Abstract
Free full text

Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis
Abstract
Trehalose is a naturally occurring disaccharide that has gained attention for its ability to induce cellular autophagy and mitigate diseases related to pathological protein aggregation. Despite decades of ubiquitous use as a nutraceutical, preservative, and humectant, its mechanism of action remains elusive. Here, we showed that trehalose inhibited members of the SLC2A (also known as GLUT) family of glucose transporters. Trehalose-mediated inhibition of glucose transport induced AMPK (adenosine 5′-monophosphate-activated protein kinase)-dependent autophagy regression of hepatic steatosis in vivo, and a reduction in the accumulation of lipid droplets in primary murine hepatocyte cultures. Our data indicated that, by inhibiting glucose transport, trehalose triggers beneficial cellular autophagy.
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in the world (1). Indeed, more than one billion individuals worldwide has this highly morbid disease, which is characterized in its earliest stages by excessive hepatic fat deposition and in its latest stages by steatohepatitis, fibrosis, cirrhosis, liver failure, and progression to hepatocellular carcinoma (1). Unfortunately, few efficacious treatments exist.
We elucidated a role for the solute carrier 2A (SLC2A) family (also referenced as the glucose transporter, or GLUT family) of hexose transporter homologs in mediating NAFLD (2). The GLUT family consists of fourteen 12-transmembrane domain-containing proteins that mediate uptake of hexose and other substrates across cellular membranes (3), thus, these carrier proteins are central regulators of cellular energetics. GLUT2 and GLUT8 are the most abundant hepatic GLUTs (2), both of which transport glucose and fructose (3). GLUT8 deletion in mice blunts hepatic membrane hexose flux and prevents fructose-induced hepatic steatosis (2). Defects in cellular energetics can activate nutrient-sensing pathways that stimulate macroautophagy (hereafter, “autophagy”) – tightly regulated cellular ‘self-eating’, which occurs in response to amino acid or glucose starvation – to counteract acutely diminished energy substrates. The AMP-activated protein kinase (AMPK) pathway is among the signaling cascades activated in this context (4), and this kinase stimulates autophagy by phosphorylating unc52-like kinase-1 (ULK1) at Ser317 in response to increased ratio of cellular AMP to ATP (5, 6).
Sugars that are not transported can act as competitive inhibitors for the GLUT transporters (7). For example, the disaccharide maltose inhibits uptake of a glucose analog by GLUT2 with a Ki of ~100 mM (7). Thus, exogenous sugars may impair cellular energetics and induce autophagy. One particular disaccharide, trehalose, is a glucose-glucose disaccharide linked by an α,α, 1,1-glycoside bond. Trehalose is ubiquitous to a wide variety of organisms, including bacteria, yeast, fungi, insects, and invertebrates, but trehalose is absent in vertebrate animals (8).. Trehalose has garnered attention for its ability to mitigate protein aggregate accumulation by stimulating cellular autophagy in several cell types. In particular, trehalose has reduced toxic protein aggregates in murine models of the neurological diseases Huntington’s disease (9), amyotrophic lateral sclerosis (10), and prion disease (11) and in the pancreatic β cells of a mouse model of type 2 diabetes (12). The mechanism by which trehalose activates autophagy, however, is not fully described. Because autophagic insufficiency may underlie the pathogenesis of NAFLD (13), mechanistic insights into how trehalose induces hepatic autophagy may aid in the development of novel NAFLD therapeutics.
Therefore, we tested the hypothesis that trehalose induces hepatocyte autophagy by inhibiting hexose uptake. Morever, because the efficacy of trehalose in treating or preventing NAFLD in pre-clinical models has not been addressed, we tested whether this autophagy-inducing agent mitigates hepatic steatosis. Our data indicated that trehalose induces hepatocyte autophagy and mitigates hepatocyte lipid droplet accumulation – at least in part – by inhibiting hexose uptake and subsequently activating the AMPK to ULK1 pathway.
RESULTS
Trehalase and the trehalose receptor, Tre1, share homology with SLC2A family members
We performed phylogenetic analysis with Clustal Omega multiple sequence alignment to examine evolutionary relationships among SLC2A members and trehalose-specific enzymes - trehalase and the Drosophila trehalose receptor, Tre1. Phylogenic analysis revealed evolutionary commonality between trehalase, Tre1, and the mouse SLC2A family member, SLC2A8, with more distant common evolutionary branches connecting SLC2A2 and SLCA3 to Tre1 and trehalase (Fig. 1A). SLC2A1 and SLC2A4 were the most distant, lacking any shared evolutionary branches. We quantified domain-specific homology using pair-wise alignment between Tre1 and SLC2A1 using the crystal structure data for SLC2A1 (14). Regions of highest conservation occurred between residues 55–79 of Tre1 and residues 257 – 281 of SLC2A1 and between residues 46–85 in Tre1 and residues 297–332 to SLC2A8 (Fig. 1B). In both SLC2A1 and SLC2A8,, the residues 257–281 (GLUT1) and residues 297–332 (GLUT8) include a conserved region that corresponds to the pore-forming α-helical seventh transmembrane domain, which confers SLC2A family member substrate specificity (15).

Trehalose inhibits glucose transport through SLC2A transporters. A. Common evolutionary ancestry for trehalase, Tre1, and SLC2A family members by ClustalW2 phylogeny analysis. B. Tre1 pairwise alignment with the substrate-selecting 7th-transmembrane domains in SLC2A1 and SLC2A8. C. 2-DG uptake in response to increasing trehalose concentrations in 293 cells expressing the indicated GLUT protein. Data are expressed as the mean % uptake relative to control cultures not treated with trehalose ± SEM from N = 3 independent experiments, n = 3 replicates per experiment. D. [3H]-2-DG and [14C]-Fructose uptake in primary hepatocytes. Data are expressed as the mean uptake relative to trehalose-untreated control groups ± SEM from N = 3 independent experiments, n = 3 replicates per experiment. E. [3H]-2-DG and [14C]-Fructose uptake in HepG2 cells.. Data are shown as mean uptake relative to trehalose-untreated control groups ± SEM from N = 3 independent experiments, n = 3 replicates per experiment., ***, P < 0.001 versus vehicle-treated by two-tailed T-Test. N.S., not significant versus vehicle-treated.
Trehalose inhibits hexose uptake in multiple cell types
We examined whether trehalose modulated SLC2A-mediated uptake of [3H]-2-deoxy-D-glucose (2DG), a glucose analog that is transported but not metabolized. We stably transfected 293 cells that also stably expressed GLUT1 shRNA to knockdown the endogenous GLUT1 transporter (16) with empty vector or with plasmids expressing one of the four class I GLUTs (GLUT1-4), which are ubiquitous (3), or GLUT8, which is abundant in hepatocytes (2). We also tested the effect of trehalose on GLUT8-mediated 2DG uptake, because blocking GLUT8 prevents the development of hepatic steatosis (2). We assayed uptake of 2DG in each cell line over a range of substrate concentrations to determine the half-maximal inhibitory concentration (IC50) in each GLUT isoform-enriched system. Trehalose inhibited [3H]-2DG uptake in each of the class I GLUT-overexpressing and the GLUT8-overexpressing 293 lines (Fig. 1C).
We also evaluated the effects of trehalose on [3H]-2DG uptake and [14C]-fructose uptake in cultured primary murine hepatocytes and the human hepatocyte cell line HepG2. Because GLUT8 is abundant in hepatocytes and the overexpression study indicated an IC50 of 126 mM, we exposed the hepatocyte cells to 100 mM trehalose. In primary hepatocyte cultures, 100 mM trehalose inhibited 2DG uptake without significantly affecting [14C]-fructose uptake (Fig. 1D). Therefore, for all subsequent experiments involving primary hepatocytes, we used 100 mM trehalose. In HepG2 hepatocytes, 100 mM trehalose significantly blunted both [3H]-2DG and [14C]-fructose uptake when compared with vehicle-treated cultures (Fig. 1E). The inhibition of fructose uptake may reflect greater dependence of HepG2 cells on GLUT8-mediated fructose uptake in comparison with primary hepatocytes (2).
