Abstract
Free full text

Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms
Abstract
A process of global importance in carbon cycling is the remineralization of algae biomass by heterotrophic bacteria, most notably during massive marine algae blooms. Such blooms can trigger secondary blooms of planktonic bacteria that consist of swift successions of distinct bacterial clades, most prominently members of the Flavobacteriia, Gammaproteobacteria and the alphaproteobacterial Roseobacter clade. We investigated such successions during spring phytoplankton blooms in the southern North Sea (German Bight) for four consecutive years. Dense sampling and high-resolution taxonomic analyses allowed the detection of recurring patterns down to the genus level. Metagenome analyses also revealed recurrent patterns at the functional level, in particular with respect to algal polysaccharide degradation genes. We, therefore, hypothesize that even though there is substantial inter-annual variation between spring phytoplankton blooms, the accompanying succession of bacterial clades is largely governed by deterministic principles such as substrate-induced forcing.
eLife digest
Small algae in the world's oceans remove about as much carbon dioxide from the atmosphere as land plants. These algae do not grow continuously, but often surge in numbers during temporary blooms. Such blooms can be large enough to be seen from space by satellites. The lifespan of algae within such blooms is short, and when they die, marine bacteria feed on the remnants, which releases much of the stored carbon dioxide.
Much of an algal cell consists of different types of polysaccharides. These large molecules are essentially made from sugars linked together. Polysaccharides are varied molecules and can contain many different sugars that can be linked in a number of different ways. During algae blooms bacteria proliferate that are specialized in the degradation of these polysaccharides. In 2012, researchers reported how over the progression of an algae bloom different groups of marine bacteria bloomed in rapid succession. However, it remained unknown whether the same or different groups of bacteria respond to algae blooms at the same place from year to year, and whether or not these bacteria use the same enzymes to degrade the polysaccharides.
Teeling, Fuchs et al. – who include many of the researchers from the 2012 study – now report on the analysis of a series of algae blooms that occurred in the southern North Sea between 2009 and 2012. The analysis is based on samples collected every week during the spring seasons, and shows that certain groups of related bacteria, known as clades, became common during each bloom. Teeling, Fuchs et al. also found indications that the clades that repeatedly occurred had similar sets of genes for degrading algal polysaccharides, but that the sets were different between the clades.
These data suggest that there is a specialized bacterial community that together can degrade the complex mixture of algal polysaccharides during blooms. This community reappears each year with an unexpectedly low level of variation. Since different species of algae made up the blooms in each year, this finding suggests that the major polysaccharides in these algae are similar or even identical.
Future work will focus on the specific activities of bacterial enzymes that are needed to degrade polysaccharides during algae blooms. Study of these enzymes in the laboratory will help to resolve, which polysaccharides are attacked in which manner, and to ultimately help to identify the most abundant algal polysaccharides. This will improve our current understanding of the carbon cycle in the world’s oceans.
Introduction
Pelagic zones of the world's oceans seemingly constitute rather homogenous habitats, however, they feature enough spatial and temporal variation to support a large number of species with distinct niches. This phenomenon has been termed 'paradox of the plankton' by G. Evelyn Hutchinson (Hutchinson, 1961). Interactions within planktonic microbial communities are manifold and complex (see Amin et al., (2012) and Worden et al. (2015) for reviews). Still, planktonic microbial communities are simple in comparison to benthic or terrestrial soil communities and thus particularly suitable for the study of microbial community composition dynamics. In recent years, continuous biodiversity studies at long-term sampling stations have started to reveal discernible deterministic patterns within marine microbial plankton communities (see Fuhrman et al. (2015) for a recent review). This is particularly true for less dynamic oligotrophic oceanic regions that are dominated by the members of the alphaproteobacterial Pelagibacteriaceae (SAR11 clade) and the cyanobacterial Prochlorococcaceae (Prochlorococcus marinus). By contrast, more dynamic eutrophic coastal regions are subject to frequent system perturbations and thus seldom in a state of equilibrium. This can lead to apparently stochastic changes in bacterioplankton community composition. To capture recurrence of biodiversity patterns in such coastal areas, sampling must occur at the order of weekly to sub-weekly time scales over multiple years. Owing to the lack of such intensively sampled long-term time series data, our current understanding of the extent and predictability of recurring microbial biodiversity patterns for such marine habitats is still limited.
A particularly important connection in the marine carbon cycle exists between marine microalgae as primary producers and heterotrophic bacteria that feed on algal biomass. Global photosynthetic carbon fixation is estimated to exceed 100 Gigatons yearly, of which marine algae contribute about half (Falkowski et al., 1998; Field et al., 1998; Sarmento and Gasol, 2012). Planktonic uni- to pluricellular algae such as diatoms, haptophytes, and autotrophic dinoflagellates are the most important marine primary producers. Diatoms alone are estimated to contribute 20–40% to global carbon fixation (Nelson et al., 1995; Mann, 1999; Armbrust, 2009).
Primary production by planktonic microalgae differs from primary production by sessile macroalgae or land plants as it is much less constant, but culminates in blooms that are often massive, as occurs worldwide during spring blooms from temperate to polar regions. These blooms are highly dynamic phenomena that are time-limited by nutrients, predator grazing and viral infections. Bloom termination results in a short-lived massive release of algal organic matter that is consumed by dedicated clades of heterotrophic bacterioplankton. This trophic connection leads to synchronized blooms of planktonic bacteria during phytoplankton blooms, as has been described in various studies (Bell and Kuparinen, 1984; Niu et al., 2011; Tada et al., 2011; Teeling et al., 2012; Yang et al., 2015; Tan et al., 2015).
The activities of these heterotrophic bacteria impact the proportion of algal biomass that is directly mineralized and released back into the atmosphere mostly as carbon dioxide, and the algae-derived biomass that sinks out to the bottom of the sea as carbonaceous particles. These are further remineralized by particle-associated bacteria while sinking and by benthic bacteria when reaching the sediment, even in the deep sea (e.g. Ruff et al., 2014). The remainder is buried for a long time as kerogen and forms the basis for future oil and gas reservoirs. The ratio between bacterial mineralization and burial of algae-derived organic matter thus has a profound influence on the atmospheric carbon dioxide concentration (Falkowski et al., 1998). However, the bulk of bacteria during phytoplankton blooms are free-living and not attached to particles or algae. These bacteria play a pivotal role in the mineralization of algae-derived non-particulate dissolved organic matter (DOM).
The bacterial clades that respond most to phytoplankton blooms belong to the classes Flavobacteriia (phylum Bacteroidetes) and Gammaproteobacteria, and the Roseobacter clade within class Alphaproteobacteria (Buchan et al., 2014). This response is typically not uniform, but consists of a series of distinct clades that bloom one after another. In the year 2009, we investigated the response of bacterioplankton to a diatom-dominated spring phytoplankton bloom in the German Bight (Teeling et al., 2012). Within the free-living bacteria (0.2 to 3 µm) we observed a swift succession of bacterial clades that were dominated by Flavobacteriia and Gammaproteobacteria, with consecutively blooming Ulvibacter (Flavobacteriia), Formosa (Flavobacteriia), Reinekea (Gammaproteobacteria), Polaribacter (Flavobacteriia) genera and SAR92 (Gammaproteobacteria) as prominent clades.
Using time-series metagenome and metaproteome analyses, we demonstrated that the substrate-spectra of some of these clades were notably distinct. The succession of bacterioplankton clades hence constituted a succession of distinct gene function repertoires, which suggests that changes in substrate availability over the course of the bloom were among the forces that shaped the bacterioplankton community. Dominance of bottom-up over top-down control is assumed to be characteristic for the initial phases of spring phytoplankton blooms. After winter, inorganic nutrients are aplenty, and the overall abundance of microbes is low. When suitable temperature and sunlight conditions are met in spring, algae and subsequently bacteria can enter an almost unrestricted proliferation. In contrast, predators such as flagellates, protists and zooplankton can only start proliferating when their food sources are available in larger numbers. Hence, top-down control by predation sets in only during later bloom phases. This situation is distinct from summer and fall phytoplankton blooms.
Pronounced differences between blooming clades were found in the gene frequencies and protein expression profiles of transporters and carbohydrate-active enzymes (CAZymes; [Cantarel et al., 2009; Lombard et al., 2014]), such as glycoside hydrolase (GH), polysaccharide lyase (PL), carbohydrate esterase (CE), or carbohydrate-binding module (CBM) containing genes. The latter indicates a pronounced niche partitioning with respect to algal polysaccharide degradation. Marine algae produce large quantities of distinct polysaccharides, for example storage, cell matrix and cell wall constituents, or as part of extracellular transparent exopolymer particles (TEP). It has been recently shown that in particular Flavobacteriales and Rhodobacterales respond to TEP availability (Taylor et al., 2014). The diversity of algal polysaccharides is too high for a single bacterial species to harbor all the genes required for the complete degradation of all naturally occurring variants. Thus, polysaccharide-degrading bacteria specialize on dedicated subsets of polysaccharides, which is why the decomposition of algal polysaccharides during and after algal blooms is a concerted effort among distinct bacterial clades with distinct glycan niches (e.g. Xing et al., 2015).
In this study, we provide evidence that the succession of bacterioplankton clades that we reported for the 2009 North Sea spring phytoplankton bloom re-occurred during the spring blooms from 2010 to 2012. We tested whether the bacterioplankton clades and their associated CAZyme repertoires differ from year to year or exhibit recurrent patterns. We analyzed spring bacterioplankton community composition via 16S rRNA catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) and 16S rRNA gene tag sequencing, as well as gene function repertoires by deep metagenome sequencing. Our efforts have culminated into the as of yet highest resolved dataset capturing the response of planktonic bacteria to marine spring phytoplankton blooms and have allowed identification of recurring patterns that might ultimately lead to an explanatory model for bacterioplankton succession dynamics during spring algae blooms.
Results
Sampling site characteristics
The samples were taken at Helgoland Island about 40 km offshore in the southeastern North Sea in the German Bight at the station 'Kabeltonne' (54° 11.3' N, 7° 54.0' E; Figure 1) between the main island and the minor island, Düne (German for 'dune'). Water depths at this site fluctuate from 6 to 10 m over the tidal cycle. During most of the year, a westerly current transports water from the English Channel alongside the Dutch and Frisian coast to Helgoland, but water around the island is also influenced by nutrient inputs from the rivers Weser and Elbe and from the northern North Sea (Wiltshire et al., 2010). During the 2009 to 2012 study period, the lowest water temperatures were measured in mid to late February (min. 2010: 1.1°C; max. 2009: 3.4°C), followed by a continuous increase until a peak in August (min. 2011: 18.0°C; max. 2009: 18.7°C) (Supplementary file 1).
m over the tidal cycle. During most of the year, a westerly current transports water from the English Channel alongside the Dutch and Frisian coast to Helgoland, but water around the island is also influenced by nutrient inputs from the rivers Weser and Elbe and from the northern North Sea (Wiltshire et al., 2010). During the 2009 to 2012 study period, the lowest water temperatures were measured in mid to late February (min. 2010: 1.1°C; max. 2009: 3.4°C), followed by a continuous increase until a peak in August (min. 2011: 18.0°C; max. 2009: 18.7°C) (Supplementary file 1).
Phytoplankton - diversity and bloom characteristics
Spring phytoplankton blooms in the North Sea typically develop during March and reach highest intensities during April and May. The highest chlorophyll a concentrations are usually observed at the coastlines including the area around Helgoland Island (Figure 2). North Sea spring blooms are thus large-scale phenomena that are, however, influenced by local conditions, such as riverine inputs. At Helgoland island, spring phytoplankton blooms started around mid March when water temperatures surpassed 3 to 5°C (Figure 3A–H; Supplementary file 1). The diatoms Chaetoceros debilis and Chaetoceros minimus, Mediopyxis helysia, Rhizosolenia styliformis and Thalassiosira nordenskioeldii, the silicoflagellate Chattonella, the haptophyte Phaeocystis and dinoflagellates dominated these blooms in terms of cell numbers (Figure 3I–L; Supplementary file 2). Relative abundances of these algae varied in no apparent order during the observed blooms, and we have yet to understand the factors that determine these variations. The sizes of the dominant algae taxa are different, with Chaetoceros minimus and in particular Phaeocystis spp. featuring the smallest cells and Mediopyxis helysia and Rhizosolenia styliformis featuring the largest cells. Spherical Phaeocystis spp. cells for example have estimated biovolumes of ~50 to 250 µm3, whereas elipsoid cylindrical Mediopyxis helysia cells have a biovolume of ~82,000 µm3 and for Rhizosolenia styliformis even a biovolume of ~282,000 µm3 has been reported (Olenina, 2006; Loebl et al., 2013). Considering biomass, the blooms were largely dominated by the diatoms T. nordenskioeldii and M. helysia and the silicoflagellate Chattonella. Blooms of these three algae were bimodal in all years with dominance of first T. nordenskioeldii followed by Chattonella in 2009 (Figure 3I), T. nordenskioeldii followed by M. helysia in 2010 (Figure 3J), M. helysia followed by Chattonella in 2011 (Figure 3K) and a pronounced bimodal bloom of Chattonella species in 2012 (Figure 3L). All blooms were accompanied by a notable decrease of silicate (Figure 3A–D; Supplementary file 1), which diatoms use for frustule formation (see Yool and Tyrrell (2003) for the controlling effect of silicate on diatom abundance).

Data are shown for the southern North Sea for the months February to May (monthly averages) of the years 2009 to 2012. Images were retrieved from the GlobColour website using the ‘extended Europe area’ at full resolution (1 km) as merged products of weighted averages from the following sensors: MERIS, MODIS AQUA, SeaWIFS and VIIRS. See GlobColour website for details (http://hermes.acri.fr). The position of Helgoland Island is indicated by a blue dot.
