Abstract
Free full text

BCR-ABL1-like cases in pediatric acute lymphoblastic leukemia: a comparison between DCOG/Erasmus MC and COG/St. Jude signatures
Patients with pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL) with the BCR-ABL1 fusion gene form a small, high-risk group with a poor prognosis.1 Approximately 15% of cases of BCP-ALL are characterized by a gene expression signature similar to that of BCR-ABL1-positive disease and an unfavorable prognosis.2,3 Two independent gene expression signatures were described that identify BCR-ABL1-like ALL, alternatively called Philadelphia-like ALL.2,3 The BCR-ABL1-like signature described by Mullighan et al. is based on the prediction analysis of microarrays (PAM) classifier consisting of 257 gene probe sets trained on BCR-ABL1-positive cases.4,5 Genetic alterations activating kinase or cytokine receptor signaling are a hallmark of this subtype, which also has frequent deletions of IKZF1, and poor outcome.4,5 The signature developed by Den Boer et al. is based on hierarchical clustering (HC) of 110 gene probe sets identified to predict the major pediatric ALL subtypes (T-cell ALL, ETV6-RUNX1, high-hyperdiploidy, TCF3 or MLL-rearranged, BCR-ABL1).2,6 This group of BCR-ABL1-like patients had frequent deletions in B-cell development genes (e.g. IKZF1), dic(9;20), and intrachromosomal amplification of chromosome 21 (iAMP21).2,6 The two signatures have nine overlapping probe sets (Online Supplementary Table S1). Here, we compared BCR-ABL1-like cases identified by each of these two signatures in two independent, previously published pediatric cohorts. We found that the two BCR-ABL1-like signatures identify molecularly distinct but overlapping groups of patients with a poor prognosis. The tyrosine kinase fusion genes affecting ABL1, PDGFRB and JAK2 were exclusively found in the BCR-ABL1-like group; no fusion-positive patients were found in the remaining cases negative by either signature.
This signature comparison study comprised children with newly diagnosed BCP-ALL negative for the known aberrations BCR-ABL1, ETV6-RUNX1, MLL rearrangement, TCF3 rearrangement, and high-hyperdiploidy in consecutive Dutch Childhood Oncology Group (DCOG) and German Cooperative ALL trials (146 cases),2,6 and the P9906 study of the US Childhood Oncology Group (COG) (143 cases).3–5,7,8 The patients’ characteristics are summarized in Online Supplementary Table S2. Affymetrix U133 Plus 2.0 microarray files were exchanged between Erasmus MC and St. Jude Children’s Research Hospital, and BCR-ABL1-like cases were identified by both HC and PAM signatures following described methods (see Online Supplementary Methods).2,4–6 Molecular aberrations were determined as described in the Online Supplementary Methods. Cumulative incidence of relapse probabilities were estimated using a competing risks model and the equality of these cumulative incidences was tested with the Gray test.
In the population-based DCOG/COALL cohort, 79 out of 146 B-other cases were identified as BCR-ABL1-like by HC, 33 by PAM, and 25 cases by both signatures (henceforth called double-positive cases; Figure 1A). The cases identified by each of the signatures were predominantly (72–76%) NCI-Rome9 high-risk (Online Supplementary Table S3, Figure 1B). We compared the frequencies of molecular aberrations among the BCR-ABL1-like identified cases with the 59 B-other BCP-ALL cases negative for either of the two BCR-ABL1-like signatures and negative for BCR-ABL1, ETV6-RUNX1, MLL rearrangement, TCF3 rearrangement, and high-hyperdiploidy (Figure 1D, Online Supplementary Figure S2A). The double-positive samples showed significant enrichment of IKZF1 deletion (P=0.01), CRLF2 high expression (P<0.001), iAMP21 (P<0.001), RB1 deletion (P=0.03), and kinase-activating fusions (P<0.001). Among the cases that were only identified by HC, dic(9;20) was enriched (P<0.001). Among the cases that were only identified by PAM, IKZF1 deletion, high CRLF2 expression, and JAK2 mutations (P≤0.01) were enriched. Deletions of B-cell development genes CDKN2A/B, PAX5, EBF1, and ETV6 were frequent in all groups, including non-BCR-ABL1-like B-others, and not enriched in specific groups (Online Supplementary Table S4). Similar results were obtained when analyzing the 92 NCI-HR cases (Figure 1E, Online Supplementary Table S5).