Trehalose rapidly induces AMPK and ULK1 activation in cultured primary hepatocytes
Preventing glucose uptake into hepatocytes is predicted to induce a starvation-like state and activate cellular autophagy. We therefore determined whether trehalose depleted hepatocyte intracellular ATP by incubating primary hepatocyte cultures with or without trehalose for 30 minutes prior to lysis and then performing an enzymatic-fluorimetric ATP concentration assay (17, 18). ATP concentration in trehalose-treated hepatocyte cultures was 20% lower when compared with untreated cultures (Fig. 2A). Reduced ATP concentration should increase the AMP:ATP ratio, leading to the phosophorylation at Thr172 and activation of AMPK, an energy-sensing kinase that stimulates autophagy (19). We monitored phosphorylated Thr172 of AMPK by Western blotting in primary hepatocyte cultures exposed to vehicle or trehalose for up to 4 hours. Trehalose stimulated an increase in AMPK (Thr172) phosphorylation within 30 minutes, and this increase was sustained throughout the 4-hour timecourse (Fig. 2B). In contrast, AMPK activation was at basal levels in cells that had not been exposed to trehalose (0, in Fig. 2B) and were analyzed after 4 hours.
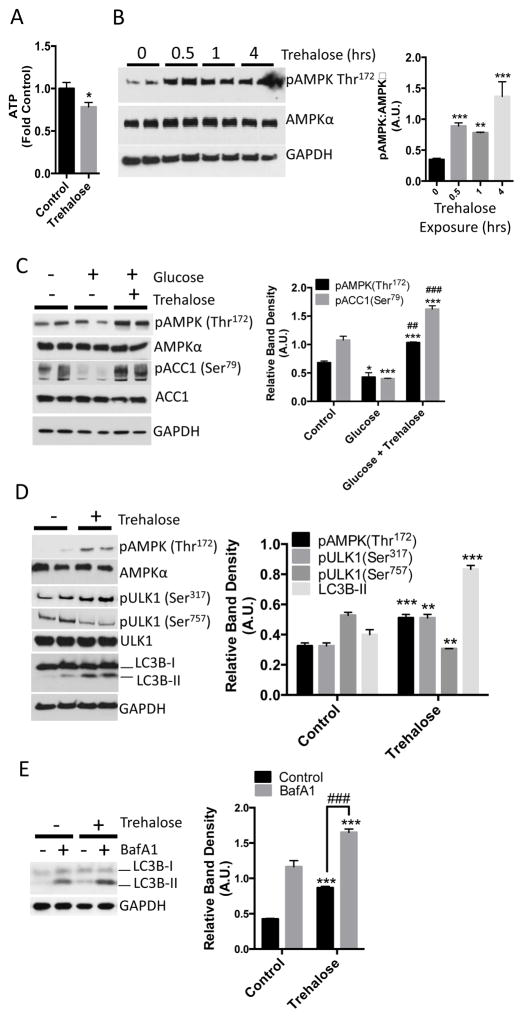
Trehalose decreases hepatocyte ATP and activates AMPK and autophagic flux. A. ATP quantification in hepatocytes treated with or without 100 mM trehalose for 30 minutes. Data are shown as mean fold of control (growth-media-treated) ATP ± SEM for, N= 3 independent experiments, n = 3 replicates per experiment. *, P < 0.05 by two-tailed T-Test. B. Left, immunoblot demonstrating AMPK (Thr172) phosphorylation in trehalose-treated hepatocytes (0–4hrs). Right, immunoblot quantification of pAMPK (Thr172) normalized to AMPKα band density. Data are shown as mean ± SEM for, N= 3 independent experiments, n = 2–3 replicates per experiment. **, P < 0.01 ***, P < 0.001 versus untreated control (“0 hour” data) (one-way ANOVA and Sidak’s post hoc analysis). C. Left, immunoblot depicting AMPK and ACC1 phosphorylation in HBSS-starved hepatocytes, followed by refeeding with unsupplemented HBSS (denoted as “−” group on blot and “control” on graph) or HBSS containing 25 mM glucose in the presence or absence of 100 mM trehalose for 30 minutes. Right, quantification of AMPK and ACC1 phosphorylation at Thr172 and Ser79 normalized to AMPKα and ACC1 band density, respectively. Data are shown as mean ± SEM for, N = 3 independent experiments, n = 2 replicates per experiment, In the glucose-treated group * and *** represent P < 0.05 and P < 0.001 versus controls (one-way ANOVA and Sidak’s post hoc analysis). In the glucose + trehalose-treated group, *** represents P < 0.001 versus cells treated with glucose alone (one-way ANOVA and Sidak’s post hoc analysis). ## and ###, P < 0.01 and P < 0.001 versus control group (one-way ANOVA and Sidak’s post hoc analysis) D. Left, immunoblot depicting ULK1 phosphorylation (Ser317 and Ser757) and LC3B-II accumulation after 1 hour incubation without (control) or with 100 mM trehalose. Right, quantification of AMPK and ULK1 phosphorylation, and LC3B-II band density normalized to AMPKα, ULK1, and GAPDH, respectively. Data are shown as mean ± SEM for, N = 3 independent experiments, n = 2 replicates per experiment, **, P < 0.01 versus control and ***, P < 0.001 versus control by 2-tailed T-Test. E. LC3B-I and LC3B–II immunoblotting in hepatocytes following treatment with or without 100 mM trehalose and 100 nM bafilomycin A1 for 1 hr. Control groups were treated with DMSO. Right, LC3B-II band density, normalized to GAPDH density. Data are shown as mean ± SEM for, N = 5 independent experiments, n = 1–3 replicates per experiment ***, P < 0.001 versus control groups. ###, P < 0.001 between bracketed groups by one-way ANOVA and Sidak’s post-hoc analysis for multiple comparisons.
To test if trehalose blocked glucose-mediated suppression of AMPK activity, we deprived primary hepatocytes of glucose, then replaced the starvation solution with unsupplemented Hank’s balanced salt solution (HBSS, denoted as “control”, which contains 5.6 mM glucose) or with HBSS containing 25 mM glucose (denoted as “glucose”) in the presence or absence of trehalose (denoted as “Glucose + trehalose”) for 30 minutes. For these experiments, we monitored not only AMPK activation as phosphorylation at Thr172, but also AMPK activity as measured by phosphorylation of its substrate Actyl-CoA carboxylase 1 (ACC1) at Ser79. Compared to control hepatocytes, glucose refeeding suppressed phosphorylation of AMPK and ACC1 (Fig. 2C). In contrast, hepatocytes subjected to glucose refeeding in the presence of trehalose exhibited marked ACC1 (Ser79) and AMPK (Thr172) phosphorylation when compared with control cultures or when compared with cultures subjected to 25 mM glucose refeeding alone (Fig. 2C), suggesting that glucose refeeding in the presence of trehalose does not simply recapitulate the fed state.
AMPK activates autophagy by phosphorylating the autophagy-promoting protein ULK1 at Ser317 (6). To determine whether trehalose activates ULK1 and autophagy, we treated primary hepatocytes with or without trehalose and then monitored ULK1 phosphorylation and the abundance of the autophagosome marker protein, LC3B-II (12), by Western blot. Consistent with induction of autophagy, trehalose significantly enhanced AMPK (Thr172) and ULK1 (Ser317) phosphorylation when compared with untreated primary hepatocytes (Fig. 2D). Consistent with a nutrient-deprivation response, we also observed that ULK1 phosphorylation at Ser757 – a site phosphorylated by the mammalian target of rapamycin complex 1 (mTORC1) (6)– was significantly lower in trehalose-treated cultures after 1 hour (Fig. 2D).
These changes in the autophagy-activating AMPK signaling pathway correlated with LC3B-II accumulation. To directly test whether LC3B-II accumulation was indeed due to trehalose induction of autophagic flux – and not secondary to defects in LC3B-II degradation – we incubated primary hepatocytes with trehalose for one hour in the presence or absence of bafilomycin A1 (BafA1), a vATPase inhibitor (12) that prevents LC3B-II degradation in autophagosomes. Trehalose-treated hepatocytes exhibited increased LC3B-II when compared with untreated cultures (Fig. 2E), and BafA1 further enhanced LC3B-II accumulation (Fig. 2E).
Our data indicated that trehalose acted as a glucose transport inhibitor that induced hepatocyte autophagic flux. Therefore, we evaluated whether other structurally unrelated glucose transport inhibitors could induce autophagy. The HIV protease inhibitors are a class of compounds that inhibit SLC2A family members with distinct isoform specificity (20). The HIV protease inhibitor, Lopinavir, is a class I SLC2A glucose transporter inhibitor (IC50 range: 4–32μM for GLUT1, GLUT2, GLUT3, and GLUT4) (16). To test whether lopinavir blocked glucose uptake in hepatocytes, we incubated primary hepatocyte cultures with solvent (DMSO) or with lopinavir (40 μM) 5 minutes prior to performing 2DG uptake measurements. Lopinavir reduced 2DG uptake in primary hepatocytes by approximately 50% when compared with solvent-treated cultures fig. S1A). Moreover, within 15 minutes we detected AMPK (Thr172) and ULK1 (Ser317) phosphorylation and LC3B-II accumulation in lopinavir-treated primary hepatocytes (fig. S1B). In contrast, the disaccharide sucrose (100 mM) did not induce AMPK signaling or LC3B-II accumulation at any time point through 4 hours of incubation in primary hepatocytes (fig. S2).