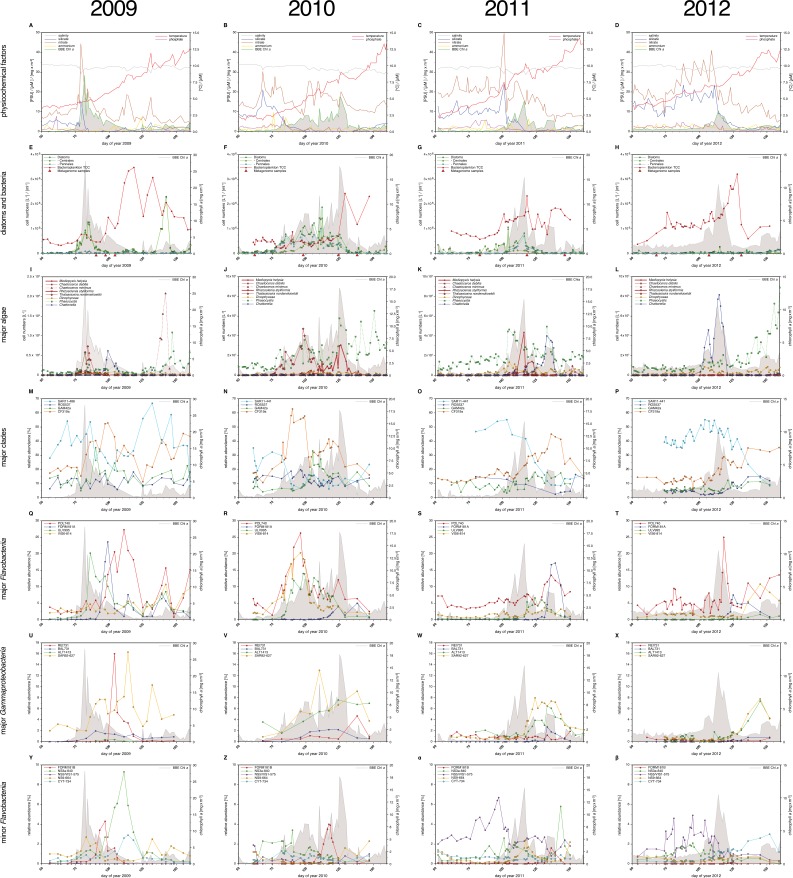
Sampling: Surface seawater samples were taken at the North Sea island Helgoland between the main island and the minor island 'Düne' (station 'Kabeltonne', 54°11'03''N, 7°54'00''E) using small research vessels (http://www.awi.de/en/expedition/ships/more-ships.html) and processed in the laboratory of the Biological Station Helgoland within less than two hours after sampling. Cells for microscopic visualization methods were first fixed by the addition of formaldehyde to sampled seawater, which was then filtered directly onto 0.2 µm pore sized filters. Physicochemical and phytoplankton data: Physicochemical parameters and phytoplankton data were assessed in subsurface water on a weekday basis as part of the Helgoland Roads LTER time series as described in Teeling et al. (2012). The Helgoland Roads time series is accessible via the public database Pangaea (http://www.pangaea.de) and can be used to assess long-term changes of the North Sea pelagic ecosystem. Left-hand side legends correspond to ordinates on the left, and right-hand side legends to ordinates on the right. A-D: Physicochemical measurements including measurements of BBE Chl a (chlorophyll a fluorescence by algal group analyzer sensor). Left ordinate: salinity [PSU], silicate [µM], nitrate [µM], ammonium [µM] and chlorophyll a [mg/m3]; right ordinate: temperature [°C] and phosphate [µM]. E-H: Counts of the diatom groups. I-L: Microscopic cell counts of the most abundant algae genera (red: diatoms; orange: dinoflagellates: green: haptophytes; blue: silicoflagellates). Algae with large cells and thus large biovolumes are depicted by bold solid lines and algae with small cells are represented by dotted lines. Rhizosolenia styliformis and Mediopyxis helysia feature large cells, whereas Chaetoceros minimus and in particular Phaeocystis species feature small cells. The latter typically reaches lengths of below 10 µm and Phaeocystis spp. biovolumes therefore typically are hundreds to thousands fold smaller than those of R. styliformis and M. helyisa cells (Olenina, 2006; Loebl et al., 2013). Physicochemical data are summarized in Supplementary file 1, and data on the major phytoplankton clades in Supplementary file 2. Total cell counts and CARD-FISH of bacterioplankton: E-H: TCC (total cell counts); red triangles depict sampling of metagenomes. M-β: Recurrent bacterioplankton clades as assessed by CARD-FISH (Catalyzed Reporter Deposition-Fluorescence in situ Hybridization) with the following probes: M-P (major bacterial groups): SAR11-486 and SAR11-441: alphaproteobacterial SAR11-clade; ROS537: alphaproteobacterial Roseobacter clade; GAM42a: Gammaproteobacteria; CF319a: Bacteroidetes. Q-T (major Flavobacteriia clades): POL740: genus Polaribacter; FORM181A: genus Formosa; ULV995: genus Ulvibacter; VIS6-814: genus-level clade VIS6 within the family Cryomorphaceae-Owenweeksia; U-X (major Gammaproteobacteria clades): REI731: genus Reinekea; BAL731: genus Balneatrix; ALT1413: families Alteromonadaceae and Colwelliaceae; SAR92-627: genus-level clade SAR92. Y-β (minor Bacteroidetes clades): FORM181B: species-specific for Formosa sp. Hel1_33_131; NS3a-840: NS3 marine group; NS5/VIS1-575: VIS1 genus-level clade within the NS5 marine group; NS9-664: NS9 marine group; CYT-734: Cytophagia clade Marinoscillum. Total and CARD-FISH cell counts are summarized in Supplementary file 3 and the corresponding probes in Supplementary file 4.
Bloom maximum intensities decreased from 2009 to 2012 with chlorophyll a maxima (measured by fluorescence) reaching 28 mg m-3 (day 82), 18 mg m-3 (day 125), 15 mg m-3 (day 116), and 11 mg m-3 (day 114) in each respective year (Figure 3A–D; Supplementary file 1). Maximum bacterioplankton cell counts were observed either close to or after the chlorophyll maxima (Supplementary file 3). In 2009, the bacterioplankton peaked at 3.5x106 cells ml-1 (day 118), 36 days after the Chl a maximum. In 2010, the peak in Chl a was broader and only four days ahead of the bacterioplankton peak abundance of 2.4x106 cells ml-1 (day 129). In 2011, the Chl a peak was only two days ahead of the bacterioplankton peak abundance of 2.3x106 cells ml-1 (day 118), whereas it was nine days ahead in 2012, where the bacterioplankton peaked at 3.2x106 cells ml-1 (day 123).
Bacterioplankton - diversity and bloom characteristics
DAPI and CARD-FISH cell staining (Supplementary files 3 and 4) showed that SAR11 dominated the bacterioplankton community in winter, but with the onset of each spring bloom relative abundances of Bacteroidetes followed by Gammaproteobacteria increased and finally surpassed those of the SAR11 (Figure 3M–P). Bacteroidetes reached higher maximum relative abundances than Gammaproteobacteria, 40% (2012) to 60% (2010) as compared to 10% (2012) to 30% (2010), respectively.
Bacteroidetes genera Polaribacter, Formosa, and VIS6, a genus-level clade within the family Cryomorphaceae (Gómez-Pereira et al., 2010), peaked each year with relative abundances well above 5%, reaching relative abundances of up to 25% sometimes within less than a week (Figure 3Q–T). Ulvibacter reached similar peak abundances with the exception of 2012, where this genus never surpassed 2% and ranged below 1% most of the time. Within Gammaproteobacteria the genus-level SAR92 clade responded notably in all years increasing from background levels below 1% to relative abundance of 8% to 16%. Members of the Alteromonadales families Alteromonadaceae and Colwelliaceae bloomed in three (2010: 8%; 2011: 6%; 2012: 7%), and genus Reinekea in two (2009: 16%; 2010: 5%) of the years (Figure 3U–X). Some less abundant, but nevertheless recurrent taxa included the genus Balneatrix within Gammaproteobacteria with relative abundances up to 2% (Figure 3U–X). Minor recurring groups of Bacteroidetes (Figure 3Y-β) included the NS3a marine group (9% in 2009 and 6% in 2011), the genus-level VIS1 clade within the NS5 marine group detected before the Chl a peaks of 2011 (7%) and 2012 (5%), and the Cytophagia clade Marinoscillum that reached 1–3% abundance most years after initial blooms.
We used complementary 16S rRNA gene tag sequencing for the detection of bacterioplankton clades that were not recovered by CARD-FISH probes (Supplementary file 5). Relative proportions of 16S tags from distinct clades correlated for the most part those inferred from CARD-FISH cell counts (Figure 4A–P), but members of SAR11 were substantially underreported - a known limitation of the 806R primer used in V4 amplification for 2010 to 2012 samples (Apprill et al., 2015). Additional abundant clades detected in the 16S amplicon data comprised the Flavobacteriia genus Tenacibaculum (Figure 4U–X) that bloomed in 2010 (read frequencies of max. ~12%) and 2011 (max. ~5%). Within Gammaproteobacteria, clades with read frequencies ≥5% in at least one year comprised the genera Aeromonas, Glaciecola, Pseudoalteromonas, Pseudomonas, Psychrobacter and the SAR86 and ZD0405 clades (Figure 4Q–T). Within the alphaproteobacterial Rhodobacteriaceae, high abundances of 'Candidatus Planktomarina temperata' (DC5-80-3 lineage) and the NAC11-7 clade were detected reaching 6–21% and 7–19% of the tag data, respectively (Figure 4U–X). Also within Alphaproteobacteria the genus Sulfitobacter peaked with a read frequency of ~7% in 2010 (Figure 4V), and within Betaproteobacteria the order Methylophilales (dominated by OM43 clade members) was detected with high relative abundances of up to ~10% before blooms, which decreased with bloom progression. Verrucomicrobia (Figure 4U–X) were detected with decreasing peak read frequencies of 7.7% (2010), 5.0% (2011) and 2.9% (2012). This decrease corresponds to decreasing bloom intensities, which supports a proposed role of Verrucomicrobia in polysaccharide decomposition (e.g. Martinez-Garcia et al., 2012).
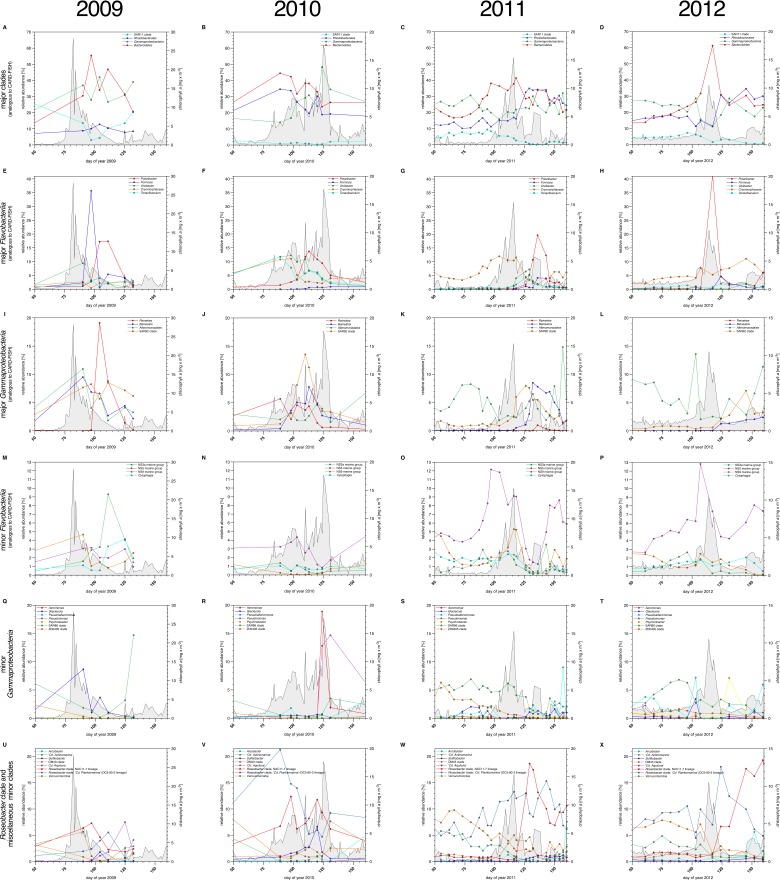
Sampling: Surface seawater samples were taken at the North Sea island Helgoland between the main island and the minor island 'Düne' (station 'Kabeltonne', 54°11'03''N, 7°54'00''E) using small research vessels (http://www.awi.de/en/expedition/ships/more-ships.html) and processed in the laboratory of the Biological Station Helgoland within less than two hours after sampling. Biomass of free-living bacteria for DNA extraction was harvested on 0.2 µm pore sized filters after pre-filtration with 10 µm and 3 µm pore sized filters to remove large debris and particle-associated bacteria. Biomass of the 0.2–3 µm bacterioplankton fraction was used for DNA extraction and subsequent 16S rRNA gene tag sequencing. 16S rRNA gene tag sequencing: A total of 142 samples were collected for the years 2010 to 2012. After DNA extraction, the V4 region of the 16S rRNA gene was amplified using the primers 515F (5' GTGCCAGCMGCCGCGGTAA 3') and 806R (5' GGACTACHVGGGTWTCTAAT 3') (Caporaso et al., 2011). Sequencing was carried out on an Illumina (San Diego, CA, USA) MiSeq sequencer with and 2x250 bp chemistry. This dataset was complemented by 16S rRNA gene tags from 7 samples from our initial study on the 2009 spring bloom (Teeling et al., 2012). DNA of these samples was amplified using the primers 314F (5’ CCTACGGGNGGCWGCAG 3') and 805R (5’ GACTACHVGGGTATCTAATCC 3') (Herlemann et al., 2011) and sequenced on the 454 FLX Ti platform. Data analysis: All tag data were analyzed using the SILVAngs pipeline with the SILVA (Quast et al., 2013) v119 database. The SAR92 clade was subsequently reclassified to comply with the recently released SILVA v123, where the SAR92 no longer belong to the order Alteromonadales. The corresponding abundance data is summarized in Supplementary file 5. Time points from days 50 to 160 were plotted for all years. Panel A-P depict data that are analogous to the CARD-FISH data presented in Figure 3, with addition of the Flavobacteriia genus Tenacibaculum (E-H). Panels Q-X show minor Gammaproteobacteria clades (Q-T) and Roseobacter clades together with miscellaneous other minor clades (U-X) that were not tested by CARD-FISH probes.
Within Bacteroidetes, Gammaproteobacteria and Rhodobacterales a total of eleven clades peaked during at least two of the four spring blooms with relative cell abundances or, for those clades that were not assessed by CARD-FISH, relative read frequencies ≥5%. These were six Flavobacteriia clades (Formosa, Polaribacter, NS3a marine group, Tenacibaculum, Ulvibacter, VIS6 clade Cryomorphaceae), three Gammaproteobacteria clades (Alteromonadaceae/Colwelliaceae, Reinekea and SAR92), and two Roseobacter clades (DC5-80-3 and NAC11-7).