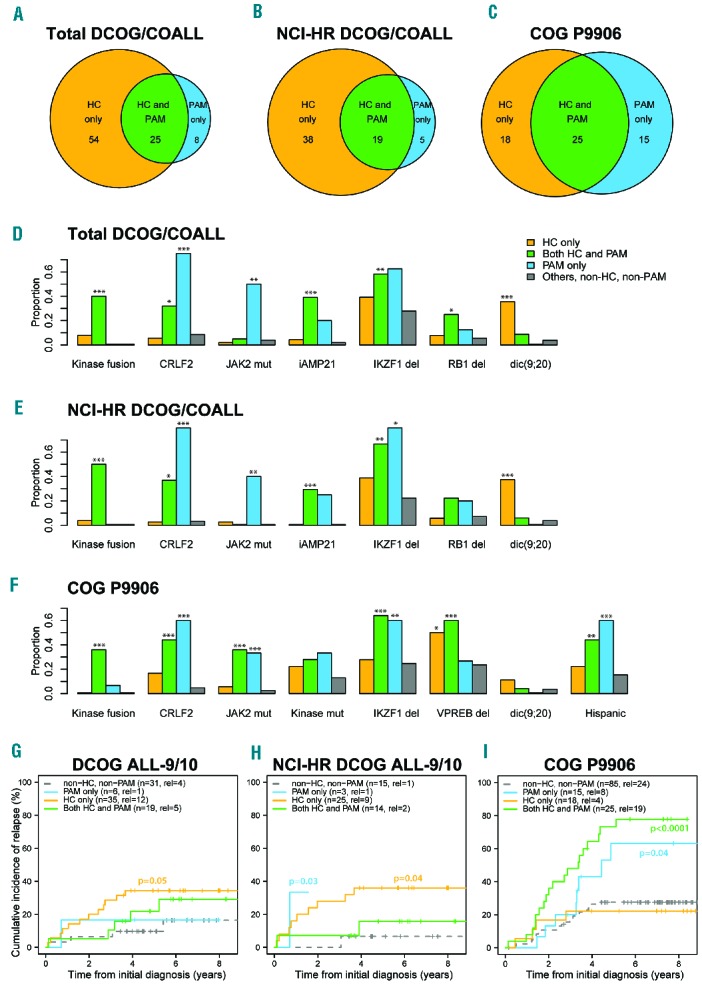
Comparison of BCR-ABL1-like cases identified by the HC and PAM BCR-ABL1-like signatures. (A–C) Venn diagrams showing the overlap in BCR-ABL1-like cases identified by the HC and PAM signatures in (A) the total DCOG/COALL cohort (B), the NCI-Rome high risk cases of the DCOG/COALL cohort, and (C) the high risk COG P9906 cohort. (D–F) Bar plots for (D) the total DCOG/COALL cohort (D), the NCI-Rome high risk cases of the DCOG/COALL cohort (E), and the high risk COG P9906 cohort (F) showing the proportions of the indicated aberrations in, respectively, the samples identified only by the HC signature (orange), double-positive samples identified by both signatures (green), samples identified only by the PAM signature (blue), and the remaining, non-BCR-ABL1-like B-other cases (gray). Fisher exact test results comparing proportions in each group to the remaining B-other cases are indicated as *P-value ≤0.05; **P-value ≤0.01 and ***P-value ≤0.001. mut: mutation; del: deletion. For aberration frequencies and P-values see Online Supplementary Tables S4–6. (G-I) Cumulative incidences of relapse for (G) the DCOG cases who were treated on ALL-9/10 protocols, (H) the NCI-Rome high risk cases treated on DCOG ALL-9/10 protocols, and (I) the high-risk cases treated on the COG P9906 protocol. The total numbers of patients and relapses and Gray P-values ≤0.1 are indicated in each plot. Additional outcome results are presented in Online Supplementary Table S7.
We also compared the overlap between HC- and PAM-identified BCR-ABL1-like cases in 143 B-other BCP-ALL cases negative for the known aberrations BCR-ABL1, ETV6-RUNX1, MLL rearrangement, TCF3 rearrangement, and high-hyperdiploidy in the high-risk COG P9906 cohort. The PAM classification and molecular aberrations for these samples were described previously.4 We identified 43 BCR-ABL1-like cases using HC, 40 cases using PAM, with 25 double-positive cases (Figure 1C). We compared the frequencies of molecular aberrations among the BCR-ABL1-like identified cases with 85 B-other BCP-ALL cases that were not identified by either of the two BCR-ABL1-like signatures (Figure 1F, Online Supplementary Figure S2B). Among the double-positive cases, translocations resulting in kinase-activating fusions, IKZF1 deletion, CRLF2 high expression, JAK2 mutation, Hispanic or Latino background, VPREB, EBF1, and BTG1 deletion were most enriched (P<0.01). IKZF1 deletion, high CRLF2 expression, JAK2 mutation, Hispanic or Latino background and EBF1 deletion were enriched in cases only identified by PAM (P≤0.01), while VPREB1 deletion (P=0.04) was enriched in cases only identified by HC (Online Supplementary Table S6).
