally, numerous reports, using different approaches, including those using blocking antibodies, an antisense strategy, RNA interference, or specific tyrosine kinase inhibitors, have highlighted IGF-1/IGF-1R signaling as an extremely attractive target for the development of new therapeutics against cancer. More specifically, IGF-1R has been shown to be overexpressed in Ewing tumors (25) and to be required for EWS/FLI-1-induced transformation of mouse fibroblasts (33). Impeding the IGF-1R pathway in Ewing cells with IGF-1R-blocking antibodies (23) or using an IGF-1R antisense strategy (24) has been shown to inhibit cell growth and tumorigenic properties. The results of the present study indicate that the constitutive activation of the IGF-1 pathway in Ewing tumors is also a consequence of the transcriptional repression of IGFBP-3 by EWS/FLI-1. Indeed, luciferase analyses indicate that EWS/FLI-1 can down-regulate the IGFBP-3 promoter. However, this inhibition is lower than that observed with immunofluorescence experiments. The transfection efficiency may account for a difference between expression analysis in single cells (as shown in Fig. Fig.3A)3A) or populations of transfected cells (Fig. (Fig.3B).3B). In addition, although our study identifies EWS/FLI-1 regulatory sequences within the IGFBP-3 promoter, it is possible that they are not the unique EWS/FLI-1-responsive elements and that additional sites, located in exonic or intronic regions, may also play a role in IGFBP-3 regulation. Additionally, the full repressive effect of EWS/FLI-1 may require stable chromatin integration. The balance between EWS/FLI-1, FLI-1, and other ETS family proteins, which share similar, if not identical, DNA binding sites, may account for the variable levels of IGFBP-3 observed in different Ewing cell lines (27).
The silencing of IGFBP-3 can revert, at least in part, the increased apoptosis linked to EWS/FLI-1 inhibition. Moreover, treating Ewing cells with recombinant IGFBP-3 leads to a strong induction of apoptotic cells associated with a dramatic decrease in the amounts of phosphorylated AKT and ERK and of mitochondrial RAF-1, which inactivates the IGF-1 pathway.
In addition to IGF-1- and IGF-1R-dependent effects, recent reports have shown that IGFBP-3 may also act through distinct receptors and through nuclear translocation (14, 15, 20, 21, 29). Such IGF-1- and IGF-1R-independent mechanisms may play a role in the IGFBP-3 induction of apoptosis in Ewing cells, since in the absence of serum, wild-type IGFBP-3 is still able to trigger apoptosis, whereas a noninternalizable version of IGFBP-3 with a mutated NLS is inefficient. In that respect, it will be of strong interest to determine whether such IGF-1-independent mechanisms may account for the observed resistance of Ewing cells to therapies interfering with the function of the IGF1-R (22, 24). Therefore, IGFBP-3-based treatments, targeting both IGF-1-dependent and -independent pathways, may be interesting alternative strategies.
It is noteworthy that Ewing tumors mainly arise in bone, at puberty, at the time of a strong activation of IGF-1 signaling induced by the spurt of growth hormone. The occurrence of an EWS/FLI-1 fusion gene in the Ewing parental cell, which may be a bone marrow stromal cells as suggested by a recent report (32), could have a synergistic effect with growth hormone activation and cause an uncontrolled IGF1-induced proliferation through the loss of expression of IGFBP-3. In that respect, this loss of control could also be related to the decreased expression of other negative regulators of this pathway, including IGFBP-5 and IGFBP-6, which are also strongly down-regulated by EWS/FLI-1.
Finally, the oncogenic properties of EWS/FLI-1 certainly do not rely exclusively on inhibition of IGFBPs. Indeed, we could show that blocking IGFBP-3 induction does not fully revert the apoptotic phenotype induced by EWS/FLI-1 knockdown. Other targets, including negative regulators of the Wnt, MAPK, and STAT pathways and proteins involved in the remodeling of the ECM, may also contribute to the malignant phenotype of Ewing cells.
Acknowledgments
We thank Jacques Ghysdael for the ΔEB expression vectors used in this study.
This work was supported in part by the Institut National de la Santé et de la Recherche Médicale and by grants from the Ligue Nationale contre le Cancer (Laboratoire Associé), the Institut Curie, and the Réseau National des Génopoles. A. Prieur is a recipient of a fellowship from the Ligue Nationale contre le Cancer, and F. Tirode is a recipient of a fellowship from the Centre National de la Recherche Scientifique. P. Cohen was supported in part by National Institutes of Health grants AG20954, CA100938, and P50CA92131.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.24.16.7275-7283.2004
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc479730?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.24.16.7275-7283.2004
Article citations
Functional Classification of Fusion Proteins in Sarcoma.
Cancers (Basel), 16(7):1355, 29 Mar 2024
Cited by: 1 article | PMID: 38611033 | PMCID: PMC11010897
Review Free full text in Europe PMC
Ewing sarcoma from molecular biology to the clinic.
Front Cell Dev Biol, 11:1248753, 11 Sep 2023
Cited by: 3 articles | PMID: 37752913 | PMCID: PMC10518617
Review Free full text in Europe PMC
Targeted Therapy for EWS-FLI1 in Ewing Sarcoma.
Cancers (Basel), 15(16):4035, 09 Aug 2023
Cited by: 3 articles | PMID: 37627063 | PMCID: PMC10452796
Review Free full text in Europe PMC
EHD1-dependent traffic of IGF-1 receptor to the cell surface is essential for Ewing sarcoma tumorigenesis and metastasis.