Each year a succession was observed within the Flavobacteriia and Gammaproteobacteria clades. The succession in the Flavobacteriia was more pronounced than in the Gammaproteobacteria, but the sequence of clades varied. Spearman rank correlation analyses revealed that the abundances of the most prominent Flavobacteriia clades were for the most part correlated with multiple algae groups and physiochemical factors (Supplementary file 6). According to linear regression analyses, the strongest abiotic predictors were temperature, salinity, silicate and nitrate, and the strongest biotic predictors were Phaeocystis spp. haptophytes, Rhizosolenia spp., Chaetoceros debilis, and Chaetoceros minimus diatoms and the silicoflagellate Chattonella (Supplementary file 7). It should be noted though that linear regressions were computed based on log-transformed abundance data and not algae volumes (which were not measured). Thus, the influence of the rather small cell-sized algae such as Chaetoceros minimus is likely overestimated. Such limitations notwithstanding it is noteworthy that in no case a simple one-to-one relationship between specific algae and specific bacterioplankton groups was detected. The strongest significant (p<0.05) correlations were obtained for the Ulvibacter clade that was positively correlated with diatoms and haptophytes and negatively correlated with silicoflagellates. Further results comprised an opposite trend for the VIS1 clade of the NS5 marine group, and a correlation of Polaribacter and Chattonella abundances (see Supplementary file 6 for details).
Bacterioplankton - genetic repertoires
In total, 16 metagenomes of free-living bacterioplankton (0.2–3 µm) were generated from time points before, during and after spring phytoplankton blooms, six during 2009 using the 454 FLX Ti platform that were published previously (Teeling et al., 2012) and ten during 2010–2012 using the Illumina HiSeq2000 platform. Most of the 454 (0.5–4 pico titer plates / metagenome) and all of the Illumina metagenomes (1 lane / metagenome; 2x150 bp) were deeply sequenced (Supplementary file 8) with final assembled contigs of up to 96 kbp and 458 kbp, respectively.
Taxonomic classification of the metagenome contigs resulted in identification of major bloom-associated clade sequence bins (Supplementary file 9), including Formosa, Polaribacter, the NS3a and NS5 marine groups, and Cryomorphaceae of the Flavobacteriia and Alteromonadales, Reinekea, Glaciecola and the SAR92 clade of the Gammaproteobacteria. Classification was poor, however, for Ulvibacter (Flavobacteriia) and Balneatrix (Gammaproteobacteria), most likely since the only available reference genomes (unidentified eubacterium SCB49; Balneatrix alpica) were too distant from North Sea representatives. Clone libraries from 2009 (Teeling et al., 2012) indicated 16S rRNA similarities of only 94% and 91%, respectively, for these two clades. Other abundant clades comprised the betaproteobacterial Burkholderiales and Methylophilales (including the OM43 clade), the alphaproteobacterial SAR116 and Roseobacter NAC11-7 clade, and the gammaproteobacterial SAR86 and ZD0405 clades. Lower abundant clades comprised, amongst others, the OM60 (NOR5) group, the AEGEAN-169 group, and Sulfitobacter.
We plotted contig GC contents versus coverage to evaluate our taxonomic classification, which in some cases allowed to assess the coherence of some of the clades (Figure 5). For example, Reinekea (Figure 5D,H,I) and the NS5 marine group (Figure 5H,L,M,O) were mostly represented by distinct clusters, whereas Polaribacter (Figure 5D,H,I) was almost always represented by at least two clusters indicating the presence of sub-populations. In general, the number of clusters increased from pre-bloom to mid-bloom situations and decreased slightly towards late bloom situations and notably towards post-bloom situations. This tendency was more evident in 2009, the year with the highest bloom intensity and the largest number of metagenome samples spanning a broader timespan (Figure 5A–F). It is noteworthy that high in situ abundance did not always correlate with good metagenome assemblies. SAR11 for example, while highly abundant in all metagenome datasets, yielded few large contigs, possibly due to population heterogeneity and presence of hyper-variable regions described in sequenced SAR11 genomes (Wilhelm et al., 2007).
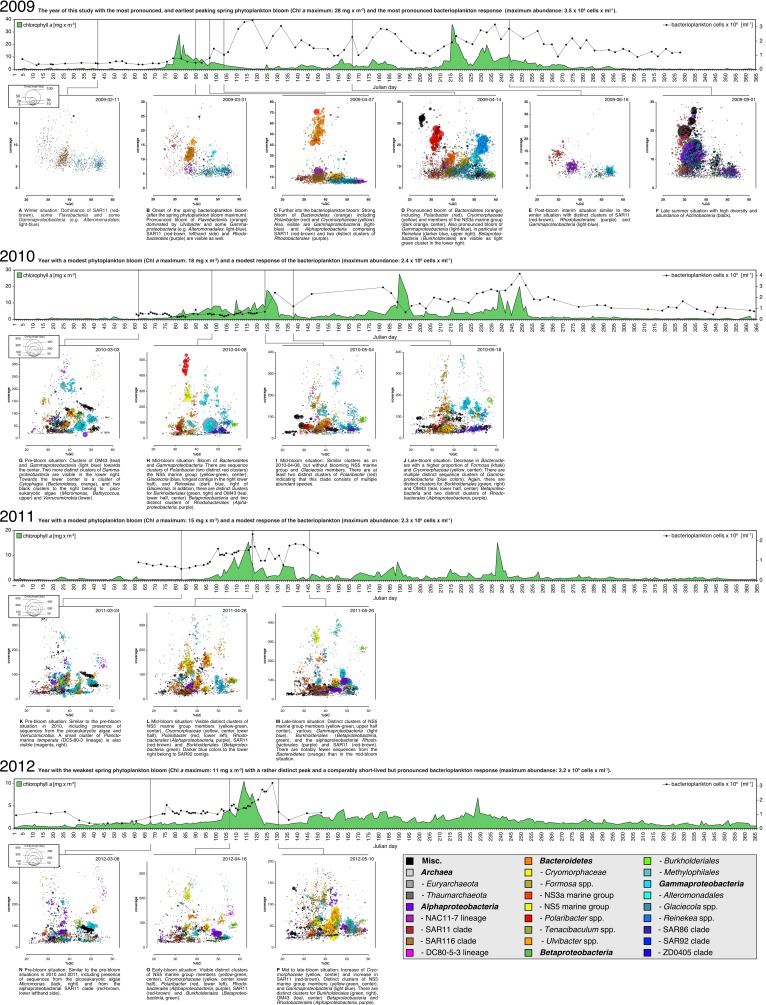
Sampling: Surface seawater samples were taken at the North Sea island Helgoland between the main island and the minor island 'Düne' (station 'Kabeltonne', 54°11.03' N, 7°54.00' E) and processed in the laboratory of the Biological Station Helgoland within less than two hours after sampling. Biomass of free-living bacteria was harvested on 0.2 µm pore sized filters after pre-filtration with 10 µm and 3 µm pore sized filters to remove large debris and particle-associated bacteria. Sequencing: Community DNA was extracted and sequenced; 2009 samples were sequenced on the 454 FLX Ti platform, and 2010-2012 samples on the Illumina HiSeq2000 platform (16 metagenomes in total). Reads were assembled using Newbler (2009) or a combination of SOAPdenovo and Newbler (2010-2012) and the resulting contigs were taxonomically classified (Supplementary file 9). Visualization: The resulting metagenome contigs are visualized as bubbles with radii that are proportional to their lengths and colors that indicate their predicted taxomomic affiliations. These bubbles are drawn in planes that are defined by the contig's GC contents and coverage values. Colors are restricted to selected abundant taxa (see legend below) to highlight distinct clusters, mostly from the Bacteroidetes, Alphaproteobacteria, Betaproteobacteria and Gammaproteobacteria. Likewise only contigs are shown that exceed a minimum length of 2750 bp for pyrosequencing data (2009) and 15,000 bp for llumina data (2010-2012), respectively. Sparse contigs with very high coverage or GC contents below 20% or above 60% were also excluded from visualizations. The 16 metagenomes are shown arranged in order on yearly timescales that depict chlorophyll a contents as proxies for phytoplankton abundance. Metagenome sizes*: 2009-02-11: 49.1 Mbp / 2009-03-31: 44.9 Mbp / 2009-04-07: 52.7 Mbp / 2009-04-14: 96.0 Mbp / 2009-06-16: 29.8 Mbp / 2009-09-01: 79.2 Mbp 2010-03-03: 537.3 Mbp / 2010-04-08: 325.8 Mbp / 2010-05-04: 453.0 Mbp / 2010-05-18: 512.3 Mbp 2011-03-24: 629.1 Mbp / 2011-04-28: 541.8 Mbp / 2011-05-26: 604.0 Mbp 2012-03-08: 574.0 Mbp / 2012-04-16: 543.9 Mbp / 2012-05-10: 614.1 Mbp *sums of assembled bases
Analyses of functional genes identified in the bacterioplankton metagenomes revealed that increases in Flavobacteriia relative abundance during blooms was always accompanied by an increase in community-wide CAZyme gene frequency as well as an increase in the diversity of CAZyme families (Figure 6A–C). As blooms subsided, CAZyme frequencies also declined. This was most pronounced for more densely sampled years in 2009 and 2010. In 2012 the decline in CAZymes was not captured as the last metagenome sample was taken before the bloom decline ( Figure 6A, x-axis).
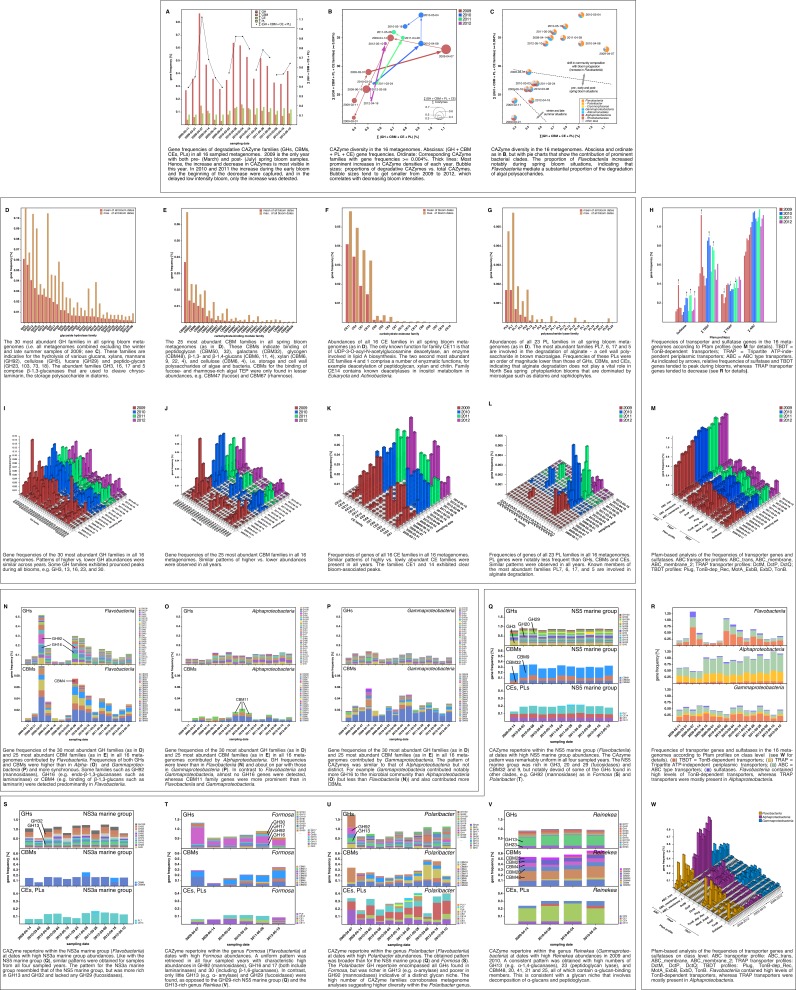
Sampling: Surface seawater samples were taken at the North Sea island Helgoland between the main island and the minor island Düne' (station 'Kabeltonne', 54°11'03''N, 7°54'00''E) and processed in the laboratory of the Biological Station Helgoland within less than two hours after sampling. Biomass of free-living bacteria was harvested on 0.2 µm pore sized filters after pre-filtration with 10 µm and 3 µm pore sized filters to remove large debris and particle-associated bacteria. Sequencing: Community DNA was extracted and sequenced. 2009 samples were sequenced on the 454 FLX Ti platform, and 2010–2012 samples on the Illumina HiSeq2000 platform (16 metagenomes in total). Reads were assembled using Newbler (2009) or a combination of SOAPdenovo and Newbler (2010–2012) and the resulting contigs were taxonomically classified (Supplementary file 9). Metagenome sizes*: 2009-02-11: 49.1 Mbp / 2009-03-31: 44.9 Mbp / 2009-04-07: 52.7 Mbp / 2009-04-14: 96.0 Mbp / 2009-06-16: 29.8 Mbp / 2009-09-01: 79.2 Mbp 2010-03-03: 537.3 Mbp / 2010-04-08: 325.8 Mbp / 2010-05-04: 453.0 Mbp / 2010-05-18: 512.3 Mbp 2011-03-24: 629.1 Mbp / 2011-04-28: 541.8 Mbp / 2011-05-26: 604.0 Mbp 2012-03-08: 574.0 Mbp / 2012-04-16: 543.9 Mbp / 2012-05-10: 614.1 Mbp *sums of assembled bases Data Analysis: CAZymes were predicted as consensus of searches against the CAZy, dbCAN and Pfam databases with custom E-value cutoffs (Supplementary file 11). Sulfatase and transporter genes were predicted based on HMMER searches against the Pfam databases with an E-value cutoff of E-5. Gene frequencies were computed as [(sum of average coverage of target genes) *100 / (sum of average coverage of all genes)]. All dates in the graphs are in the format [yyyy-mm-dd].
Figure 6—figure supplement 1.
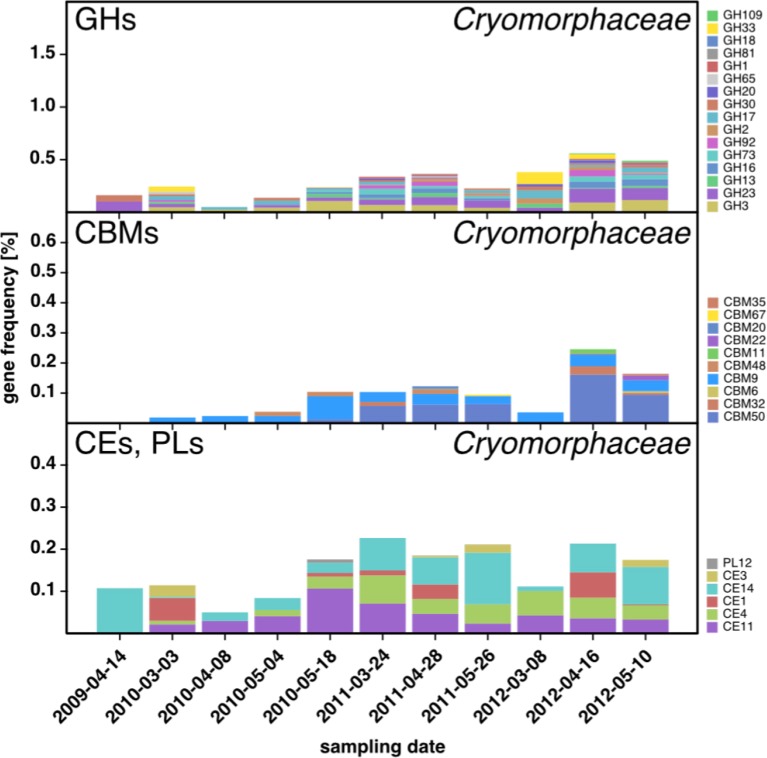
GH and CBM frequencies were low compared to those of other abundant Flavobacteriia clades (see Figure 6Q,S,T,U), indicating that these Cryomorphaceae might have a distinct ecophysiological niche in which polysaccharide degradation plays a lesser role.