We evaluated the prognostic value of the BCR-ABL1-like signatures in DCOG cases from two recent, similar treatment protocols. In the DCOG ALL-9/10, the cases identified by HC only (P<0.05) and the total HC group (P=0.07) showed higher relapse rates compared with the non-BCR-ABL1-like B-other reference group (Figure 1G, Online Supplementary Figure S3A-C, Online Supplementary Table S7). Outcome analysis of the DCOG ALL-9/10 cases at high risk of treatment failure according to NCI-Rome criteria (white blood cell count ≥50×109/L and/or age ≥10 years) showed a similar unfavorable outcome (Figure 1H, Online Supplementary Table S7). In the high-risk P9906 protocol, total HC (P=0.005), total PAM (P<0.0001), PAM-only (P=0.04) and HC-and-PAM (P<0.0001) BCR-ABL1-like cases showed higher relapse rates compared with non-BCR-ABL1-like B-other (Figure 1I, Online Supplementary Figure S3D–F, Online Supplementary Table S7).
Several differences between the (genetic) composition of the discovery cohorts and the methodology to develop the signatures may contribute to the partial overlap in BCR-ABL1-like cases identified by the HC and PAM signatures.2,4 Firstly, the 110 probe sets used for HC were previously selected to discriminate the major subtypes of pediatric ALL and as such those 110 probe sets were not selected to only identify BCR-ABL1-like ALL. In contrast, the 257 probe sets used for PAM were chosen for high discriminative value to identify BCR-ABL1-positive cases. Secondly, the COG discovery cohort consisted of high-risk patients according to the COG criteria, whereas the DCOG/COALL discovery cohort represented all risk groups.4,5 Thirdly, differences in genetic ancestry between the American COG and European DCOG/COALL cohorts could have affected the signatures. Several of the hallmarks of BCR-ABL1-like ALL identified in the COG high-risk cohorts, including increased frequencies of CRLF2 aberration and JAK2 mutation, have been associated with Hispanic/Latino ethnicity.7,10 The lack of Hispanic/Latino patients in the DCOG/COALL cohort may have contributed to the lower frequency of CRLF2 and JAK2 abnormalities in this cohort, and the lower number of BCR-ABL1-like cases identified by the PAM signature. Likely due to these cohort differences, the prognostic value of the HC and PAM signatures is most discriminative in the cohort of patients in whom these signatures were identified, DCOG/COALL and COG P9906, respectively.
The biggest challenge is how to treat BCR-ABL1-like cases in clinical practice. BCR-ABL1-like ALL identified by the HC signature comprise approximately 15% of BCP-ALL cases in the population-based DCOG/COALL cohorts, and constitute approximately 50% of the B-other cases negative for BCR-ABL1, MLL or TCF3 rearrangements, ETV6-RUNX1, and high-hyperdiploidy.6 BCR-ABL1-like ALL identified by the PAM signature reflected 11% of BCP-ALL and 29% of B-other cases in a US population-based cohort.11 Both in the St. Jude Total Therapy XV study and in the DCOG ALL-10 study, patients with BCR-ABL1-like ALL were more likely to have higher levels of minimal residual disease after induction therapy.6,12 In the Total Therapy XV study, BCR-ABL1-like cases were responsive to risk-oriented increase of treatment intensity, showing similar outcomes to other BCP-ALL cases.12 However, in the ALL-10 study the majority of BCR-ABL1-like cases had intermediate minimal residual disease levels at the end of induction therapy, indicating that these patients are not identified as high-risk by minimal residual disease diagnostics and need additional molecular diagnostics.6
In the P9906 cohort, sequencing analyses identified kinase-activating gene fusions including ABL1, PDGFRB and JAK2.4,5 These fusions were present in 25% of PAM-identified BCR-ABL1-like cases, and in 21% of HC-identified cases. Kinase-activating gene fusions resulted in a constitutive activation of kinase signaling, and gave sensitivity to kinase inhibitors in ex vivo-cultured primary leukemic blasts and xenograft mouse models.4,11 Recent studies in patients with tyrosine kinase gene rearrangements and a poor response to induction chemotherapy showed rapid and sustained responses upon tyrosine kinase inhibitor therapy.11,13,14 A substantial part of the BCR-ABL1-like patients (75–80%) are negative for ABL and JAK class fusions whereas their prognosis is still poor. Within the P9906 cohort, the majority of these cases had abnormalities in CRLF2 and/or activating kinase mutations, thus differing from the DCOG/COALL cohort, which may be explained by the higher frequency of Hispanic/Latino ancestry because this coincides with a higher frequency of CRLF2/JAK2 aberrations.3–5,7,8 Studies addressing the pathobiology of the remaining cases are, therefore, of great importance to find other targets for treatment in the European population of patients.