Trehalose activates AMPK signaling and autophagic flux in mouse liver
We assessed the effect of trehalose on AMPK signaling and autophagic flux in mice. To first determine whether orally delivered trehalose was detectable in peripheral venous circulation, we administered by gavage saline or trehalose (3 g/kg) for 0.5–4 hours to wild-type mice, collected peripheral blood and performed liquid chromatogaphy-mass spectrometric analysis on separated serum. Within 30 minutes, we detected trehalose in the serum of the mice (Fig. 3A). To ascertain whether AMPK and ULK1 were activated in response to trehalose administration, we excised liver from the mice and immunoblotted for AMPK (Thr172) and ULK1 (Ser317) phosphorylation. Livers from mice administered trehalose exhibited increased AMPK Thr172 within 30 minutes, increasing ULK1 Ser317 phosphorylation that plateaued after 2 hours, and a peak of LC3B-II accumulation after 1 hour (Fig. 3B). Because LC3B-II could accumulate in liver tissue due to increased autophagic flux or due to decreased LC3B-II degradation, mice were administered saline or trehalose or trehalose (3 g/kg) 1 hour after intraperitoneal injection with saline or leupeptin (40 mg/kg) to block LC3B-II degradation in autophagosomes (12, 21). Consistent with an increase in autophagic flux, leupeptin enhanced the accumulation of LC3B-II in the trehalose-treated animals (Fig. 3C).
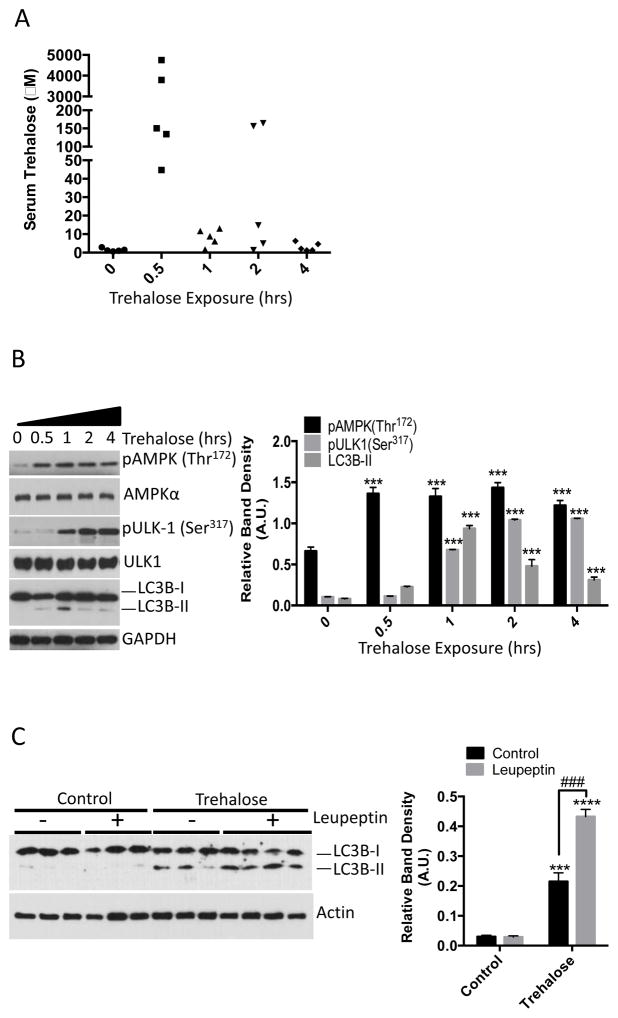
Orally administered trehalose rapidly accumulates in peripheral circulation and induces hepatic autophagy. A. Liquid chromatography-mass spectrometric analysis of serum isolated from mice administered 3 g/kg trehalose by gavage and analyzed after 0.5, 1, 2, or 4 hours. Serum analyzed at the “0 hour” trehalose-treatment time point was derived from mice 30 minutes after gavage with 0.9% NaCl. N = 5 mice per treatment group. B. Left, immunoblot analysis of crude liver lysates from mice administered 3 g/kg trehalose and analyzed after 0.5, 1, 2 or 4 hrs. Liver lysates analyzed at the “0 hour” trehalose-treatment time point were derived from mice 30 minutes after gavage with 0.9% NaCl. Right, Quantification of pAMPK, pULK-1 and LC3B-II, normalized to AMPKα, ULK1, and GAPDH, respectively. Data are shown as mean ± SEM for, N = 5 independent mice per treatment group. ***, P < 0.001 versus 0-hour treatment group (one-way ANOVA and Sidak’s post hoc analysis) C. Left, immunoblot analysis of LC3B-II in liver lysates from mice treated with or without 40 mg/kg leupeptin 1 hr prior to oral gavage with 3 g/kg trehalose. Right, quantification of LC3B-II:Actin band density ratio. Data are shown as mean ± SEM for, N = 3–4 independent mice per treatment group, ***, P < 0.001 and ****, P < 0.0001 versus saline-treated controls (2-tail T-Test). ***, P < 0.001 between trehalose-treated groups treated with leupeptin versus leupeptin-untreated group (denoted by brackets) by 2-tail T-Test.
Trehalose reduces triglyceride accumulation in cultured hepatocytes
Autophagic insufficiency may underlie the pathogenesis of NAFLD (22), a disorder of excessive hepatic fat accumulation that ranges in phenotype from simple steatosis to steatohepatitis, cirrhosis, and liver failure (1). The accumulation of triglycerides in cultured hepatocytes serves as an in vitro model of fatty liver disease and can be induced by exposing the cells to the monosaccharide fructose (2, 23). We tested whether trehalose mitigates hepatocyte triglyceride accumulation in this cell culture model by incubating serum-deprived primary hepatocytes in the presence or absence of fructose (5 mM for 48 hr) with or without co–administration of trehalose. We extracted the lipids and performed enzymatic-colorimetric triglyceride analysis. Fructose significantly induced triglyceride accumulation in the primary hepatocyte cultures, and trehalose blocked the accumulation of triglyceride (Fig. 4A). To quantify key fructose-responsive regulatory factors, we quantified the abundance of transcripts encoding carbohydrate response-element binding protein (ChREBP) and glycerol-3-phosphate acyltransferase (GPAT). Trehalose partially blocked the fructose-mediated increase in ChREBP and GPAT mRNA abundance (Fig. 4B). Trehalose also significantly reduced fatty acid-induced triglyceride accumulation in primary hepatocytes exposed to BSA-conjugated oleic, palmitic, stearic, and linoleic acids conjugated to bovine serum albumin (BSA) at a ratio of 1:1:1:1 for a total of 500μM fatty acid for 48 hours (Fig. 4C).