Figure 6—figure supplement 2.
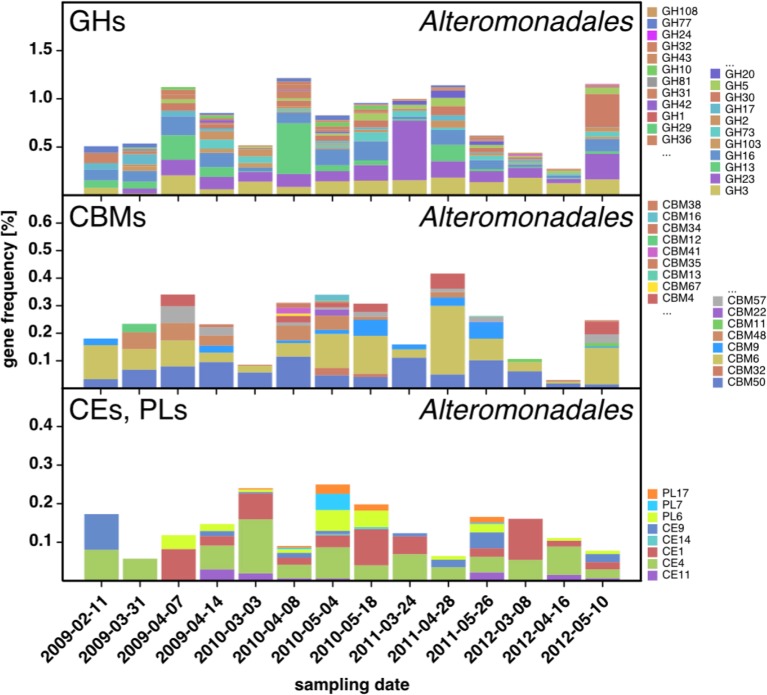
As expected at the taxomomic level of order, the CAZyme repertoire was diverse. Nonetheless, the obtained pattern was relatively consistent across all four studied years and at some dates notably enriched in CAZymes that play a role during phytoplankton blooms such as GH13 and GH16
The 20 glycoside hydrolase families with the highest mean abundances during bloom dates were, in descending order, GH3, 23, 13, 16, 103, 73, 92, 2, 17, 30, 5, 20, 36, 65, 29, 1, 42, 31, 81, and 18 (Figure 6D). While most of these families comprise a diverse range of functions, they are indicative for the hydrolysis of certain glucans, xylans, mannans (GH92), cellulose (GH5), fucans (GH29), and peptidoglycan (GH23, 103, 73, and 18). The abundant families GH3, 16, 17 and 5 comprise β-1,3-glucanases and the family GH30 β-1,6-glucanases. Both enzymes are involved in the cleavage of chrysolaminarin. Chrysolaminarin, a mostly linear β-1,3-glucan with occasional β-1,6 branches, is the storage polysaccharide in diatoms and thus one of the most abundant polysaccharides on Earth.
The ten families of carbohydrate-binding modules with the highest mean abundances during blooms were, in descending order, CBM50, 32, 6, 9, 48, 11, 22, 57, 20, and 4 (Figure 6E). These CBM domains are indicative for the binding of peptidoglycan (CBM50), galactans (CBM32), glycogen (CBM48), β-1,3- and β-1,4-glucans (CBM6, 11, 4), xylan (CBM6, 9, 22, 4), and cellulose (CBM6, 4). This suggests a pronounced specialization of the bacterial community in the acquisition of storage polysaccharides (β-1,3-glucans such as chrysolaminarin; α-1,4-glucans such as starch and glycogen) and cell wall polysaccharides (xylose, cellulose and peptidoglycan) of both algae and bacteria. By contrast, CBMs for the binding of algal TEP, which consists predominantly of fucose- and rhamnose-rich anionic sulfated heteropolysaccharides (Passow, 2002), were only found in lower abundances, e.g. CBM47 (fucose) and CBM67 (rhamnose).
Among carbohydrate esterase families, CE11, 4, 1, and 14 exhibited the highest mean abundances during blooms (Figure 6F). The only known function for family CE11 is that of the UDP-3-O-acyl-N-acetylglucosamine deacetylase, an enzyme involved in lipid A biosynthesis. The second and third most abundant CE families 4 and 1 comprise a number of enzymatic functions including deacetylation of peptidoglycan, xylan, and chitin.
Finally, the polysaccharide lyase families PL6, 7, 17 and 5 constituted the most abundant PL families during bloom dates (Figure 6G). Known members of these PL families are all involved in the usage of alginate. However, gene frequencies of these PL families were an order of magnitude below those of abundant GHs, CBMs, and CEs, indicating that alginate degradation does not play a vital role in North Sea spring phytoplankton blooms. Alginate is a cell wall constituent in brown macroalgae, however, the microalgae that dominate North Sea spring blooms are devoid of alginate.
For all of these GH, CBM, PL, and CE families, we observed remarkably similar gene frequency patterns during all four spring blooms, often with peaks in the same families (Figure 6I–L). Alongside CAZymes, sulfatase and TonB-dependent transporter (TBDT) gene frequencies also increased during blooms while tripartite ATP-independent periplasmic (TRAP) transporter genes showed an almost opposite trend (Figure 6H,M). A class-level analysis revealed that sulfatases and TBDT genes were predominantly present in Bacteroidetes, whereas TRAP transporters were mostly present in Alphaproteobacteria (Figure 6R,W). The observed shifts in sulfatase, TBDT and TRAP transporter frequencies hence reflect the shifts in relative abundance between these two classes. This is in agreement with our study on the 2009 spring bloom (Teeling et al., 2012) and furthermore demonstrates recurrence of this phenomenon during four consecutive years.
Class-level analyses of the most abundant GH and CBM families (Figure 6N–P) showed that Flavobacteriia not only contributed more total CAZymes to the microbial community than Alpha- and Gammaproteobacteria, but also exhibited a tighter coupling between GH and CBM genes with highly similar abundance profiles (Figure 6N). The distribution of families was also more uneven in Flavobacteriia, indicative of a more pronounced substrate specialization compared to Alpha- and Gammaproteobacteria. GH92 (mannosidase) and GH16 (including β-1,3-glucanase) genes, for example, were predominantly found in Flavobacteriia, possibly indicating more readiness to decompose mannans and chrysolaminarin.
Metagenome taxonomic classification provided sufficient data for analysis of CAZyme repertoires of the flavobacterial NS5 marine group, NS3a marine group, Formosa, Polaribacter, and Cryomorphaceae, and the gammaproteobacterial Alteromonadales and Reinekea clades (Supplementary file 10). For most of these clades, the analyses revealed fingerprint-like patterns, which corroborates the hypothesis that these clades have distinct glycan niches that are relatively stable across years. For example, the NS5 marine group (Figure 6Q) was rich in GH3, 20 and 29 (fucosidases), but notably devoid of GH92 (mannosidases). By contrast, Formosa and Polaribacter clades (Figure 6T,U) contained higher abundances of GH92 genes. The Formosa CAZyme profile was also characterized by high proportions of GH16, 17, and 30 families, which all contain enzymes that can decompose chrysolaminarin. Polaribacter contained a broader set of CAZymes that included all families found in Formosa, however, Polaribacter was richer in GH13 (e.g. α-amylase) and poorer in GH92 (mannosidases) than Formosa. Likewise, the GH repertoires of the NS3a and NS5 marine groups were similar (Figure 6Q,S), but the NS3a marine group was richer in GH13 and 32 and devoid of GH29 family fucosidases. The high number of CAZyme families in Polaribacter corroborated metagenome bin analyses that suggested a higher diversity within this clade. CAZyme gene frequencies were much lower in the Cryomorphaceae than in the other investigated Flavobacteriia clades with GH frequencies barely exceeding 0.5% (Figure 6—figure supplement 1). This suggests a different ecophysiological niche and a distinct role of the Cryomorphaceae during phytoplankton blooms.
For Gammaproteobacteria, recurring patterns were detected for the prominent Alteromonadales and Reinekea clades. Alteromonadales contained some of the GH families that play important roles during phytoplankton blooms, such as GH13 and 16, but were notably poor in or even devoid of others, such as GH29 and GH92, respectively (Figure 6—figure supplement 2). In contrast to other prominent clades, we did not obtain sufficient metagenome sequences for Reinekea for all four years, but only for 2009 and 2010 (Figure 6V). However, the Reinekea CAZyme patterns of 2009 and 2010 were well conserved with high proportions of GH23 and 13, and CBM48, 20, 41, 21, and 25. The GH23 family comprises peptidoglycan lyases and the GH13 family contains α-1,4-glucanases (e.g. α-amylase). CBM48, 20, 41, 21, and 25 all bind α-1,4-glucans such as starch and glycogen, and the ubiquitous CBM50 contains peptidoglycan-binding members. These results are consistent with a glycan niche that involves decomposition of external α-1,4-glucans and possibly peptidoglycan.
Discussion
Nutrient-poor marine surface waters are dominated by clades such as SAR11 and Prochlorococcus. Both feature small, reduced genomes and can use sunlight and small organic molecules (e.g. Gómez-Pereira et al., 2013). The otherwise heterotrophic SAR11 use proteorhodopsin for supplemental phototrophy (Giovannoni et al., 2005) and phototrophic Prochlorococcus cyanobacteria are capable of supplemental uptake of amino acids (Zubkov et al., 2003) and glucose (Muñoz-Marín et al., 2013). Microbial communities in nutrient-rich 'green' surface oceans by contrast feature higher proportions of heterotrophic species that feed on more complex organic substrates. In particular during phytoplankton blooms, the release of algae-derived organic matter selects for fast growing species with genomic adaptations towards algal biomass remineralization. These are typically members of the Flavobacteriia and Gammaproteobacteria classes and the alphaproteobacterial Roseobacter clade. Similarly adapted species from these clades compete for substrates during phytoplankton blooms with variation in which species prevail. Despite this stochastic effect, the most well-adapted species will be successful more often and thus exhibit patterns of annual recurrence.
During spring phytoplankton blooms at Helgoland in the North Sea, we observed recurrent bloom-associated abundance peaks of in particular flavobacterial clades, namely Formosa, Polaribacter, the NS3a marine group, Tenacibaculum, Ulvibacter, and the Cryomorphaceae VIS6 clade. Within Gammaproteobacteria Alteromonadaceae/Colwelliaceae, Reinekea, and the SAR92 clade were clearly bloom-associated and recurrent as was Methylophilales within Betaproteobacteria, and 'Cd. Planktomarina temperata' from the DC5-80-3 lineage (a.k.a Roseobacter clade affiliated = RCA group) and the NAC11-7 lineage within the Roseobacter clade. It has already been shown that the abundant North Sea isolate 'Cd. Planktomarina temperata' RCA23T is associated with decaying phytoplankton (Giebel et al., 2011) and high abundances and in particular high activity have been reported during a spring phytoplankton bloom event in the North Sea of 2010 (Voget et al., 2015; Wemheuer et al., 2015). High activities of members of the RCA and the SAR92 clades during North Sea spring phytoplankton blooms have also been reported in 2009 (Klindworth et al., 2014) and 2010 (Wemheuer et al., 2015), just as an increase of Bacteroidetes of the genera Marinoscillum and Polaribacter during 2010 (Wemheuer et al., 2015).
Many of the other clades we report here (including some low abundance groups) have been found during blooms of dinoflagellates, including AEGEAN-169, Alteromonadales, NS3a marine group, NS5 marine group, OM43, OM60 (NOR5), SAR116, SAR86, and ZD0405 (Yang et al., 2015) or Cryomorphaceae, Glaciecola and Sulfitobacter (Tan et al., 2015). The OM43 clade (order Methylophilales) comprises methylotrophs known to feed on algae C1 compounds (Halsey et al., 2012), and it has been reported that Sulfitobacter species SA11 shares a mutually beneficial exchange of compounds with the diatom species Pseudo-nitzschia multiseries (Amin et al., 2015).
There is of course unaddressed diversity in all these clades. Some of the genera might be dominated by a single species while others might be more diverse with considerable variation in competitive success between bloom events. Nevertheless, the high level of recurrence in particular of flavobacterial clades indicates a strong selection of few clades highly adapted for the manipulation and uptake of specific and complex polysaccharides (and likely other biopolymers) and disagrees with substantial levels of sloppy-feeding by these bacteria that would allow other less adapted clades to arbitrarily reach high abundances via cross-feeding. This might be attributed to the capability of Bacteroidetes for very efficient macromolecule uptake as it has been recently shown for uptake of α-mannan by the human gut bacterium Bacteroides thetaiotaomicron (Cuskin et al., 2015). This bacterium binds α-mannan macromolecules to its surface, followed by rapid cleavage into larger oligomers that are immediately imported via a TonB-dependent transporter (TBDT) into the periplasm without detectable loss. The bulk of the degradation into smaller molecules takes place in the periplasm where the substrate is secure from outside competitors before transport into the cytoplasm.
TonB-dependent transporters are not specific to Bacteroidetes, but it seems that only Bacteroidetes have evolved a functional coupling of SusC-like TBDT porins with SusD-like TonB-dependent receptors (TBDRs) that bind and guide the substrate to the porin. At least so far, only Bacteroidetes genomes feature characteristic susCD gene tandems. Within Bacteroidetes genomes such tandems are frequently found in so-called polysaccharide utilization loci (PULs; [Sonnenburg et al., 2010]). PULs are operons or regulons where one or more susCD gene tandems are co-located with CAZymes. Further frequent accessory genes in PULs encompass transcriptional regulators, proteases, transporter components and sulfatases. The latter are required for the desulfation of sulfated polysaccharides, which marine algae produce in large quantities. The diversity of PULs in marine Bacteroidetes genomes is high and largely unexplored and so far only few PULs have been linked to dedicated algal polysaccharides (e.g. Hehemann et al., 2012; Kabisch et al., 2014; Xing et al., 2015). The large, complex and efficient PUL uptake systems might explain why Flavobacteria consistently outcompeted Gammaproteobacteria during the onset of all blooms.
It is noteworthy that we observed a shift in bacterioplankton biodiversity alongside a shift in functional gene repertoires for the major clades in all four years of this study. The abundance of CAZymes and sulfatases increased from pre- to mid-bloom situations and leveled off post-bloom. Likewise, similar abundance patterns were observed for the most abundant CAZyme families in all studied years.