The scope of the current study was to compare the two BCR-ABL1-like signatures. In the USA, a newly developed low-density array measuring the expression of 15 genes representative of the heterogeneous genomic lesions associated with BCR-ABL1-like ALL is currently being used in a first step, combined with genomic aberrations, to identify BCR-ABL1-like cases.15 This assay is not currently available for world-wide use, nor can gene expression signatures be easily obtained and interpreted in ordinary diagnostic laboratories. We, therefore, expect that the identification of genomic aberrations strongly associated with BCR-ABL1-like ALL is crucial for diagnostic procedures. To this aim it is also important to understand the overlap and differences between the two sentinel studies on BCR-ABL1-like ALL.2,3
Footnotes
Funding: This work was supported by the Pediatric Oncology Foundation Rotterdam (KOCR), the European Union’s Seventh Framework Programme (FP7/2007–2013) under the ENCCA project (HEALTH-F2-2011-261474), the Center for Translational Molecular Medicine BioChip program, NIH grant R37-CA36401, and NCI Comprehensive Cancer Center grant CA21765. Kathryn Roberts and Charles Mullighan are gratefully acknowledged for performing the PAM identification of the DCOG/COALL cohort. The TARGET group, led by Cheryl Willman, Stephan Hunger, Mignon Loh and Charles Mullighan, are acknowledged for sharing the P9906 patients’ characteristics and clinical outcome data, and constructive discussions on the data presented in this paper.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
Articles from Haematologica are provided here courtesy of Ferrata Storti Foundation
Full text links
Read article at publisher's site: https://doi.org/10.3324/haematol.2015.124941
Read article for free, from open access legal sources, via Unpaywall:
http://www.haematologica.org/content/100/9/e354.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Philadelphia chromosome-like acute lymphoblastic leukemia with concomitant rearrangements of CRLF2 and ABL1: a pediatric case report.
BMC Pediatr, 24(1):517, 10 Aug 2024
Cited by: 0 articles | PMID: 39127642 | PMCID: PMC11316372
The Gene Expression Classifier ALLCatchR Identifies B-cell Precursor ALL Subtypes and Underlying Developmental Trajectories Across Age.
Hemasphere, 7(9):e939, 25 Aug 2023
Cited by: 11 articles | PMID: 37645423 | PMCID: PMC10461941
Integrating copy number data of 64 iAMP21 BCP-ALL patients narrows the common region of amplification to 1.57 Mb.
Front Oncol, 13:1128560, 23 Feb 2023
Cited by: 1 article | PMID: 36910655 | PMCID: PMC9996016
Whole genome sequencing provides comprehensive genetic testing in childhood B-cell acute lymphoblastic leukaemia.
Leukemia, 37(3):518-528, 19 Jan 2023
Cited by: 27 articles | PMID: 36658389 | PMCID: PMC9991920
Characterization of Philadelphia-like Pre-B Acute Lymphoblastic Leukemia: Experiences in Mexican Pediatric Patients.
Int J Mol Sci, 23(17):9587, 24 Aug 2022
Cited by: 5 articles | PMID: 36076986 | PMCID: PMC9455471
Go to all (43) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL.
Blood, 122(15):2622-2629, 23 Aug 2013
Cited by: 176 articles | PMID: 23974192 | PMCID: PMC3795461
Tyrosine kinase fusion genes in pediatric BCR-ABL1-like acute lymphoblastic leukemia.
Oncotarget, 8(3):4618-4628, 01 Jan 2017
Cited by: 39 articles | PMID: 27894077 | PMCID: PMC5354859
A novel targeted RNA-Seq panel identifies a subset of adult patients with acute lymphoblastic leukemia with BCR-ABL1-like characteristics.
Blood Cancer J, 10(4):43, 24 Apr 2020
Cited by: 8 articles | PMID: 32332702 | PMCID: PMC7182567
BCR-ABL1-like B-lymphoblastic leukemia/lymphoma: Review of the entity and detection methodologies.
Int J Lab Hematol, 41 Suppl 1:126-130, 01 May 2019
Cited by: 2 articles | PMID: 31069976
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R37-CA36401
Grant ID: R37 CA036401
Grant ID: CA21765
Grant ID: P30 CA021765