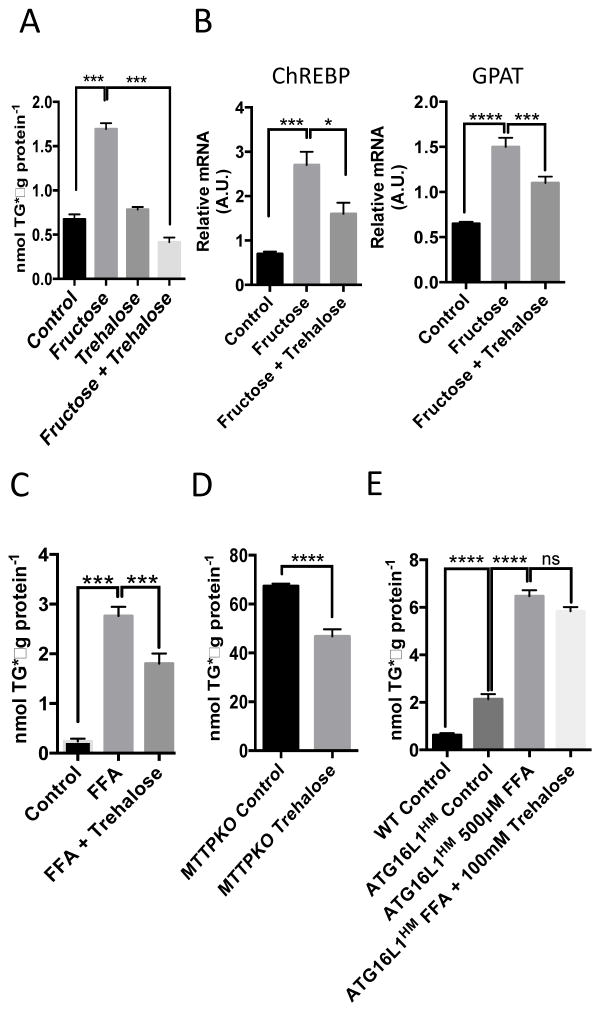
Trehalose mitigates triglyceride accumulation in multiple in vitro models of steatosis. A. Triglyceride (TG) quantification in hepatocytes treated with or without 5 mM fructose in the presence or absence of 100 mM trehalose. Data are shown as mean ± SEM for, N = 3 independent experiments, n = 3 replicates per treatment group per experiment. ***, P < 0.001 (one-way ANOVA and Sidak’s post hoc analysis). B. Quantitative, real-time RT-PCR (qPCR) analysis of carbohydrate response element binding protein (ChREBP) and glycerol phosphate acyltransferase (GPAT) mRNA in hepatocytes treated with fructose in the presence or absence of 100mM trehalose. Target abundances are normalized to α-actin expression within each sample. Data are shown as mean ± SEM for, N = 3 independent experiments, n = 3 replicates per treatment group per experiment. ****, P < 0.0001, ***, P < 0.001, and *, P < 0.05 (one-way ANOVA and Sidak’s post hoc analysis). C. TG quantification in hepatocytes treated with or without BSA-conjugated fatty acids (500μM) in the presence or absence of 100mM trehalose (48hr). Data are shown as mean ± SEM for, N = 3 independent experiments, n = 2–3 replicates per treatment group per experiment. ***, P < 0.001, (one-way ANOVA and Sidak’s post hoc analysis). D. TG quantification in microsomal triglyceride transfer protein-deficient primary hepatocytes treated with or without 100mM trehalose (48hr). ****, P < 0.0001 by 2-tail T-Test. Data are shown as mean ± SEM for, N = 3 independent experiments, n = 3 replicates per treatment group per experiment. E., TG quantification in WT and ATG16L1HM hepatocytes treated with BSA alone (control) or with BSA-conjugated fatty acids in the presence or absence of trehalose (100mM, 48hr). Data are shown as mean ± SEM for, N = 3 independent experiments, n = 3 replicates per treatment group per experiment. ****, P < 0.0001 by one-way ANOVA and Sidak’s post-hoc test.
To determine whether the protective effects of trehalose effects on hepatocyte triglyceride accumulation extended to other models of hepatic triglyceride accumulation, we evaluated the effect of trehalose on hepatocytes from mice genetically deficient in hepatic microsomal triglyceride transfer protein (MTTP), hepatocytes from mice expressing knock-in hypomorphic alleles of the gene encoding the autophagy-promoting complex protein ATG16L1 [ATGH16L1HM (24)], and HepG2 cells. MTTP-knockout mice develop hepatic steatosis in the absence of dietary stress (25). Trehalose reduced the accumulation of triglycerides in cultured hepatocytes from MTTP-knockout mice compared with the abundance of triglycerides in the untreated MTTP-knockout hepatocytes (Fig. 4D).
Compared to hepatocytes from wild-type mice, the hepatocytes from the ATGH16L1HM mice exhibit increased triglyceride accumulation even in growth medium (Fig. 4E). However, exposure to the BSA-conjugated fatty acids induced a significant increase in triglyceride accumulation that trehalose did not reduce (Fig. 4E), indicating that the ability of trehalose to prevent triglyceride accumulation required autophagy, which was compromised in the cells from the ATG16L1HM mice.
HepG2 cells are a cell line derived from a human hepatocellular carcinoma. Similar to the primary hepatocyte response to trehalose, HepG2 cultures exposed to trehalose exhibited significantly greater LC3B-II accumulation, and this response was enhanced by BafA1 (fig. 3A). Furthermore, BSA-conjugated fatty acids induced triglyceride accumulation in the HepG2 cells and trehalose reduced this response (fig. S3B).
Feeding mice trehalose prevents NAFLD and dyslipidemia
High fructose diet (60% of calories from fructose, HFrD) induces hepatic lipid droplet accumulation and dyslipidemia in mice (2, 26–28) and thus is a model of NAFLD. The data in the cultured hepatocytes suggested that trehalose may be liver protective. To investigate this possibility, we fed mice 3% trehalose water ad libitum 48 hours prior to placing the mice on the HFrD for ten days. Biochemical analysis of plasma from fasted mice revealed significantly reduced circulating triglycerides and cholesterol without significant changes in free fatty acids (Fig. 5A) in trehalose-treated mice fed HFrD. Histological analysis and quantitation of lipid vesicles in liver tissue by Oil Red-O neutral lipid staining revealed marked steatosis in HFrD-fed mice, which was significantly reduced by trehalose (Fig. 5B). Biochemical analysis of liver tissue showed that liver triglycerides, cholesterol, and free fatty acids were increased in the HFrD-fed mice and that each was significantly reduced in livers from the trehalose-treated mice fed HFrD (Fig. 5C). Consistent with histological and biochemical improvements, animals fed HFRD and trehalose exhibited significant reductions in transcripts encoding acyl-coA carboxylase 1 (ACC1), stearyl-coA desaturase-1 (SCD1), glycerol phosphate acyltransferase-1 (GPAT), and peroxisome proliferator-activated receptor γ (PPARγ) (Fig. 5D), compared with their abundance in animals fed HFrD alone.
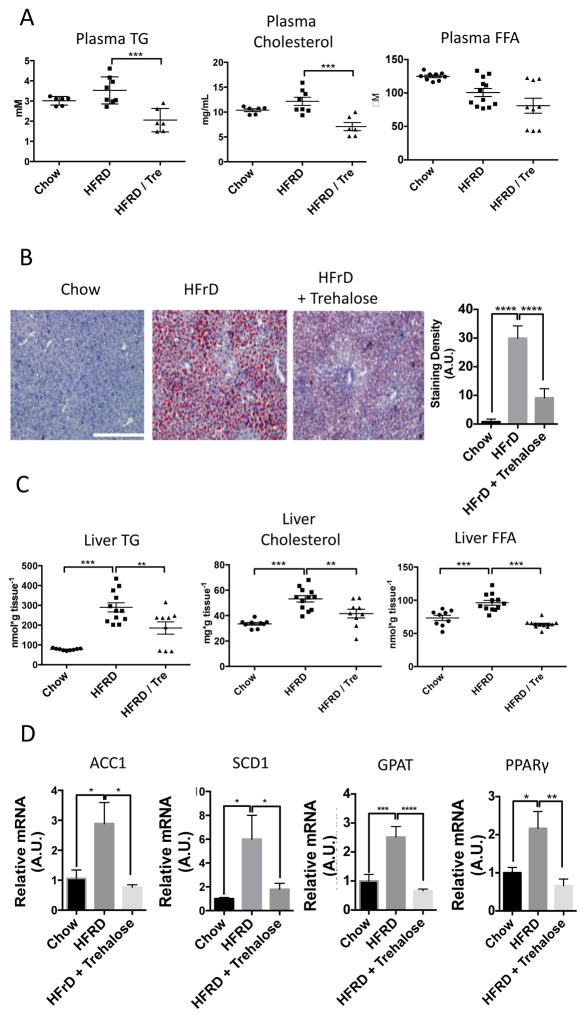
Trehalose reduces HFrD-induced hepatic steatosis in vivo. A. 8 week-old mice were fed chow, 10-day 60% fructose diet or 10-day 60% fructose diet initiated two days after initiating 3% trehalose fed ad libitum in drinking water. Fasting plasma triglycerides, cholesterol and free fatty acids were quantified. Data are shown as mean ± SEM for, N = 6–12 indepedendent mice per treatment group. ***, P < 0.001 (one-way ANOVA and Sidak’s post hoc analysis). B. Left, Oil Red-O staining in frozen liver sections from mice described above. Scale bar, 200μm. Right, Blinded-observer quantification of staining red staining density (minimum 3 random fields from three cryosections obtained from three different mice per group) by ImageJ (version 1.47) densitometry software. Data are shown as mean ± SEM for, N = 9–12 mice per treatment group. ****, P < 0.0001 (one-way ANOVA and Sidak’s post hoc analysis). C. Hepatic TG, cholesterol and FFA quantification in 8 week-old mice fed chow, 10-day 60% fructose diet or 10-day 60% fructose diet initiated two days after initiating 3% trehalose fed ad libitum in drinking water. Data are shown as mean ± SEM for, N = 9–12 mice per treatment group. ****, P < 0.0001, ***, P < 0.001, and **, P < 0.01 (one-way ANOVA and Sidak’s post hoc analysis). D.Left to right, qPCR analysis of acyl-CoA carboxylase-1 (ACC1), stearyl-coA desaturase-1 (SCD1), glycerol phosphate acyltransferase (GPAT) and peroxisome proliferator antigen receptor-gamma (PPARγ) mRNA in liver tissue mRNA extracted from mice fed 10-day HFrD with or without 3% trehalose water. Target abundances are normalized to α-actin expression within each sample. Data are shown as mean ± SEM for, N = 9–12 mice per treatment group. ****, P < 0.0001, ***, P < 0.001, **, P < 0.01 and *, P < 0.05 (one-way ANOVA and Sidak’s post hoc analysis).