We did observe an increase in the abundance of TBDRs during bloom situations, and we have shown previously that TBDRs are among the most abundantly expressed proteins during the bacterial mineralization of algae biomass (Teeling et al., 2012). The relevance of TBDRs in nutrient-rich oceanic regions has been also supported by in situ metaproteome studies of samples from the South Atlantic Ocean (in particular at coastal upwelling zones; [Morris et al., 2010]) and from the Antarctic Southern Ocean (Williams et al., 2013).
The recurrent patterns in bacterioplankton diversity and functional repertoires during the four studied spring blooms are remarkable in view of the variation among algae taxa. For example, even though algal biomass of the 2012 spring bloom was dominated by silicoflatellate Chattonella spp. and not by diatoms as was true for the three preceding years, the respective bacterioplankton communities were strikingly similar. Likewise only few bacterioplankton taxa seemed to be weakly correlated with dedicated phytoplankton taxa, and no clear correlation was found between distinct bacterioplankton taxa and individual distinct diatom clades in statistical analyses (Supplementary files 6 and 7). It seems that phytoplankton community composition did not exert a strong effect on the composition of the free-living non-phycosphere bacterioplankton community. Instead, the dominating algae (in terms of biomass) seemed to produce similar or perhaps identical types of substrates for specifically adapted clades of heterotrophic bacterioplankton. It is therefore conceivable that recurrence is more pronounced on the functional level than on the taxonomic level, since species from different taxa with similar ecophysiological niches might functionally substitute each other in different years. This hypothesis is supported by the bacterioplankton communities' similar CAZyme gene repertoires in the 2009 to 2012 spring blooms and in particular the consistency on class level that was almost unaffected by distinct blooming clades, yet needs to be further tested by deep metatranscriptome sequencing and metaproteomics during multiple spring phytoplankton blooms in future studies. Considering the extent of recurrence, our combined metagenome data (>5 million predicted proteins) should provide sufficient search space for such an analysis.
The existence of recurrent key players during North Sea spring phytoplankton blooms suggests that the bacterioplankton community composition during and after such blooms is governed by deterministic effects. We have shown before that temperature exerts a strong effect on North Sea bacterioplankton as it selects for temperature-dependent guilds, for example when comparing spring and summer blooms (Lucas et al., 2015). Within short-lived spring blooms, however, the supply of algae-derived organic matter is among the main factors that shape the bacterioplankton composition. In particular the different types of structurally distinct polysaccharides that algae produce in large quantities seem to exert such substrate-induced forcing. Since Flavobacteriia are more specialized on polysaccharides than Gammaproteobacteria, this would also explain, why Flavobacteriia dominated the recurrent clades (Figure 3Q–T, Figure 4E–J) and Gammaproteobacteria clades exhibited more stochastic peaks (Figure 4Q–T).
At the beginning of a bloom, most available polysaccharides will be exopolysaccharides, but as the bloom commences and algae become senescent, more and more cellular algal substrates are released, culminating in the bloom's final die off phase. Bacteria will naturally consume the more degradable substrates such as storage polysaccharides (e.g. chrysolaminarin) first, and more recalcitrant substrates (e.g. branched and sulfated polysaccharides) later. TEP for example seems to undergo such selective feeding, as it has been suggested that in particular fucose-rich TEP is less readily degraded than mannose and galactose rich TEP (for review see Passow, 2002). Such selective feeding creates an additional change in substrate availability and leads to a succession of substrate niches for specifically adapted bacterioplankton clades to grow.
Concluding remarks
Bacterioplankton communities during spring phytoplankton blooms in the coastal North Sea undergo swift and dynamic composition changes and thus are difficult to investigate. Nonetheless, we found clades that recurrently reached high abundances among Flavobacteriia (Formosa, Polaribacter, NS3a marine group, Ulvibacter, VIS6 clade Cryomorphaceae, Tenacibaculum), Gammaproteobacteria (Alteromonadaceae/Colwelliaceae, SAR92, Reinekea) and Roseobacter clade Alphaproteobacteria (DC5-80-3, NAC11-7). Recurrence was not only detectable on the taxonomic but also on the functional level with a highly predictable increase in TonB-dependent polysaccharide uptake systems and distinct CAZyme patterns. The niches of abundant bacterioplankton clades are more complex and manifold than the glycan niches that we explore in this study. CAZymes, however, have the advantage that they allow linking of gene repertoires and possible environmental functions in a way currently not feasible for other macromolecules such as proteins and lipids. Our results suggest that besides stochastic also deterministic effects influence phytoplankton-bacterioplankton coupling during blooms. They indicate that during spring phytoplankton blooms similar principles of resource partitioning and specialization are at play as within human gut microbiota that decompose fiber-rich plant material, albeit at a much larger scale. Rather the availability of substrates commonly occurring in microalgae than one-to-one interactions of particular phytoplankton and bacterioplankton species caused the succession of free-living bacterioplankton clades.
Materials and methods
Phytoplankton and physicochemical data
Physicochemical parameters (Supplementary file 1) and phytoplankton data (Supplementary file 2) were assessed in subsurface water on a weekday basis as part of the Helgoland Roads LTER time series. Details on the acquisition of these data have been described previously (Teeling et al., 2012). The Helgoland Roads time series is accessible via the public database Pangaea (http://www.pangaea.de).
Bacterioplankton
Sampling of bacterioplankton was carried out as described previously (Teeling et al., 2012). In brief, surface seawater samples were taken at the long-term ecological research station 'Kabeltonne' (54° 11.3' N, 7° 54.0' E) at the North Sea island Helgoland using small research vessels (http://www.awi.de/en/expedition/ships/more-ships.html) and processed in the laboratory of the Biological Station Helgoland within less than two hours after sampling.
Biomass of free-living bacteria for DNA extraction was harvested on 0.2 µm pore sized filters after pre-filtration with 10 µm and 3 µm pore sized filters to remove large debris and particle-associated bacteria. By contrast, cells for microscopic visualization methods were first fixed by the addition of formaldehyde to sampled seawater, which was then filtered directly onto 0.2 µm pore sized filters. All filters were stored at -80°C until further use.
Microscopy: total cell counts, CARD-FISH
Assessment of absolute cell numbers and bacterioplankton community composition was carried out as described previously (Thiele et al., 2011). To obtain total cell numbers, DNA of formaldehyde fixed cells filtered on 0.2 µm pore sized filters was stained with 4',6-diamidino-2-phenylindole (DAPI). Fluorescently labeled cells were subsequently counted on filter sections using an epifluorescence microscope. Likewise, bacterioplankton community composition was assessed by catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) of formaldehyde fixed cells on 0.2 µm pore sized filters. DAPI and CARD-FISH cell counts are summarized in Supplementary file 3 and the corresponding probes in Supplementary file 4.
16S rRNA V4 gene tag sequencing
Surface seawater samples were collected on bi-monthly to bi-weekly time scales from January 2010 to December 2012 at Helgoland roads. 500 ml of each sample were subjected to fractionating filtration as described above using 10, 3 and 0.2 µm pore size polycarbonate membrane filters (Millipore, Schwalbach, Germany). DNA of the 0.2–3 µm fraction was extracted from filters as described previously (Sapp et al., 2007) and quantified using the Invitrogen (Carlsbad, CA, USA) Quant-iT PicoGreen dsDNA reagent as per manufacturer's instructions. Concentrations ranged from <1 to 20 µg DNA/ml.
ml of each sample were subjected to fractionating filtration as described above using 10, 3 and 0.2 µm pore size polycarbonate membrane filters (Millipore, Schwalbach, Germany). DNA of the 0.2–3 µm fraction was extracted from filters as described previously (Sapp et al., 2007) and quantified using the Invitrogen (Carlsbad, CA, USA) Quant-iT PicoGreen dsDNA reagent as per manufacturer's instructions. Concentrations ranged from <1 to 20 µg DNA/ml.
50 µl aliquots of each sample were pipetted into 96-well plates and sent to the Department of Energy (DOE) Joint Genome Institute (JGI, Walnut Creek, CA, USA) for amplification and sequencing as follows: Sample prep was done on a PerkinElmer (Waltham, MA, USA) Sciclone NGS G3 Liquid Handling Workstation capable of processing 96 plate-based samples in parallel, utilizing the 5 PRIME (Gaithersburg, MD 20878, USA) HotMasterMix amplification kit and custom amplification primers targeting the V4 region of the 16S rRNA gene using 515F (5' GTGCCAGCMGCCGCGGTAA 3') and 806R (5' GGACTACHVGGGTWTCTAAT 3') (Caporaso et al., 2011). Primers also contained Illumina adapter sequences and a barcode index. PCR reactions were set up in 75 µl with 1x HotMasterMix (5 PRIME) with final concentrations of 0.4 µg/µl BSA and 0.2 µM of each primer. This volume was split into triplicate 25 µl reactions for independent amplification and then pooled to reduce PCR bias. Prepared amplicon libraries were normalized, multiplexed into a single pool per plate and quantified using the KAPA Biosystems (Wilmington, MA, USA) next-generation sequencing library qPCR kit on a Roche (San Francisco, CA, USA) LightCycler 480. Libraries were sequenced on an Illumina (San Diego, CA, USA) MiSeq sequencer using the Reagent Kit v3 and 2x250 bp chemistry. The resulting sequences are available from the DOE-JGI GOLD database (Reddy et al., 2015) as part of the COGITO project (Gp0056779) and from the NCBI short read archive (SRA) (SRA278189).
16S rRNA gene tag analysis
Roche 454 16S rRNA gene tags from 2009 MIMAS (Microbial Interactions in Marine Systems) project (Teeling et al., 2012) were reanalyzed for comparison with the Illumina-based COGITO extension project from subsequent years 2010–2012. The 2009 datasets was generated using the primers Bakt_314F (5' CCTACGGGNGGCWGCAG 3') and Bakt_805R (5’ GACTACHVGGGTATCTAATCC 3') (Herlemann et al., 2011). The forward primers of both datasets target distinct, but the reverse primers target the same region. Hence, only those 454 reads sequenced from the 805 direction were reanalyzed for comparison. For 2010–2012, raw MiSeq paired-end reads (2x250 bp) were merged and filtered using illumina-utils (https://github.com/meren/illumina-utils) to retain only read pairs without mismatches in the overlapping regions. These high-quality Illumina tags and the 454 tags were then processed separately but with the same methods via the SILVAngs pipeline (Quast et al., 2013), which includes additional quality filtering steps via alignment as well as length, ambiguity and homopolymer filters. Sequences were dereplicated at 100% identity and then globally clustered at 98%. Representative OTUs were classified to genus level against the SILVA (Quast et al., 2013) v119 database using BLAST with a similarity threshold = (sequence identity + alignment coverage) / 2 >=93%. The SAR92 clade was reclassified according to SILVA v123. Reads were mapped against representative OTUs to obtain final abundance counts. For the purpose of this study, OTUs were collapsed based on shared taxonomy no higher than the genus level.
For MIMAS samples, we retained a total of 110,995 454 reads across 7 samples with an average of 16,000 per sample. After SILVAngs quality filtering, 110,866 remained for clustering. 6102 representative OTUs were identified and 107,708 total sequences were assigned to a relative in the database during classification within the 93% similarity threshold. The final abundance matrix collapsed on shared taxonomic classification contained 500 unique taxa.
In total, 20,869,432 paired raw MiSeq reads were obtained across 142 samples from 2010–2012 COGITO samples. 15,016,350 merged reads with no mismatches in the overlapping region were retained with an average of 106,000 per sample. Reads were randomly sub-sampled to 40,000 tags per sample to reduce computational demands. In total 6,120,000 tags were submitted to the SILVAngs pipeline. After additional quality filtering, 6,116,021 sequences were clustered at 98% and the resulting 935,006 representative OTUs were classified. A total of 5,676,259 sequences were assigned to a relative in the database within the 93% similarity threshold. The final abundance matrix collapsed on shared taxonomy no higher than the genus level contained 1995 unique taxa (Supplementary file 5).
Metagenome sequencing
Total community DNA of 2009 samples (02/11/09; 03/31/09; 0407/09; 04/14/09; 06/16/09) was sequenced on the 454/Roche FLX Ti platform as described previously (Teeling et al., 2012). Metagenome sequencing of 2010–12 samples (03/03/2010; 04/08/10; 05/04/10; 05/18/10; 03/24/11; 04/28/11; 05/26/11; 03/08/12; 04/16/12; 05/10/12) was performed at the DOE Joint Genome Institute on the Illumina HiSeq2000 platform. Libraries were created from 100 ng environmental DNA per sample that was sheared to 270 bp using a Covaris E210 (Covaris, Woburn, MA, USA) and size selected using SPRI beads (Beckman Coulter, Indianapolis, IN, USA). The fragments were treated with end-repair, A-tailing, and ligation of Illumina compatible adapters (IDT, Coralville, IA, USA) using the KAPA-Illumina library creation kit. The libraries were quantified using KAPA Biosystem’s next-generation sequencing library qPCR kit and run on a Roche LightCycler 480 real-time PCR instrument. The quantified libraries were then prepared for sequencing on the Illumina HiSeq sequencing platform utilizing a TruSeq PE Cluster Kit v3, and Illumina’s cBot instrument to generate a clustered flowcell for sequencing. Sequencing was performed on the Illumina HiSeq2000 sequencer using TruSeq SBS sequencing Kits, v3, following a 2x150 bp indexed run recipe.
Raw reads were screened against Illumina artifacts with kmer size of 28, step size of 1. Reads were subsequently trimmed from both ends using a minimum quality cutoff of 3; reads with three or more N's or with average quality score <Q20 were removed. In addition, reads <50 bp were removed. The remaining quality-filtered Illumina reads were assembled using SOAPdenovo v1.05 (Luo et al., 2012) at a range of kmers (81, 85, 89, 93, 97, 101) with default settings (options: -K 81 -p 32 -R -d 1). Contigs generated by each assembly (6 total contig sets), were de-replicated using JGI in house Perl scripts. Contigs were then sorted into two pools based on length. Contigs <1800 bp were re-assembled using Newbler (Life Technologies, Carlsbad, CA, USA) in attempt to generate larger contigs (options: -tr, -rip, -mi 98, -ml 80). Contigs >1800 bp as well as the contigs from the Newbler assembly were combined using minimus 2 (options: -D MINID=98 -D OVERLAP=80) from the AMOS package (http://sourceforge.net/projects/amos). Read depths were estimated based on read mapping with bbmap (http://bio-bwa.sourceforge.net/). The metagenome study information is available from the DOE-JGI GOLD database (study: Gs0000079). The unassembled reads are available from the NCBI SRA (see Supplementary file 8), and the assembled and annotated metagenome datasets from the IMG/M system (Markowitz et al., 2014).