Commun Biol, 6(1):758, 20 Jul 2023
Cited by: 2 articles | PMID: 37474760 | PMCID: PMC10359273
SIX1 and EWS/FLI1 co-regulate an anti-metastatic gene network in Ewing Sarcoma.
Nat Commun, 14(1):4357, 19 Jul 2023
Cited by: 3 articles | PMID: 37468459
Go to all (266) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 38 of 38)
- (1 citation) ENA - M31159
- (1 citation) ENA - J04177
- (1 citation) ENA - AU148057
- (1 citation) ENA - AL050025
- (1 citation) ENA - D13889
- (1 citation) ENA - M80927
- (1 citation) ENA - U58111
- (1 citation) ENA - AK026420
- (1 citation) ENA - M15330
- (1 citation) ENA - BC003096
- (1 citation) ENA - AF043337
- (1 citation) ENA - AB005043
- (1 citation) ENA - U16307
- (1 citation) ENA - BC001422
- (1 citation) ENA - BF752277
- (1 citation) ENA - AY029208
- (1 citation) ENA - M11734
- (1 citation) ENA - AU144167
- (1 citation) ENA - AW149417
- (1 citation) ENA - AA292373
- (1 citation) ENA - L03203
- (1 citation) ENA - BC005047
- (1 citation) ENA - AW242315
- (1 citation) ENA - M19267
- (1 citation) ENA - M57731
- (1 citation) ENA - AL574096
- (1 citation) ENA - AY029180
- (1 citation) ENA - AI264196
- (1 citation) ENA - AA576961
- (1 citation) ENA - AC005378
- (1 citation) ENA - S69738
- (1 citation) ENA - L27560
- (1 citation) ENA - BC003355
- (1 citation) ENA - M18767
- (1 citation) ENA - BF575514
- (1 citation) ENA - BE620457
- (1 citation) ENA - AI346835
- (1 citation) ENA - AI653981
Show less
RefSeq - NCBI Reference Sequence Database (Showing 48 of 48)
- (1 citation) RefSeq - NM_017458.1
- (1 citation) RefSeq - NM_003186.2
- (1 citation) RefSeq - NM_005824.1
- (1 citation) RefSeq - NM_005532.1
- (1 citation) RefSeq - NM_000204.1
- (1 citation) RefSeq - NM_002309.2
- (1 citation) RefSeq - NM_000859.1
- (1 citation) RefSeq - NM_002178.1
- (1 citation) RefSeq - NM_001124.1
- (1 citation) RefSeq - NM_005651.1
- (1 citation) RefSeq - NM_006741.1
- (1 citation) RefSeq - NM_004591.1
- (1 citation) RefSeq - NM_002966.1
- (1 citation) RefSeq - NM_014320.1
- (1 citation) RefSeq - NM_002658.1
- (1 citation) RefSeq - NM_004414.2
- (1 citation) RefSeq - NM_012242.1
- (1 citation) RefSeq - NM_006158.1
- (1 citation) RefSeq - NM_002045.1
- (1 citation) RefSeq - NM_016352.1
- (1 citation) RefSeq - NM_007286.1
- (1 citation) RefSeq - NM_002084.2
- (1 citation) RefSeq - NM_001432.1
- (1 citation) RefSeq - NM_012288.1
- (1 citation) RefSeq - NM_002852.1
- (1 citation) RefSeq - NM_002526.1
- (1 citation) RefSeq - NM_001511.1
- (1 citation) RefSeq - NM_000050.1
- (1 citation) RefSeq - NM_002317.1
- (1 citation) RefSeq - NM_002318.1
- (1 citation) RefSeq - NM_015577.1
- (1 citation) RefSeq - NM_000610.1
- (1 citation) RefSeq - NM_013451.1
- (1 citation) RefSeq - NM_000064.1
- (1 citation) RefSeq - NM_005978.2
- (1 citation) RefSeq - NM_006472.1
- (1 citation) RefSeq - NM_000104.2
- (1 citation) RefSeq - NM_004369.1
- (1 citation) RefSeq - NM_000358.1
- (1 citation) RefSeq - NM_005195.1
- (1 citation) RefSeq - NM_001423.1
- (1 citation) RefSeq - NM_014600.1
- (1 citation) RefSeq - NM_018444.1
- (1 citation) RefSeq - NM_014778.1
- (1 citation) RefSeq - NM_003358.1
- (1 citation) RefSeq - NM_002993.1
- (1 citation) RefSeq - NM_000963.1
- (1 citation) RefSeq - NM_005382.1
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing's sarcoma.
Mol Cancer Res, 4(11):851-859, 01 Nov 2006
Cited by: 136 articles | PMID: 17114343
Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing's sarcoma cells.
Cancer Res, 66(20):9937-9947, 01 Oct 2006
Cited by: 95 articles | PMID: 17047056
PDGF-C is an EWS/FLI induced transforming growth factor in Ewing family tumors.
Oncogene, 20(5):626-633, 01 Feb 2001
Cited by: 73 articles | PMID: 11313995 | PMCID: PMC3860748
Identification of target genes in their native cellular context: an analysis of EWS/FLI in Ewing's sarcoma.
Cell Cycle, 5(18):2049-2053, 15 Sep 2006
Cited by: 34 articles | PMID: 16969112
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: CA10098
Grant ID: R01 CA100938
Grant ID: P50CA92131
Grant ID: P50 CA092131
NIA NIH HHS (2)
Grant ID: AG20954
Grant ID: R01 AG020954
Worldwide Cancer Research (1)
Grant ID: 03-0129