Overexpression of either GLUT8 or AMPK reverses the beneficial effects of trehalose on hepatic steatosis
The data from the hepatocyte cultures and mouse model of NALFD suggested that trehalose inhibits hepatocyte hexose transport, activates AMPK and ULK1, and subsequently stimulates autophagic flux. To confirm that inhibition of hexose transport was essential to this mechanism of trehalose action, we tested whether trehalose-induced autophagy was reversed by expression of a SLC2A family member in primary hepatocytes. We transfected primary hepatocytes with adenovirus encoding green fluorescent protein (GFP), GLUT8, or GLUT2 and then exposed the cells to trehalose for 1 hour. Adenoviral overexpression of GLUT8, but not GLUT2, abolished trehalose-stimulated LC3B-II accumulation (Fig. 6A). GLUT8 overexpression also blunted the reduction in fructose-induced triglyceride accumulation in response to trehalose (Fig. 6B).
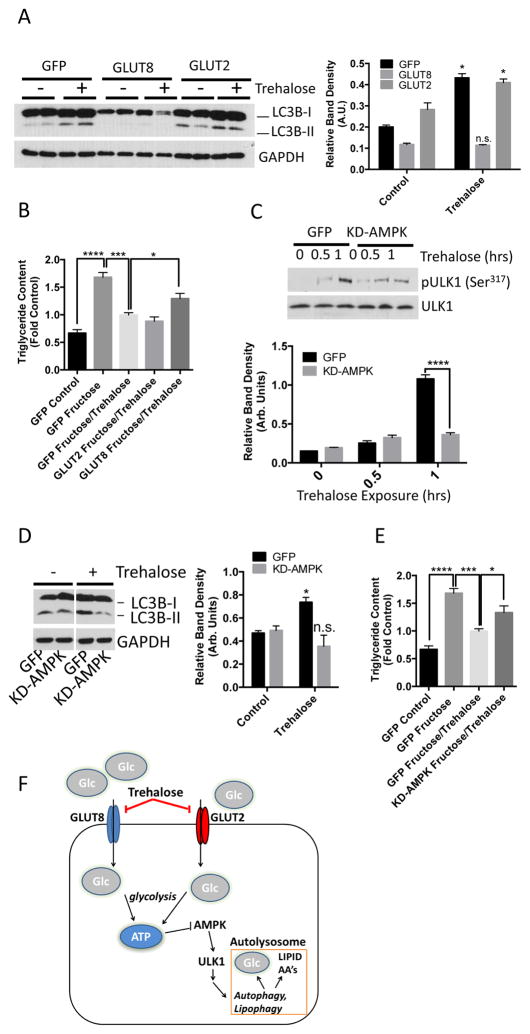
GLUT8 and AMPK mediate trehalose-induced autophagy and protection from triglyceride accumulation. A. LC3B-II immunoblots in hepatocyte lysates following adenoviral GFP, GLUT8 or GLUT2 expression. B. Quantitative triglyceride measurements in hepatocytes treated with or without 5mM fructose in the presence or absence of 100mM trehalose after adenoviral expression of GFP, GLUT8 or GLUT2. Data are shown as mean ± SEM for, N = 3 independent experiments, n = 2 replicates per experiment. ****, P < 0.0001, ***, P < 0.001, and *, P < 0.05 (one-way ANOVA and Sidak’s post hoc analysis). C. Above, immunoblot depicting phosphorylated ULK1 (Ser317) in trehalose-treated hepatocytes expressing a kinase-dead AMPK mutant (KD-AMPK). Below, pULK1 band density quantification, normalized to ULK1 band density. Data are shown as mean ± SEM for, N = 3 independent experiments, n = 1–3 replicates per experiment. ****, P < 0.0001 between bracketed groups (2-tailed T-Test). D. Above, immunoblot of LC3B-II accumulation in 100mM trehalose-treated hepatocytes (1hr) expressing KD-AMPK. Below, quantification of LC3B-II band normalized to GAPDH. *, P < 0.05 and N.S., not significantly different versus growth media group (2-tailed T-Test). E. Quantitative triglyceride measurements in hepatocytes treated with or without 5 mM fructose in the presence or absence of 100mM trehalose (48hr) after adenoviral expression of GFP or KD-AMPK. Data are shown as mean ± SEM for, N = 3 independent experiments, n = 3 replicates per experiment. ****, P < 0.0001, ***, P < 0.001 *, P < 0.05 (one-way ANOVA and Sidak’s post hoc analysis). F. Proposed model depicting trehalose blockade of GLUT2 and GLUT8, resulting in AMPK and ULK1 activation, and enhanced autophagic flux.
To test that trehalose required AMPK signaling to induce hepatocyte autophagy and mitigate fructose-induced triglyceride accumulation, we expressed GFP or kinase-dead AMPK (KD-AMPK) in the primary hepatocytes and exposed the cells to trehalose. Trehalose treatment resulted in robust ULK1 (Ser317) phosphorylation in GFP-expressing hepatocytes but not in the KD-AMPK-expressing hepatocytes (Fig. 6C). Moreover, KD-AMPK expression attenuated LC3B-II accumulation in cells exposed to trehalose (Fig. 6D). Hepatocytes expressing KD-AMPK also exhibited significantly less effective trehalose-mediated reduction in triglyceride accumulation in response to fructose (Fig. 6E).
DISCUSSION
The data herein provide evidence that trehalose inhibits glucose transport to induce hepatic autophagy and prevent hepatic steatosis in an SLC2A- and AMPK-dependent manner (Fig. 6F). Trehalose may therefore create a functionally “starved” state that activates AMPK signaling in the hepatocyte, even in the presence of abundant glucose. The beneficial effects of hepatic AMPK activation in increasing fat oxidation and insulin sensitivity are well-documented (29).
Although the data support a role for AMPK signaling hepatocyte autophagy and limiting triglyceride accumulation, we also found decreased ULK1 Ser757 phosphorylation, which is a target of mTORC1. Thus, we cannot rule out the possibility that trehalose also results in inhibition of mTORC1 signaling and that mTORC1 inhibition is required for the full effects of trehalose in promoting autophagy. There is precedent for AMPK-mTORC1 crosstalk. In response to starvation, mTORC1-dependent sites on ULK1 and ULK2 are rapidly dephosphorylated (30). Moreover, AMPK phosphorylates the mTORC1 subunit raptor to inhibit mTORC1 activity (30). Therefore, trehalose-activated autophagy may involve mTORC1 suppression – perhaps through crosstalk from the AMPK pathway. Future mechanistic work should address the specific role of mTORC1 in trehalose-induced autophagy.
Compared with the rapid LC3B-II accumulation in hepatocytes within minutes of trehalose exposure that we found, prior reports describing the effects of trehalose reported a protracted (24 hours) time course required for trehalose to induce LC3B-II accumulation in COS-7 cells (9). The timing of the autophagic response to apparent starvation is likely a tissue-specific phenomenon. Physiologically, a rapid response of hepatocytes to nutrient stress is reasonable, given an organism’s requirement for rapid hepatic adaptation to “starvation” to quickly mobilize energy stores and avert hypoglycemia. The observed differences could also reveal distinct acute (minutes) and longer term (hours to days) mechanisms of trehalose action that remain to be fully elucidated. For example, Rubinsztein and colleagues demonstrated LC3B-II accumulation 24 hours after trehalose exposure in COS-7 cells. This LC3B-II accumulation was recapitulated by inducing trehalose synthase expression in T-REx 293 cell lines, which synthesize intracellular trehalose in response to tetracycline (9). Trehalose access to the cytoplasm through fluid-phase endocytosis, pinocytosis, or other transport mechanisms may underlie any distinctions between acute and longer-term cellular actions of trehalose. Despite these intriguing differences, our work suggested that trehalose – or potentially other glucose transport modulators – may be useful clinically to prevent or treat diseases of autophagic insufficiency (31), including NAFLD.
MATERIALS AND METHODS
Mouse models
Wild-type C57BL/6J males and females (Jackson Laboratory, Bar Harbor, ME) were used for all experiments. ATG16L1HM (24) mice were obtained from Herbert W. Virgin’s laboratory (Washington University School of Medicine). Hepatic MTTP-deficient mice (25) were obtained from Nicholas Davison’s laboratory (Washington University School of Medicine). All Experiments were performed in accordance with the Washington University Animal Studies Committee.