Metagenome analysis
The DOE-JGI MAP v.4 annotation pipeline (Huntemann et al., 2015) was used for initial metagenome gene prediction and annotation. The annotated metagenomes were loaded in the IMG/M system as of mid 2014, and subsequently imported into a GenDB v2.2 annotation system (Meyer et al., 2003) for taxonomic classification and data mining.
All genes were searched against the NCBI non-redundant protein database (as of June 17th, 2014) using USEARCH v6.1.544 (Edgar, 2010), against the Pfam v25 database (Finn et al., 2014) using HMMER v3 (Punta et al., 2012), for signal peptides using SignalP v3.0 (Nielsen et al., 1999) and for transmembrane helices using TMHMM v2.0c (Krogh et al., 2001). CAZymes were automatically annotated based on HMMER searches against the Pfam v25 and dbCAN (Yin et al., 2012) databases and BLAST (Altschul et al., 1990) searches against the CAZy database (Cantarel et al., 2009; Lombard et al., 2014) using E-value cut-offs that were specifically adjusted for each CAZyme family (Supplementary file 11). Genes were only annotated as CAZymes when at least two of the search results were congruent, and CAZymes were only analyzed for contigs ≥500 bp.
Taxonomic classification of the metagenome sequences into taxonomically coherent bins ('taxobins') was carried out with a modified version of the Taxometer approach described in (Teeling et al., 2012). Taxometer consolidates predictions of a set of individual sequence classification tools into a consensus using a weighted assessment on seven selected ranks (superkingdom, phylum, class, order, family, genus, species) of the NCBI taxonomy (http://www.ncbi.nlm.nih.gov/Taxonomy/). We combined taxonomic information inferred from (i) Pfam hits using the CARMA3 approach (Gerlach and Stoye, 2011), (ii) BLASTp hits using the KIRSTEN approach (Teeling et al., 2012; supp. data), and (iii) mapping of quality-filtered (illumina-utils; https://github.com/meren/illumina-utils/) Illumina reads to selected reference sequences. In contrast to the original Taxometer approach we omitted signature-based classification with Self-Organizing Maps and mapping of reads containing partial 16S rRNA gene sequences. The prediction tools that were used are outlined below:
We used the HMMER-based module of CARMA3 (not the BLAST-based module) that infers taxonomy of sequences by post-processing genes with HMMER3 hits to the Pfam database. The basic principle is to apply a reciprocal search technique to reduce the number of identified matches and thereby to improve taxonomic classification quality.
KIRSTEN (Kinship Relationship Reestablishment) infers taxonomy of sequences by post-processing BLASTp hits to the NCBI nr database by means of rank-based evaluations on all levels of the NCBI taxonomy with an increasing stringency from the superkingdom down to the species level. On each taxonomic level, all occurring taxa are weighted by the sum of their BLASTp bit scores. When the taxon with the highest weight exceeds an adjustable threshold, the process continues towards the next taxonomic level. The threshold increases with each taxonomic level, i.e. the algorithm becomes more critical while it progresses. For this study, we substituted BLASTp by the UBLAST module of USEARCH 6.1 (Edgar, 2010) with an E-value cutoff of E-10 and maximum hit count of 500.
SMALT (http://www.sanger.ac.uk/resources/software/smalt/) was used to map metagenome reads on a manually compiled set of 49 reference genomes and a streamlined version of the NCBI nr database. Both, the reference genomes and the sequences selected from the NCBI nr database were selected based on habitat-specific information. This was done manually for the reference genomes and automatically for the NCBI nr database as follows: Each of the hits from the UBLAST search during KIRSTEN analysis can be associated with multiple taxa, since in nr redundant sequences from different taxa are merged. We used this information to extract all taxa associated with a given hit, then combined the taxa of all hits and finally extracted all sequences of these taxa from Genbank. This way a sample-specific streamlined subset of Genbank was generated that greatly sped up the mapping process. Only metagenome reads with at least 95% identity to any sequence in the sample-specific sequence database were used. Reads were subsequently back-mapped to contigs. Since contigs consist of many reads, this way, contigs were associated with multiple taxonomic paths. Taxonomic paths resulting from classification with reference genomes and the habitat-specific streamlined nr were concatenated. Paths representing less than 1% of the reads of a given contig were discarded.
Finally, Taxometer was used to combine all gene-based predictions from CARMA3 and KIRSTEN and the mapping results for each contig, and to infer a consensus taxonomy. Taxonomic predictions were possible for 94.6% of the contigs above 1 kbp. The results are summarized in Supplementary file 9.
Statistical analyses
The Spearman rank correlation test (Supplementary file 6) was used to test for correlations between Bacteroidetes clade abundances and environmental variables (chlorophyll a, temperature, salinity, silicate, phosphate, nitrate, nitrite, ammonia) and phytoplankton abundances (classes: diatoms, dinoflagellates, coccolithophorids, silicoflagellates, flagellates, ciliates, green algae; species: Mediophyxis helysia, Thalassiosira nordenskioeldii, Chaetoceros debilis and C. minimus, Rhizosolenia styliformis, Chattonella and Phaeocystis). Bacteroidetes and phytoplankton numbers were transformed to log-scale for better comparison. Linear regressions (Supplementary file 7) were done using stepwise forward regression model by using the log transformed Bacteroidetes and phytoplankton abundances. All statistical analyses were performed using the software Sigma-Plot 12 (SYSTAT, Santa Clara, CA, USA).
Acknowledgements
The expert technical assistance of Y.-L.Chen and D. Berkelmann is acknowledged. This study was funded by the Max Planck Society. The work conducted by the US. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grant:
Max-Planck-Gesellschaft to Rudolf I Amann.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
HT, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
BMF, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
CMB, Sampling at Helgoland Island, Total cell count and CARD-FISH analyses, Acquisition of data, Analysis and interpretation of data.
KK, Metagenome analyses, CAZyme analyses, Bioinformatics, Analysis and interpretation of data.
MC, Analyses of the complete 16S rRNA tag dataset, Analysis and interpretation of data.
LK, Metagenome analyses, Selection and analyses of reference genomes for metagenome taxonomic classification, Bioinformatics, Analysis and interpretation of data.
GR, Sampling at Helgoland Island, CARD-FISH analyses, Acquisition of data, Analysis and interpretation of data.
JW, Taxonomic classification of metagenomes (programming of pipeline and compilation of reference datasets), Acquisition of data, Analysis and interpretation of data.
CQ, Programming of the 16S rRNA tag data analyses pipeline, Assistance with 16S rRNA tag data analyses, Analysis and interpretation of data.
FOG, Conception and design, Analysis and interpretation of data.
JL, Sampling at Helgoland Island, Initial analyses of a part of the 16S rRNA tag data, Acquisition of data, Analysis and interpretation of data.
AW, Provision of sampling logistics and lab space at Helgoland Island, Provision of physicochemical data, Provision of algae biodiversity data, Statistical analyses of 16S rRNA tag data, Acquisition of data, Analysis and interpretation of data.
GG, Provision of sampling logistics and lab space at Helgoland Island, Provision of physicochemical data, Provision of algae biodiversity data, Statistical analyses of 16S rRNA tag data, Acquisition of data, Analysis and interpretation of data.
KHW, Provision of sampling logistics and lab space at Helgoland Island, Provision and analyses of physicochemical data, Provision and analyses of algae biodiversity data, Acquisition of data, Analysis and interpretation of data.
RIA, Conception and design, Analysis and interpretation of data, Drafting or revising the article.
Additional files
Supplementary file 1.
Physicochemical parameters.DOI: http://dx.doi.org/10.7554/eLife.11888.011
Supplementary file 2.
Cell counts of dominating phytoplankton.DOI: http://dx.doi.org/10.7554/eLife.11888.012
Supplementary file 3.
Total cell and CARD-FISH cell counts of bacterioplankton.DOI: http://dx.doi.org/10.7554/eLife.11888.013
Supplementary file 4.
Specific oligonucleotide probes used for quantification of free-living (0.2 - 3 µm) bacterioplankton populations by fluorescence in situ hybridization (FISH).DOI: http://dx.doi.org/10.7554/eLife.11888.014
Supplementary file 5.
Abundance matrix of the 16S rRNA gene tag sequence analysis.DOI: http://dx.doi.org/10.7554/eLife.11888.015
Supplementary file 6.
Spearman rank correlations of abundant Bacteroidetes clades to phytoplankton groups.Correlation values were considered only when p-value <0.05. Spearman rank correlations measure the strength of association between two ranked variables. A precondition (assumption) for this nonparametric statistical analysis is that the two variables share a monotonic relationship (i.e. when one variable increases, the other either consistently increases as well or consistently decreases). This monotonic relationship does, however, not need to be linear. While no particularly strong correlations were found between major algae groups and bacterioplankton clades, some noteworthy trends were detected. For example, Ulvibacter abundances were positively correlated with diatoms (in particular centrales) and negatively correlated with silicoflagellates, whereas an opposite trend was observed for the VIS1 clade of the NS5 marine group (i.e. negative correlation with diatoms and a positive correlation with silicoflagellates). While the explanatory power of such pairwise rank-based correlations is limited and correlation does not necessarily imply causation, these data suggest at least for Ulvibacter sp. (and to a lesser extent for Formosa clade B) that they are associated with diatoms, while the NS3a, NS5 and NS9 marine groups seemed to be rather positively correlated with flagellates such as dino- and silicoflagellates. The positive correlation between Ulvibacter sp. and diatoms was supported by Spearman rank correlation analysis between the abundances of prominent bacterioplankton clades and distinct phytoplankton groups. Ulvibacter abundances were positively correlated with the diatoms Mediopyxis helysia, Chaetoceros debilis, Chaetoceros minimus, Thalassiosira nordenskioeldii and the Phaeocystis spp. haptophytes. Correlations of other bacterioplankton groups with distinct phytoplankton groups were less pronounced. Noteworthy positive correlations were found between Chattonella spp. and Bacteroidetes abundances (probe CF319a), in particular Marinoscillum spp. (probe CYT-734), Polaribacter spp. (probe POL740) and the NS5/VIS1 and NS3a marine groups. The NS5/VIS1 marine group was negatively correlated with most diatom groups and the Phaeocystis sp. haptophytes, and only positively correlated with Chattonella species. This might indicate a preference for Chattonella spp. or simply reflect that the members of the VIS1 clade of the NS5 marine group were less competitive than other Flavobacteriia under the conditions where other algae dominated. Spearman rank correlations between major bacterioplankton clades and physicochemical parameters were most conclusive for Ulvibacter species. Ulvibacter abundances were positively correlated with chlorophyll a, consistent with the previously detected positive correlation with in particular diatoms. Ulvibacter abundances were also negatively correlated with silicate, phosphate, nitrate and ammonium. In particular the strong negative correlation with silicate, which is the limiting factor for diatom frustule formation, supports a particularly strong association of Ulvibacter spp. and diatoms. Similar but weaker trends were observed also for other bacterioplankton clades, such as Polaribacter, Formosa clade B, or VIS6 clade Cryomorphaceae. Of all genus-level clades, Polaribacter and Formosa clade A showed the strongest associations with temperature.
DOI: http://dx.doi.org/10.7554/eLife.11888.016
Supplementary file 7.
Linear regression analyses were computed in order to test, whether the abundances of major clades of Flavobacteriia were influenced by abiotic factors or by abundant algae groups.For all tested clades of Flavobacteriia, multiple abiotic factors and multiple algae groups were obtained as explanatory variables. The strongest abiotic predictors were temperature, salinity, silicate and nitrate. The strongest biotic predictors were Phaeocystis spp. haptophytes, Rhizosolenia spp., Chaetoceros debilis, and Chaetoceros minimus diatoms and the silicoflagellate Chattonella. It should be noted though that these regressions were computed based on log-transformed abundance data and not algae biovolumes (which were not measured). Insofar the in influence of the rather small cell-sized algae such as Chaetoceros miniumus is likely overestimated. Such limitations notwithstanding it is noteworthy that in no case a simple one-to-one relationship between specific algae and specific bacterioplankton groups was obtained so far.
DOI: http://dx.doi.org/10.7554/eLife.11888.017
Supplementary file 8.
Statistics of the 16 metagenomes from free-living (0.2 - 3.0 µm) North Sea bacterioplankton that were used in this study.Two additional metagenomes are listed in italics for completeness, one from a test run sampled on August 21th, 2008, and a metagenome of particle-attached (3 - 10 µm) bacterioplankton sampled on April 14th, 2009.
DOI: http://dx.doi.org/10.7554/eLife.11888.018
Supplementary file 9.
Results of the taxonomic classification of the metagenome sequences.DOI: http://dx.doi.org/10.7554/eLife.11888.019
Supplementary file 10.
Sizes in basepairs of the metagenome taxonomic bins that were used for CAZyme frequency analyses on order to genus level.Taxonomic bins with too small sizes for a sound analysis were excluded (red text).
DOI: http://dx.doi.org/10.7554/eLife.11888.020
Supplementary file 11.
E-value thresholds used for automated CAZyme family detection.Searches were performed against the CAZy database, the dbCAN database and the Pfam database using E-value thresholds that were adjusted for each family by extensive manual annotations. CAZymes were only annotated when at least two of the three database searches yielded positive results (GH = glycoside hydrolase; CBM = carbohydrate- binding module; CE = carbohydrate esterase; PL = polysaccharide lyase; GT = glycoside transferase; AA = auxiliary activities).