Cell Cultures
Methods used to generate the HEK 293 cell lines selectively overexpressing human GLUT1, GLUT2, GLUT3, GLUT4 or GLUT8 with concomitant GLUT1 shRNA overexpression were recently described (16). Briefly, HEK 293 cells obtained from the American Type Culture Collection were forced to stably express GLUT1-directed shRNA following lentiviral transfection (obtained from the RNA interference Core Laboratory at Washington University School of Medicine) in order to knock down endogenous “background’ glucose transport through GLUT1 in subsequent studies in which GLUT2, GLUT3, GLUT4 and GLUT8 were overexpressed and studied. Following puromycin selection, cells were stably transfected to express codon-optimized human GLUT2, GLUT3, GLUT4 or GLUT8 DNA in the pcDNA3.1(−)/hygro plasmid (Life Technologies, Carlsbad, CA). GLUT1 overexpression was carried out in wild-type 293 cells not expressing GLUT1 shRNA. Following hygromycin selection, colonies were grown to near confluence in 4-cm tissue culture dishes, and the highest expressing colonies for each of the GLUT isoforms were selected using [3H]-2DG uptake and quantitative reverse transcriptase (RT)-PCR. Absolute GLUT mRNA quantity for 293 cells expressing GLUT1, GLUT2, GLUT3, GLUT4 and GLUT8 are shown in table S1.
HepG2 cultures were obtained from the American Type Culture Collection (Manassas, VA). Cultures were maintained in DMEM containing 4.5 g/L glucose and 10% FBS (e.g. “regular growth media”) through passage 15.
Primary murine hepatocytes obtained from wild-type, ATG16L1HM knock-in mice and MTTPKO mice were isolated precisely as described (2). Briefly, mice were cannulated through the portal vein under anesthesia with Metophane and pentobarbitol, and the liver was perfused with oxygenated calcium- and magnesium-free Hank’s balanced salt solution, pH 7.4 (Invitrogen, Carlsbad, CA) for 5 minutes, followed by perfusion of oxygenated Dulbecco’s modified eagle medium (Sigma-Aldrich, St. Louis, MO) containing 0.05% Type IV collagenase (Sigma-Aldrich, St. Louis, MO) for 5–10 min. The perfusate was drained through an incision in the inferior vena cava. After perfusion, the liver was removed and submerged into ice-cold DMEM, and the cells were released gently. The resultant cell suspension was then filtered through a 45 μ nylon mesh, pelleted on ice for 30 minutes, then resuspended in regular growth media. Cells were plated onto tissue culture plates coated with type I collagen from (Sigma-Aldrich, St. Louis, MO) and incubated at 37 °C under 5% CO2 in regular growth media. After 1 hour of attachment, cultures were washed in regular growth media, and the cell monolayers were maintained overnight in regular growth media prior to assay.
Adenoviral constructs (KD-AMPK, GLUT2, GLUT8) were purchased from Applied Biological Materials (Richmond, BC, Canada).
Statistics
All data were analyzed using GraphPad Prism 6.0. P < 0.05 was defined as statistically significant after post hoc correction for multiple comparisons. Specific statistical tests applied are noted in the Figure Legends.
Sequence Alignment and Phylogeny
Accession numbers for sequences aligned using ClustalW2 were Trehalase (NP_067456.1), Tre1 (BAA95353.1), GLUT1 (EDL30474.1), GLUT2 (NP_112474.2), and GLUT8 (NP_062361.1). The following program-default parameters were used: Output format – Clustal without numbers; no input sequence dealignment; MBED-like clustering iteration and MBED-like clustering guide-tree; 0 combined guide-tree iterations; “default” maximum guide tree and “default” maximum HMM iterations were used; aligned order output.
[3H]-2-DG Uptake
HEK 293 cells overexpressing human GLUTs 1, 2, 3, 4, or 8 were grown in MEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 ug/ml streptomycin. [3H]-2-deoxyglucose (2DG, 50 μM, 1 μCi/ml, obtained from American Radiolabeled Chemicals, St. Louis, MO) uptake was determined at 37°C according to Tordjman et al. (32) with the following modifications. To adhere the cells to tissue culture plates, dishes were first pretreated with 25 μg/ml polyethyleneimine (Fluka, catalog number P3143) in 150 mM NaCl for 20 min followed by the removal of the solution by vacuum. Cells were then plated at 2*105 cells/ml MEM (e.g. normal growth medium containing all additives) supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 ug/ml streptomycin in 12-well plates and incubated overnight at 37°C. Following a 30-minute incubation at 37°C in glucose-free HEPES, pH 7.3, in the presence or absence of 0–500 mM trehalose, 2DG uptake was measured in glucose-free HEPES, pH 7.3 for 6 min at 37°C. Cultures were washed extensively in phosphate-buffered saline and lysed in 0.1N NaOH, 1% SDS lysis buffer and 80% of each lysate was subjected to liquid scintillation counting (Ultima Gold, Thermo Fisher, Grand Island, NY). Protein concentration was determined in the remaining lysate and total protein per well was used to normalize total 6-minute 2DG uptake in each well. Uptake data are plotted as % uptake relative to trehalose-unexposed HEK293 cultures.
Primary hepatocytes and HepG2 cells were plated at 5*105 cells per well in 12-well plate and maintained at 37°C overnight in regular growth media until 2DG uptake was assayed. On the day of assay, cultures were incubated (30 minutes at 37°C) in glucose-free HEPES, pH 7.3, in the presence or absence of 100 mM trehalose or 40 μM lopinavir. Cultures were then incubated with 1 μCi/mL [3H]-2DG (50 μM) in glucose-free HEPES, pH 7.3, at 37°C for 6 minutes in the presence or absence of 100 mM trehalose or 40 μM lopinavir prior to washing, lysis and analysis as described above. DMSO (vehicle) was used as the vehicle control in 2DG uptake assays in which lopinavir was used to inhibit 2DG uptake.
[14C]-Fructose uptake was performed as described (2). Briefly, primary hepatocyte and HepG2 cultures were plated at 5*105 cells per well in 12-well plates and maintained at 37°C overnight in regular growth media until assay. On the day of assay, cultures were incubated (30 minutes at 37°C) in glucose-free HEPES, pH 7.3, in the presence or absence of 100mM trehalose. Cultures were then incubated with 1 μCi/mL (4 μM) [14C]-Fructose (American Radiolabeled Chemicals, St. Louis, MO) for 1 minute, prior to washing, lysis and analysis as described above.
Hepatocyte ATP Measurement
ATP concentrations in cultured hepatocytes were measured as described previously (17, 18). Briefly, primary hepatocytes grown in six-well plates in DMEM containing 4.5 g/L glucose and 10% FBS were treated in the presence or absence of 100mM trehalose for 30 minutes. Cells were trypsinized and the cell pellet was washed in phosphate-buffered saline prior to homogenization in 0.1 mL 0.1 N NaOH, and protein denaturation by incubation at 80°C for 20 minutes. The homogenate was neutralized with 50 μL neutralization buffer (0.15 N HCl, 0.1 M Tris–HCl, pH 6.6) to produce a final 34 mM Tris–HCl, pH 8.1 cell extract.
The enzymatic-fluorimetric ATP assay was carried out using 20 μL of cell extract. 34 mM Tris–HCl, pH 8.1 was used as a “blank” reaction to quantify background fluorescence. ATP standards, and cell extracts were added to 50 μL ATP reagent (60 mM Tris–HCl pH 8.1, 0.03% BSA, 1.5 mM MgCl2, 1 mM dithiothreitol, 450 μM D-glucose, 100 mM nicotinamide adenine dinucleotide phosphate (NADP+), 3 μg/mL hexokinase without (NH4)2SO4, and 1 μg/mL glucose-6-phosphate dehydrogenase without (NH4)2SO4). The reaction was carried out at 60°C for 20 minutes and terminated after 20 minutes by adding 10 mL 0.5 N NaOH. 340nm wavelength emission was subsequently measured on a Packard BF10000 Fluorocount microtitre plate fluorometer following addition of 1mL of 6 N NaOH, 10 mM imidazole, and 0.01% H2O2 to each reaction. ATP concentrations in each experiment were normalized to total protein (determined by bicinchoninic acid assay (BCA assay, Thermo Scientific, Grand Island, NY) in the neutralized cell extract and were calculated as (nmol ATP)*(mg total protein)−1. Data from three independent experiments (n = 2–3 samples per condition per experiment) are expressed relative to untreated culture [ATP] obtained in each experiment.
Immunoblotting
Immunoblotting was performed as recently described (33). Samples for immunoblot analysis were prepared as follows: Cultures for HepG2 and primary hepatocytes were seeded at 1 X 106 cells per well, grown in six-well plates were placed on ice following experimental manipulations. Each sample was lysed in 125 μL ice-cold RIPA (50 mM Tris-HCl, pH 8.0, with 150 mM sodium chloride, 1.0% Igepal CA-630 (NP-40), 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate). Cell debris was cleared from each lysate by microcentrifugation (16,200 RCF) prior to protein concentration determination by BCA assay (Thermo Fisher, Grand Island, NY) to normalize protein loading. Samples were diluted 1:1 in 2X Laemmli sample buffer (BioRad, Hercules, CA). 10–25 μg protein were resolved by 10% bis-acrylamide SDS-PAGE and transferred to 0.2 μm-pore nitrocellulose membranes (VWR, Batavia, IL) by semi-dry electrophoresis (Bio-Rad, Hercules, CA). Non-specific antibody binding to nitrocellulose membranes was blocked by 60-minute incubation in 5% milk in Tris-buffered saline, tween 20 (TBST, 0.05M Tris-HCl, 0.138 M NaCl, 0.0027 M KCl, TWEEN 20, pH 7.4). Following antibody blocking, primary antibodies were diluted in TBST containing 5% bovine serum albumin (Sigma-Aldrich, St. Louis, MO) overnight at 4°C. Membranes were washed in TBST and incubated (1 hour at room temperature) with horseradish peroxidase-linked anti-rabbit IgG or anti-mouse IgG (both obtained from Cell Signaling Technologies, Beverly, MA) diluted in TBST containing 5% milk. Following TBST washing, immunoreactive bands were visualized by incubating each membrane with chemiluminscent horseradish peroxidase substrate (Clarity ECL, BioRad, Hercules, CA) and exposing to autoradiographic film (VWR, Batavia, IL).
Primary antibodies were from Cell Signaling Technologies (Beverly, MA): AMPKα (#5831); phospho-AMPK(T172) (#2535); phospho-ACC1 (S79) (#11818); ACC1 (#3676); phospho-ULK1 (S317) (#2535); phospho-ULK1 (S757) (#6888); ULK1 (#8054); Actin (#4970); GAPDH (#5174); LC3B antiserum was obtained from Novus Biologicals (Littleton, CO) (#NB-100-2200).
For autophagic flux assays, primary hepatocytes and HepG2 cell lines were maintained in regular growth media prior to experiment. Cultures were then incubated in the presence or absence of dimethyl sulfoxide (DMSO) or 100 nM Bafilomycin A1 (Cayman Chemicals, Ann Arbor, MI) with or without 100 mM trehalose for 1 hour. Cultures were lysed and subjected to immunoblot analysis as above.
Lopinavir effects on autophagic flux were tested in primary hepatocytes grown in regular growth media overnight after isolation. DMSO or 40 μM lopinavir was added to regular growth media and added to cultures for 15 minutes prior to lysis and immunoblotting as above.
For HBSS starvation-refeed assays, primary murine hepatocytes were incubated in HBSS (5.3 mM KCl, 0.44 mM KH2PO4, 4.17 mM NaHCO3, 137.9 mM NaCl, 0.34 mM Na2HPO4, 5.56 mM D-glucose, Thermo Fisher, Grand Island, NY) prior to addition of HBSS alone or HBSS supplemented to a final glucose concentration of 25 mM in the presence or absence of 100mM trehalose. Cultures were lysed in ice-cold RIPA prior to protein determination and immunoblot analysis as above.
GC-MS Trehalose Quantification
13C12 trehalose (Omicron Biochemical, South Bend IN) was used as an extraction and derivatization internal standard for samples. 5nmol internal standard were added to excess 200ul isopropanol:CH3CN:H2O and the sample was vortexed centrifuged to achieve phase separation. The supernatant was dried at room temperature overnight under nitrogen gas following addition of MSTFA with 10% pyridine in CH3CN. Derivatized samples were analyzed on an Agilent 7890A gas chromatograph interfaced to an Agilent 5975C mass spectrometer. The GC column used for the study was a HP-5MS (30 m, 0.25 mm internal diameter, 0.25 μm film coating, P.J. Cobert, St. Louis, MO). A linear temperature gradient was used. The initial temperature of 80° was held for 2 minute and increased to 300 at 10°/minute. The temperature was held at 300°C for 2 minutes. The samples were run by electron ionization (EI) and the source temperature, electron energy and emission current were 200°C, 70 eV and 300 μA, respectively. The injector and transfer line temperatures were 250°C.
Quantitative Real-Time RT-PCR (qPCR)
qPCR was performed with primer sequences and method precisely as described (2). Briefly, RNA was isolated from either 1 X 106 cells (for primary hepatocyte experiments) or from 100mg dissected liver tissue (derived from in vivo chow- HFrD- and trehalose-treated animals) homogenized in Trizol reagent (Invitrogen, Carlsbad, CA) per manufacturer protocol. 1000 ng RNA per sample was reverse-transcribed to cDNA using the Quantitect Qiagen reverse transcriptase kit (Qiagen, Valencia, CA). cDNA was subjected to quantitative, real-time PCR using the SYBR Green master mix reagent (Applied Biosystems, Carlsbad, CA). Primers for quantitative real-time RT-PCR, (listed 5′ to 3′) are (2, 34–36): Acyl-CoA Carboxylase (ACC1) (sense) TGTCCGCACTGACTGTAACCA, (antisense) TGCTCCGCACAGATTCTTCA; Carbohydrate response element binding protein (ChREBP) (sense) CTGGGGACCTAAACAGGAGC, (antisense) GAAGCCACCCTATAGCTCCC; Glyceraldehyde 3-phosphate acyltransferase (GPAT) (sense) CAACACCATCCCCGACATC, (antisense) GTGACCT TCGATTATGCGATCA; Peroxisome proliferator activated receptor PPARγ (sense) CCACCAACTTCGGAATCAGCT, (antisense) TTTGTGGATCCGGCAGTTAAGA; Stearoyl-CoA desaturase (SCD)-1 (sense) CCGGAGACCCTTAGATCGA, (antisense) TAGCCTGTAAAAGATT TCTGCA AACC.
Oil Red-O Staining
Oil-Red-O Staining was performed precisely as described (2). Briefly, 10 μm frozen sections from dissected mouse livers were fixed five minutes (−20 °C) and washed in phosphate-buffered saline. Sections were stained in 0.3% Oil-Red-O in 60% isopropanol (15 minutes) followed by extensive distilled water washes until the effluate cleared. Sections were mounted in 54% aqueous glycerol mounting media prior to imaging. For staining density quantification, high-powered (40X objective) photomicrographs of stained sections were obtained while each slide’s treatment group was blinded to the research assistant. Within ImageJ software (version 1.47), pixel densities from 3–5 random fields within three distinct cryosection photomicrographs obtained from at least three different mice per group were quantified. Photomicrograph data were unblinded prior to calculating density mean and SEM, as presented in Fig. 5.
Analysis of AMPK signaling and autophagic flux in mouse liver
For in vivo autophagic flux assays, 8-week old wild-type mice were injected with 40 mg/kg leupeptin hemisulfate (21) (Sigma-Aldrich, St. Louis, MO) or with saline intraperitoneally one hour prior to orogastric gavage with saline or with 3 g/kg trehalose in saline. Mice were killed two hours after orogastric gavage, and livers were rapidly dissected and snap-frozen in liquid nitrogen prior to homogenization in ice-cold RIPA lysis buffer and immunoblot analysis. Throughout the experiment, mice were given ad libitum access to standard rodent chow and water.
For in vivo signaling assays, 8-week old wild-type mice were subjected to orogastric gavage with 3 g/kg trehalose dissolved in saline. Mice were killed 0.5–4 hours after gavage, and livers were rapidly dissected and snap frozen in liquid nitrogen prior to homogenization in ice-cold RIPA lysis buffer and immunoblot analysis. Control mice were gavaged with saline alone and killed 0.5 hours post-gavage. They are labeled as “0 hour” trehalose exposure.
Plasma and Liver Biochemical analyses
In vivo plasma and liver biochemistries were performed precisely as described previously (2, 28). Briefly, 100mg dissected liver tissue was homogenized on ice in 2 mL 2:1 chloroform:methanol (both obtained from Sigma-Aldrich, St. Louis, MO). Residual tissue debris was pelleted by centrifugation at 16,200 RCF and (20 μL supernatant was dried in chloroform-resistant microcentrifuge tubes (Thermo Fisher, Grand Island, NY) prior to addition of 200 μL Infinity TG assay reagent (1 hour at room temperature) and 490 nm optical density quantification by 96-well format plate reader (Bio-Tek, Winooski, VT). 5mM glycerol standard solutions were treated identically in parallel to obtain absolute TG quantification.