DOI: http://dx.doi.org/10.7554/eLife.11888.021
Major datasets
The following datasets were generated:
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2010/03/03,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212914,Publically available at NCBI Sequence Read Archive (accession no. SRA212914)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2010/05/04,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212889,Publically available at NCBI Sequence Read Archive (accession no. SRA212889)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,16S rRNA V4 amplicons of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads from 2010 to 2012,http://www.ncbi.nlm.nih.gov/sra/?term=SRA278189,Publically available at NCBI Sequence Read Archive (accession no. SRA278189)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2010/05/18,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212588,Publically available at NCBI Sequence Read Archive (accession no. SRA212588)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2011/03/24,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212575,Publically available at NCBI Sequence Read Archive (accession no. SRA212575)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2011/04/28,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212530,Publically available at NCBI Sequence Read Archive (accession no. SRA212530)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2011/05/26,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212476,Publically available at NCBI Sequence Read Archive (accession no. SRA212476)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2012/03/08,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212475,Publically available at NCBI Sequence Read Archive (accession no. SRA212475)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2012/04/16,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212473,Publically available at NCBI Sequence Read Archive (accession no. SRA212473)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2012/05/10,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212472,Publically available at NCBI Sequence Read Archive (accession no. SRA212472)
Hanno Teeling, Bernhard M Fuchs, Christin M Bennke, Karen Krüger, Meghan Chafee, Lennart Kappelmann, Greta Reintjes, Jost Waldmann, Christian Quast, Frank Oliver Glöckner, Judith Lucas, Antje Wichels, Gunnar Gerdts, Karen H Wiltshire, Rudolf I Amann,2016,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2010/04/08,http://www.ncbi.nlm.nih.gov/sra/?term=SRA212908,Publically available at NCBI Sequence Read Archive (accession no. SRA212908)
The following previously published datasets were used:
Hanno Teeling, Bernhard M. Fuchs, Dörte Becher, Christine Klockow, Antje Gardebrecht, Christin M. Bennke, Mariette Kassabgy, Sixing Huang, Alexander J. Mann, Jost Waldmann, Marc Weber, Anna Klindworth, Andreas Otto, Jana Lange, Jörg Bernhardt, Christine Reinsch, Michael Hecker, Jörg Peplies, Frank D. Bockelmann, Ulrich Callies, Gunnar Gerdts, Antje Wichels, Karen H. Wiltshire, Frank Oliver Glöckner, Thomas Schweder, Rudolf Amann,2012,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2009/02/11,http://www.ebi.ac.uk/ena/data/view/PRJEB2888,Publically available at the European Nucleotide Archive (study accession no. PRJEB2888, experimental accession no. ERX069460 - ERX069464)
Hanno Teeling, Bernhard M. Fuchs, Dörte Becher, Christine Klockow, Antje Gardebrecht, Christin M. Bennke, Mariette Kassabgy, Sixing Huang, Alexander J. Mann, Jost Waldmann, Marc Weber, Anna Klindworth, Andreas Otto, Jana Lange, Jörg Bernhardt, Christine Reinsch, Michael Hecker, Jörg Peplies, Frank D. Bockelmann, Ulrich Callies, Gunnar Gerdts, Antje Wichels, Karen H. Wiltshire, Frank Oliver Glöckner, Thomas Schweder, Rudolf Amann,2012,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2009/09/01,http://www.ebi.ac.uk/ena/data/view/PRJEB2888,Publically available at the European Nucleotide Archive (study accession no. PRJEB2888, experimental accession no.ERX069492 - ERX069495)
Hanno Teeling, Bernhard M. Fuchs, Dörte Becher, Christine Klockow, Antje Gardebrecht, Christin M. Bennke, Mariette Kassabgy, Sixing Huang, Alexander J. Mann, Jost Waldmann, Marc Weber, Anna Klindworth, Andreas Otto, Jana Lange, Jörg Bernhardt, Christine Reinsch, Michael Hecker, Jörg Peplies, Frank D. Bockelmann, Ulrich Callies, Gunnar Gerdts, Antje Wichels, Karen H. Wiltshire, Frank Oliver Glöckner, Thomas Schweder, Rudolf Amann,2012,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2009/06/16,http://www.ebi.ac.uk/ena/data/view/PRJEB2888,Publically available at the European Nucleotide Archive (study accession no. PRJEB2888, experimental accession no.ERX069490 - ERX069491)
Hanno Teeling, Bernhard M. Fuchs, Dörte Becher, Christine Klockow, Antje Gardebrecht, Christin M. Bennke, Mariette Kassabgy, Sixing Huang, Alexander J. Mann, Jost Waldmann, Marc Weber, Anna Klindworth, Andreas Otto, Jana Lange, Jörg Bernhardt, Christine Reinsch, Michael Hecker, Jörg Peplies, Frank D. Bockelmann, Ulrich Callies, Gunnar Gerdts, Antje Wichels, Karen H. Wiltshire, Frank Oliver Glöckner, Thomas Schweder, Rudolf Amann,2012,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2009/04/14,http://www.ebi.ac.uk/ena/data/view/PRJEB2888,Publically available at the European Nucleotide Archive (study accession no. PRJEB2888, experimental accession no.ERX069474 - ERX069481)
Hanno Teeling, Bernhard M. Fuchs, Dörte Becher, Christine Klockow, Antje Gardebrecht, Christin M. Bennke, Mariette Kassabgy, Sixing Huang, Alexander J. Mann, Jost Waldmann, Marc Weber, Anna Klindworth, Andreas Otto, Jana Lange, Jörg Bernhardt, Christine Reinsch, Michael Hecker, Jörg Peplies, Frank D. Bockelmann, Ulrich Callies, Gunnar Gerdts, Antje Wichels, Karen H. Wiltshire, Frank Oliver Glöckner, Thomas Schweder, Rudolf Amann,2012,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2009/04/07,http://www.ebi.ac.uk/ena/data/view/PRJEB2888,Publically available at the European Nucleotide Archive (study accession no. PRJEB2888, experimental accession no.ERX069470 - ERX069473)
Hanno Teeling, Bernhard M. Fuchs, Dörte Becher, Christine Klockow, Antje Gardebrecht, Christin M. Bennke, Mariette Kassabgy, Sixing Huang, Alexander J. Mann, Jost Waldmann, Marc Weber, Anna Klindworth, Andreas Otto, Jana Lange, Jörg Bernhardt, Christine Reinsch, Michael Hecker, Jörg Peplies, Frank D. Bockelmann, Ulrich Callies, Gunnar Gerdts, Antje Wichels, Karen H. Wiltshire, Frank Oliver Glöckner, Thomas Schweder, Rudolf Amann,2012,Metagenome of North Sea surface bacteriopankton (3 - 0.2 µm) from Helgoland Roads on 2009/03/31,http://www.ebi.ac.uk/ena/data/view/PRJEB2888,ERX069465 - ERX069469
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. 10.1016/S0022-2836(05)80360-2. [Abstract] [CrossRef] [Google Scholar]
- Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiology and Molecular Biology Reviews. 2012;76:667–684. 10.1128/MMBR.00007-12. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, Morales RL, Berthiaume CT, Parker MS, Djunaedi B, Ingalls AE, Parsek MR, Moran MA, Armbrust EV. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. 10.1038/nature14488. [Abstract] [CrossRef] [Google Scholar]
- Apprill A, McNally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecology. 2015;75:129–137. 10.3354/ame01753. [CrossRef] [Google Scholar]
- Armbrust EV. The life of diatoms in the world's oceans. Nature. 2009;459:185–192. 10.1038/nature08057. [Abstract] [CrossRef] [Google Scholar]
- Bell RT, Kuparinen J. Assessing phytoplankton and bacterioplankton production during early spring in Lake Erken, Sweden. Applied and Environmental Microbiology. 1984;48:1221–1230. [Europe PMC free article] [Abstract] [Google Scholar]
- Buchan A, LeCleir GR, Gulvik CA, González JM. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nature Reviews. Microbiology. 2014;12:686–698. 10.1038/nrmicro3326. [Abstract] [CrossRef] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Research. 2009;37:D233–238. 10.1093/nar/gkn663. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 Suppl 1:4516–4522. 10.1073/pnas.1000080107. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munōz-Munōz JL, Day A, Peña MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. 10.1038/nature13995. [Abstract] [CrossRef] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. 10.1093/bioinformatics/btq461. [Abstract] [CrossRef] [Google Scholar]
- Eilers H, Pernthaler J, Glöckner FO, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Applied and Environmental Microbiology. 2000;66:3044–3051. [Europe PMC free article] [Abstract] [Google Scholar]
- Eilers H, Pernthaler J, Peplies J, Glöckner FO, Gerdts G, Amann R. Isolation of novel pelagic bacteria from the German bight and their seasonal contributions to surface picoplankton. Applied and Environmental Microbiology. 2001;67:5134–5142. 10.1128/AEM.67.11.5134-5142.2001. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Falkowski PG, Barber RT, Smetacek V. Biogeochemical Controls and Feedbacks on Ocean Primary Production. Science. 1998;281:200–207. 10.1126/science.281.5374.200. [Abstract] [CrossRef] [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–240. 10.1126/science.281.5374.237. [Abstract] [CrossRef] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Research. 2014;42:D222–230. 10.1093/nar/gkt1223. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Fuhrman JA, Cram JA, Needham DM. Marine microbial community dynamics and their ecological interpretation. Nature Reviews. Microbiology. 2015;13:133–146. 10.1038/nrmicro3417. [Abstract] [CrossRef] [Google Scholar]
- Gerlach W, Stoye J. Taxonomic classification of metagenomic shotgun sequences with CARMA3. Nucleic Acids Research. 2011;39:e11888. 10.1093/nar/gkr225. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Giebel HA, Kalhoefer D, Lemke A, Thole S, Gahl-Janssen R, Simon M, Brinkhoff T. Distribution of Roseobacter RCA and SAR11 lineages in the North Sea and characteristics of an abundant RCA isolate. The ISME Journal. 2011;5:8–19. 10.1038/ismej.2010.87. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappé MS, Short JM, Carrington JC, Mathur EJ. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. 10.1126/science.1114057. [Abstract] [CrossRef] [Google Scholar]
- Gómez-Pereira PR, Fuchs BM, Alonso C, Oliver MJ, van Beusekom JE, Amann R. Distinct flavobacterial communities in contrasting water masses of the north Atlantic Ocean. The ISME Journal. 2010;4:472–487. 10.1038/ismej.2009.142. [Abstract] [CrossRef] [Google Scholar]
- Gómez-Pereira PR, Schüler M, Fuchs BM, Bennke C, Teeling H, Waldmann J, Richter M, Barbe V, Bataille E, Glöckner FO, Amann R. Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environmental Microbiology. 2012;14:52–66. 10.1111/j.1462-2920.2011.02555.x. [Abstract] [CrossRef] [Google Scholar]
- Gómez-Pereira PR, Hartmann M, Grob C, Tarran GA, Martin AP, Fuchs BM, Scanlan DJ, Zubkov MV. Comparable light stimulation of organic nutrient uptake by SAR11 and Prochlorococcus in the North Atlantic subtropical gyre. The ISME Journal. 2013;7:603–614. 10.1038/ismej.2012.126. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Halsey KH, Carter AE, Giovannoni SJ. Synergistic metabolism of a broad range of C1 compounds in the marine methylotrophic bacterium HTCC2181. Environmental Microbiology. 2012;14:630–640. 10.1111/j.1462-2920.2011.02605.x. [Abstract] [CrossRef] [Google Scholar]
- Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19786–19791. 10.1073/pnas.1211002109. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000
 km salinity gradient of the Baltic Sea. The ISME Journal. 2011;5:1571–1579. 10.1038/ismej.2011.41. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
km salinity gradient of the Baltic Sea. The ISME Journal. 2011;5:1571–1579. 10.1038/ismej.2011.41. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar] - Huntemann M, Ivanova NN, Mavromatis K, Tripp HJ, Paez-Espino D, Palaniappan K, Szeto E, Pillay M, Chen I-MA, Pati A, Nielsen T, Markowitz VM, Kyrpides NC. The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4) Standards in Genomic Sciences. 2015;10:86. 10.1186/s40793-015-0077-y. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Hutchinson GE. The Paradox of the Plankton. The American Naturalist. 1961;95:137–145. 10.1086/282171. [CrossRef] [Google Scholar]
- Kabisch A, Otto A, König S, Becher D, Albrecht D, Schüler M, Teeling H, Amann RI, Schweder T. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes 'Gramella forsetii' KT0803. The ISME Journal. 2014;8:1492–1502. 10.1038/ismej.2014.4. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Klindworth A, Mann AJ, Huang S, Wichels A, Quast C, Waldmann J, Teeling H, Glöckner FO. Diversity and activity of marine bacterioplankton during a diatom bloom in the North Sea assessed by total RNA and pyrotag sequencing. Marine Genomics. 2014;18:185–192. 10.1016/j.margen.2014.08.007. [Abstract] [CrossRef] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. 10.1006/jmbi.2000.4315. [Abstract] [CrossRef] [Google Scholar]
- Loebl M, van Beusekom JEE, Philippart CJM. No microzooplankton grazing during a Mediopyxis helysia dominated diatom bloom. Journal of Sea Research. 2013;82:80–85. 10.1016/j.seares.2012.09.010. [CrossRef] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research. 2014;42:D490–D495. 10.1093/nar/gkt1178. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Lucas J, Wichels A, Teeling H, Chafee M, Scharfe M, Gerdts G. Annual dynamics of North Sea bacterioplankton: seasonal variability superimposes short-term variation. FEMS Microbiology Ecology. 2015;91:fiv099. 10.1093/femsec/fiv099. [Abstract] [CrossRef] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. 10.1186/2047-217X-1-18. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Malmstrom RR, Straza TRA, Cottrell MT, Kirchman DL. Diversity, abundance, and biomass production of bacterial groups in the western Arctic Ocean. Aquatic Microbial Ecology. 2007;47:45–55. 10.3354/ame047045. [CrossRef] [Google Scholar]
- Mann DG. The species concept in diatoms. Phycologia. 1999;38:437–495. 10.2216/i0031-8884-38-6-437.1. [CrossRef] [Google Scholar]
- Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic Oligodeoxynucleotide Probes for the Major Subclasses of Proteobacteria: Problems and Solutions. Systematic and Applied Microbiology. 1992;15:593–600. 10.1016/S0723-2020(11)80121-9. [CrossRef] [Google Scholar]
- Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer KH. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. 10.1099/13500872-142-5-1097. [Abstract] [CrossRef] [Google Scholar]
- Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Pillay M, Ratner A, Huang J, Pagani I, Tringe S, Huntemann M, Billis K, Varghese N, Tennessen K, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Research. 2014;42:D568–573. 10.1093/nar/gkt919. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Martinez-Garcia M, Brazel DM, Swan BK, Arnosti C, Chain PS, Reitenga KG, Xie G, Poulton NJ, Lluesma Gomez M, Masland DE, Thompson B, Bellows WK, Ziervogel K, Lo CC, Ahmed S, Gleasner CD, Detter CJ, Stepanauskas R. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PloS One. 2012;7:e11888. 10.1371/journal.pone.0035314. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Pühler A. GenDB--an open source genome annotation system for prokaryote genomes. Nucleic Acids Research. 2003;31:2187–2195. [Europe PMC free article] [Abstract] [Google Scholar]
- Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS, Rocap G. Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. The ISME Journal. 2010;4:673–685. 10.1038/ismej.2010.4. [Abstract] [CrossRef] [Google Scholar]
- Muñoz-Marín MC, Luque I, Zubkov MV, Hill PG, Diez J, García-Fernández JM. Prochlorococcus can use the Pro1404 transporter to take up glucose at nanomolar concentrations in the Atlantic Ocean. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8597–8602. 10.1073/pnas.1221775110. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Nelson DM, Tréguer P, Brzezinski MA, Leynaert A, Quéguiner B. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochemical Cycles. 1995;9:359–372. 10.1029/95GB01070. [CrossRef] [Google Scholar]
- Nielsen H, Brunak S, von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Engineering. 1999;12:3–9. 10.1093/protein/12.1.3. [Abstract] [CrossRef] [Google Scholar]
- Niu Y, Shen H, Chen J, Xie P, Yang X, Tao M, Ma Z, Qi M. Phytoplankton community succession shaping bacterioplankton community composition in Lake Taihu, China. Water Research. 2011;45:4169–4182. 10.1016/j.watres.2011.05.022. [Abstract] [CrossRef] [Google Scholar]
- Olenina I. Biovolumes and size-classes of phytoplankton in the Baltic Sea. HELCOM BaltSea Environ Proc. 2006;106:144. [Google Scholar]