For plasma biochemical analysis, mice were fasted a minimum of 4 hours prior to killing. Venous blood was sampled by puncturing the facial vein at the time of killing, and plasma was separated from whole blood by centrifugation at 16,200 RCF in plasma separator tubes (Becton Dickinson, Franklin Lakes, NJ). Triglycerides, cholesterol and free fatty acids were quantified using the Infinity TG Assay Kit, Infinity Cholesterol Assay Kit (Thermo Scientific, Grand Island, NY) or the NEFA-HR(2) Fatty Acid Assay Kit (Wako Diagnostics, Richmond, VA), respectively, per manufacturer specification.
In Vitro Triglyceride accumulation assays
For in vitro biochemical analysis, primary cultured hepatocytes were grown on 10 cm-diameter culture plates and equilibrated 24 hr prior to transfection (adenoviral transfection time was 36 hrs) with or without addition of 5mM fructose or 100mM trehalose in regular growth medium for 48 hrs. Cultures were dissociated from the culture dishes by trypsinization. Pelleted cultures were washed and 10% of the resuspended pellet was removed for protein quantitation and normalization. Lipids were then extracted from the remaining cell pellet by homogenization in 2:1 chloroform:methanol. 67% of the homogenate was dried at room temperature and triglycerides were assayed using the Infinity Triglyceride Assay Reagent (Thermo Scientific, Grand Island, NY).
For the BSA-conjugated fatty acid experiments, 500 mM free fatty acid stock was conjugated to BSA by sonication in a 4:1 molar ratio in fatty-acid-free BSA in regular growth media. 500 μM mixed fatty acid mixture was comprised of palmitic acid, stearic acid, linoleic acid, and oleic acid (all obtained from Sigma Aldrich), based upon physiological circulating fatty acid constituents present in human patients (37). The final media contained 60 μM fatty acid-free BSA (Sigma-Aldrich, St. Louis, MO). Control cultures for the fatty acid exposure experiments were treated with 60 μM fatty acid-free BSA alone.
Acknowledgments
We thank Herbert Virgin, Washington University School of Medicine, for the ATG16L1HM mouse line.
Funding sources: BJD is a Scholar of the Washington University Child Health Research Center (K12HD076224), and of the Children’s Discovery Institute (MI-FR-2014-426). This work was also supported by grants from the Robert Wood Johnson Foundation, the AGA-Gilead Sciences Research Scholar Award in Liver Disease, the Pediatric Scientist Development Program (K12HD000850-29), the Nutrition & Obesity Research Center (P30DK056341), the Washington University Digestive Disease Research Core Center (P30DK52574) (to BJD and NOD, specifically the Administration and Resource Access Core, the Advanced Imaging and Tissue Analysis Core, and the Murine Models core) HL-38180 and DK-56260 (NOD), R01-DK078187 (BNF), American Heart Association Grant 14PRE18320006 (TEK), the Washington University Spencer T. Olin Fellowship (ALM), the Washington University NIGMS Institutional Training Grant in Cell and Molecular Biosciences (T32GM007067 to ALM), National Science Foundation Graduate Student Fellowship (DGE-1143954), and the Washington University Biomedical Mass Spectrometry Research Facility (P41 GM103422, P30 DK020579).
Footnotes
Data and materials availability: No restrictions on materials used in the current study.
Competing interests: KHM is on the Scientific Board of Advisors for OvaScience.
Author contributions: Designed the experiments: BJD, MRH, CBH, ALM, ZC, MC, TEK, EPN, BNF, NOD, KEY, PWH, KHM. Performed the experiments: BJD, MRH, CBH, ALM, JRC, MC, ZC, EPN, TEK. Analyzed the data: BJD, MH, CBH, ALM, ZC, MC, TEK, EPN, BNF, KEY, PWH, KHM. Wrote the manuscript: BJD. Edited and revised the manuscript: BJD, MRH, CBH, ALM, ZC, TEK, BNF, NOD, KEY, PWH, KHM.
REFERENCES AND NOTES
Full text links
Read article at publisher's site: https://doi.org/10.1126/scisignal.aac5472
Read article for free, from open access legal sources, via Unpaywall:
https://stke.sciencemag.org/content/sigtrans/9/416/ra21.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1126/scisignal.aac5472
Article citations
Oral Trehalose Intake Modulates the Microbiota-Gut-Brain Axis and Is Neuroprotective in a Synucleinopathy Mouse Model.
Nutrients, 16(19):3309, 30 Sep 2024
Cited by: 0 articles | PMID: 39408276 | PMCID: PMC11478413
Hepatocyte MMP14 mediates liver and inter-organ inflammatory responses to diet-induced liver injury.
PNAS Nexus, 3(9):pgae357, 30 Aug 2024
Cited by: 0 articles | PMID: 39282008 | PMCID: PMC11393575
Autophagy and hepatic lipid metabolism: mechanistic insight and therapeutic potential for MASLD.
NPJ Metab Health Dis, 2(1):19, 02 Aug 2024
Cited by: 0 articles | PMID: 39100919 | PMCID: PMC11296953
Review Free full text in Europe PMC
Autophagy in Disease Onset and Progression.
Aging Dis, 15(4):1646-1671, 01 Aug 2024
Cited by: 4 articles | PMID: 37962467 | PMCID: PMC11272186
Review Free full text in Europe PMC
Effect of trehalose on mortality and disease severity in ICU-admitted patients: Protocol for a triple-blind, randomized, placebo-controlled clinical trial.
Contemp Clin Trials Commun, 40:101324, 18 Jun 2024
Cited by: 0 articles | PMID: 39021672 | PMCID: PMC11252791
Go to all (155) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (1 citation) ENA - EDL30474
- (1 citation) ENA - BAA95353
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Mystery solved: Trehalose kickstarts autophagy by blocking glucose transport.
Sci Signal, 9(416):fs2, 23 Feb 2016
Cited by: 55 articles | PMID: 26905424
Review
SLC2A8 (GLUT8) is a mammalian trehalose transporter required for trehalose-induced autophagy.
Sci Rep, 6:38586, 06 Dec 2016
Cited by: 62 articles | PMID: 27922102 | PMCID: PMC5138640
TFEB-dependent induction of thermogenesis by the hepatocyte SLC2A inhibitor trehalose.
Autophagy, 14(11):1959-1975, 06 Aug 2018
Cited by: 22 articles | PMID: 29996716 | PMCID: PMC6152536
Metformin alleviates hepatosteatosis by restoring SIRT1-mediated autophagy induction via an AMP-activated protein kinase-independent pathway.
Autophagy, 11(1):46-59, 01 Jan 2015
Cited by: 168 articles | PMID: 25484077 | PMCID: PMC4502778
Funding
Funders who supported this work.
American Gastroenterological Association
American Heart Association (1)
Grant ID: 14PRE18320006
Children's Discovery Institute (1)
Grant ID: MI-FR-2014-426
Gilead Sciences Research Scholar Award in Liver Disease
Institutional Training Grant in Cell and Molecular Biosciences (1)
Grant ID: T32GM007067
NHLBI NIH HHS (3)
Grant ID: R37 HL038180
Grant ID: HL-38180
Grant ID: R01 HL038180
NICHD NIH HHS (4)
Grant ID: K12 HD076224
Grant ID: K12HD076224
Grant ID: K12 HD000850
Grant ID: K12HD000850-29
NIDDK NIH HHS (10)
Grant ID: R01 DK056260
Grant ID: DK-56260
Grant ID: R01 DK078187
Grant ID: R01-DK078187
Grant ID: P30 DK020579
Grant ID: P30 DK052574
Grant ID: P60 DK020579
Grant ID: P30 DK056341
Grant ID: P30DK056341
Grant ID: P30DK52574
NIGMS NIH HHS (3)
Grant ID: P41 GM103422
Grant ID: T32 GM007067
Grant ID: T32GM007067
NSF Graduate Student Fellowship (1)
Grant ID: DGE-1143954
Nutrition Obesity Research Center (1)
Grant ID: P30DK056341
Pediatric Scientist Development Program (1)
Grant ID: K12HD000850-29
Robert Wood Johnson Foundation
Washington University Biomedical Mass Spectrometry Research Facility (2)
Grant ID: P41 GM103422
Grant ID: P30 DK020579
Washington University Child Health Research Center (1)
Grant ID: K12HD076224
Washington University Digestive Disease Research Core Center (1)
Grant ID: P30DK52574
 1
1 