- Passow U. Transparent exopolymer particles (TEP) in aquatic environments. Progress in Oceanography. 2002;55:287–333. 10.1016/S0079-6611(02)00138-6. [CrossRef] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Research. 2012;40:D290–301. 10.1093/nar/gkr1065. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research. 2013;41:D590–596. 10.1093/nar/gks1219. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. 10.1038/nature00917. [Abstract] [CrossRef] [Google Scholar]
- Reddy TB, Thomas AD, Stamatis D, Bertsch J, Isbandi M, Jansson J, Mallajosyula J, Pagani I, Lobos EA, Kyrpides NC. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Research. 2015;43:D1099–1106. 10.1093/nar/gku950. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Ruff SE, Probandt D, Zinkann AC, Iversen MH, Klaas C, Würzberg L, Krombholz N, Wolf-Gladrow D, Amann R, Knittel K. Indications for algae-degrading benthic microbial communities in deep-sea sediments along the Antarctic Polar Front. Deep Sea Research Part II: Topical Studies in Oceanography 2014 [Google Scholar]
- Sapp M, Wichels A, Wiltshire KH, Gerdts G. Bacterial community dynamics during the winter-spring transition in the North Sea. FEMS Microbiology Ecology. 2007;59:622–637. 10.1111/j.1574-6941.2006.00238.x. [Abstract] [CrossRef] [Google Scholar]
- Sarmento H, Gasol JM. Use of phytoplankton-derived dissolved organic carbon by different types of bacterioplankton. Environmental Microbiology. 2012;14:2348–2360. 10.1111/j.1462-2920.2012.02787.x. [Abstract] [CrossRef] [Google Scholar]
- Schattenhofer M, Fuchs BM, Amann R, Zubkov MV, Tarran GA, Pernthaler J. Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environmental Microbiology. 2009;11:2078–2093. 10.1111/j.1462-2920.2009.01929.x. [Abstract] [CrossRef] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. 10.1016/j.cell.2010.05.005. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Applied and Environmental Microbiology. 2007;73:2290–2296. 10.1128/AEM.02559-06. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tada Y, Taniguchi A, Nagao I, Miki T, Uematsu M, Tsuda A, Hamasaki K. Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the western North Pacific Ocean. Applied and Environmental Microbiology. 2011;77:4055–4065. 10.1128/AEM.02952-10. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Tan SL, Xiaoshan Zhu JS, Yu S, Zhan W, Wang B, Cai Z. An association network analysis among microeukaryotes and bacterioplantkon reveals algal bloom dynamics. Journal of Phycology. 2015;5:120–132. 10.1111/jpy.12259. [Abstract] [CrossRef] [Google Scholar]
- Taylor JD, Cottingham SD, Billinge J, Cunliffe M. Seasonal microbial community dynamics correlate with phytoplankton-derived polysaccharides in surface coastal waters. The ISME Journal. 2014;8:245–248. 10.1038/ismej.2013.178. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glöckner FO, Schweder T, Amann R. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–611. 10.1126/science.1218344. [Abstract] [CrossRef] [Google Scholar]
- Thiele S, Fuchs BM, Amann RI. Identification of microorganisms using the ribosomal RNA approach and fluorescence in situ hybridization. In: Wilderer P, editor. Treatise on Water Science. vol 3. 2011. pp. 171–189. [CrossRef] [Google Scholar]
- Voget S, Wemheuer B, Brinkhoff T, Vollmers J, Dietrich S, Giebel HA, Beardsley C, Sardemann C, Bakenhus I, Billerbeck S, Daniel R, Simon M. Adaptation of an abundant Roseobacter RCA organism to pelagic systems revealed by genomic and transcriptomic analyses. The ISME Journal. 2015;9:371–384. 10.1038/ismej.2014.134. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wemheuer B, Wemheuer F, Hollensteiner J, Meyer FD, Voget S, Daniel R. The green impact: bacterioplankton response toward a phytoplankton spring bloom in the southern North Sea assessed by comparative metagenomic and metatranscriptomic approaches. Frontiers in Microbiology. 2015;6:805. 10.3389/fmicb.2015.00805. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wilhelm LJ, Tripp HJ, Givan SA, Smith DP, Giovannoni SJ. Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biology Direct. 2007;2:27. 10.1186/1745-6150-2-27. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Williams TJ, Wilkins D, Long E, Evans F, DeMaere MZ, Raftery MJ, Cavicchioli R. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environmental Microbiology. 2013;15:1302–1317. 10.1111/1462-2920.12017. [Abstract] [CrossRef] [Google Scholar]
- Wiltshire KH, Kraberg A, Bartsch I, Boersma M, Franke H-D, Freund J, Gebühr C, Gerdts G, Stockmann K, Wichels A. Helgoland Roads, North Sea: 45 Years of Change. Estuaries and Coasts. 2010;33:295–310. 10.1007/s12237-009-9228-y. [CrossRef] [Google Scholar]
- Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ. Environmental science. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science. 2015;347:1257594. 10.1126/science.1257594. [Abstract] [CrossRef] [Google Scholar]
- Xing P, Hahnke RL, Unfried F, Markert S, Huang S, Barbeyron T, Harder J, Becher D, Schweder T, Glöckner FO, Amann RI, Teeling H. Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. The ISME Journal. 2015;9:1410–1422. 10.1038/ismej.2014.225. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yang C, Li Y, Zhou B, Zhou Y, Zheng W, Tian Y, Van Nostrand JD, Wu L, He Z, Zhou J, Zheng T. Illumina sequencing-based analysis of free-living bacterial community dynamics during an Akashiwo sanguine bloom in Xiamen sea, China. Scientific Reports. 2015;5:8476. 10.1038/srep08476. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Research. 2012;40:W445–451. 10.1093/nar/gks479. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Yool A, Tyrrell T. Role of diatoms in regulating the ocean's silicon cycle. Global Biogeochemical Cycles. 2003;17:n/a–24. 10.1029/2002GB002018. [CrossRef] [Google Scholar]
- Zubkov MV, Fuchs BM, Tarran GA, Burkill PH, Amann R. High rate of uptake of organic nitrogen compounds by Prochlorococcus cyanobacteria as a key to their dominance in oligotrophic oceanic waters. Applied and Environmental Microbiology. 2003;69:1299–1304. 10.1128/AEM.69.2.1299-1304.2003. [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
Decision letter
[Editors’ note: this article was originally rejected after discussions between the reviewers, but the authors were invited to resubmit after an appeal against the decision.]
Thank you for submitting your work entitled "Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms" for consideration by eLife. Your article has been favorably evaluated by Ian Baldwin (Senior editor) and three reviewers, one of whom is a member of our Board of Reviewing Editors. Our decision has been reached after consultation between the reviewers. Based on these discussions and the individual reviews below, we regret to inform you that your work in its current format will not be considered further for publication in eLife.
We think you have submitted a very interesting study and the reviewers were collectively enthusiastic about the efforts to collect the data. However, at the same time the reviewers also were concerned about the novelty yet provided compared to the Science paper of 2012. They felt that a more rigorous analysis will yield new and interesting insights into bacterial phytoplankton interactions. Furthermore, the reviewers indicated some experiments and also experimental and methodological improvements, which might lead to a study with deeper insight into the interaction of algae and bacteria, but this cannot be in the time schedule we expect for a major revision.
Reviewer #1:
The manuscript describes the characterisation of planktonic blooms following the primary blooms of algae in the southern North Sea. Such successions during spring phytoplankton blooms were investigated for four consecutive years. An enormous amount of data was generated by dense sampling and high-resolution taxonomic analyses. These data are of very high quality. In addition, metagenomic data were provided which revealed recurring patterns at the functional level, in particular concerning polysaccharide degradation. The manuscript is very well written, the outcome is very interesting. The methods used are state of the art. I have two comments to be discussed:
1) The functional analysis is purely based on occurrence of e.g. CAZyme-encoding genes by metagenome sequencing. Of course, the findings are sound, i.e., a succession of bacteria with a distinct set of CAZyme-encoding genes was detected. I am not sure whether it would be possible to also carry out a proteome analysis to demonstrate the presence of these enzymes, many of them can be expected to be extracellular. I am not sure whether this is possible at all.
2) I think the manuscript contains sufficient novelty compared to the Science paper of 2012, where some of the authors showed that substrate-controlled succession of marine bacterioplankton populations is induced by a phytoplankton bloom. In the current manuscript a far more comprehensive analysis was provided.
In general, I think the manuscript reports very interesting data, and also functional relationships. Also, I expect the data being an extremely valuable source for further studies.
Reviewer #2:
This is a significant contribution to the understanding of how bacteria respond to spring phytoplankton blooms. It appears technically sound in design and data reporting, and provides great detail that will inform our understanding of this important phenomenon. My main concern: There are clearly consistencies in the bacterial responses yearly, however the primary conclusion that there are "recurring patterns" of bacterioplankton dynamics is not statistically supported, and frankly vague. A clear quantitative (and justified) definition is needed, otherwise one can focus on what parts repeat and say there is a repeating pattern, or focus on differences and say there isn't. The succession details in their earlier Science paper (Teeling et al., 2012) differ here. If one accepts a broad qualitative definition of "recurring patterns," the results are confirmatory of previous work by the authors and others, cited by recent reviews (Buchan et al., Nature Reviews Microbiology, for example), showing that many taxa, including those reported here, have repeated seasonality or bloom responses. Admittedly few prior studies covered the fine temporal scale measured here, and that is a considerable strength.
To quantitatively show dynamics are recurring, statistics should be applied using each year as a sort of replicate to determine which aspects of the microbial successions repeat, including relationships to algal taxa. The spring "blooms" are quite variable in their dynamics, as acknowledged by the authors, and the response differs, as expected. If bottom up controls are the dominant factor controlling the bacterial composition as suggested, the why are dynamics not similar within a year when there is a secondary peak of chlorophyll after the first bloom (see specific comments)? Could it be top-down controls?
Regarding associations (or lack thereof) with phytoplankton, I think the assessment of the phytoplankton dynamics is not universal enough to allow full conclusions. For instance, only one member an important group of phytoplankton, the haptophytes, is reported (Phaeocystis) – are there no others? Additionally, among the most numerically abundant taxa were the Green Algae, which have many significantly different species, but here they are "lumped" together. It is unlikely that particular microbial interactions would occur with whole families or orders of phytoplankton, but instead be rather very specific (e.g. see Amin et al. 2015 and references therein). Unfortunately, the manuscript only reports a relatively small number of phytoplankton species – and it would be good to see calculations of the fraction of total phytoplankton these represent (e.g. biomass totals to compare with chlorophyll-based totals to see what% may be missing. Some chlorophyll increases were not accompanied on a daily basis with counts). Furthermore, the main way the authors have looked for correlation between phytoplankton and bacteria is via Spearman's correlation for apparently all of the years together. By visual examination, many of the "responding" bacteria seem to have a delayed reaction to the increases in chlorophyll, so the authors should also look for time-delayed correlations, in addition to year-by-year studies.
CARD-FISH identified only about 70% of the bacteria, a shortcoming to understanding the full community response, addressable with their less frequent 16S observations. The broad groups appear to have little consistent response to the 'blooms.' Why no direct comparisons between CARD-FISH and 16S sequences? Some groups explored with the 16S not found in CARD-FISH are generally ignored, but CARD-FISH could represent as little as about 40% during blooms. Notable such bacteria include Verrucomicrobia (increases in May 2010, 2011,2012). Also, chloroplast data should be separated from figures of 16S tag sequencing results because they are not bacteria or archaea.
Important: Is the variability observed largely temporal (local successions) or is it a mixture of spatial and temporal variability (most likely)? Are the blooms very patchy or regional? Satellite imagery would help, even occasional. Minimally – deserves discussion.
Reviewer #3:
This manuscript describes an impressive campaign to characterize the bacterial communities in a coastally influenced environment over the course of consecutive several years. These types of long-term, high-resolution temporal studies have been previously restricted to more oligotrophic environments. Here, the authors focus their analysis on bacterial communities associated with the seasonal phytoplankton blooms that occur in their study site, with the broader goal of providing a better understanding of the underlying drivers of the recurrence of specific bacterial lineages/groups.
The scientific methods are solid, and I have no major worries regarding the quality of the data. My main concern (indeed, personal struggle) is that I am unsure of the scientific impact of this work, at least as currently presented in the manuscript. While this is a truly impressive dataset and one that should be published/disseminated, the main conclusions are that some bacterial lineages (and some of the functions they encode) show recurring seasonal and annual patterns of occurrence. Linkages between specific bacterial taxa, functions and environmental parameters (principally those associated with phytoplankton blooms) are evident. Yet, as the authors report, this has been shown previously; albeit these "elsewhere" environments are generally more oligotrophic, and perhaps subject to less environmental variability (though that is debatable) than the one characterized here. The authors frame their conclusions on the nature of the succession of bacterial groups as being one that is influenced by both "stochastic processes" and "deterministic principles", however, I did not find the manuscript to be convincingly clear on what they mean by these statements.
Given the criteria for acceptance in this journal – "highly influential research" –, I am of the position that the authors need to dig deeper to draw more novel conclusions with respects to what this time-series analysis reveals regarding environmental parameters and bacterial structure and function. The volume and nature of these data suggest that would be appropriate for some modeling efforts. This type of exercise might better support the authors conclusions and illustrate the emergent properties/recurrent patterns of specific bacterial lineages.
[Editors’ note: what now follows is the decision letter after the authors submitted for further consideration.]
Thank you for resubmitting your work entitled "Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms" for further consideration at eLife. Your revised article has been favorably evaluated by Ian Baldwin (Senior editor) and three reviewers, one of whom is a member of our Board of Reviewing Editors. The manuscript has been improved but there are some remaining issues that need to be addressed before acceptance, as outlined below:
Although we acknowledge that the revised version was considerably improved we still felt that some points made by the reviewers were valid and simply disagreed by the authors. On the other side, we also see that the dataset is very valuable and that the authors had made more new, convincing, and quantitative conclusions. There are several points which need to be addressed. In addition to those outlined by the reviewers. We ask you to tone down the language associated with the conclusions, especially those related to the presumptive underlying drivers of the observed patterns ("substrate-induced forcing"). This conclusion is based on metagenomic analysis and functions that have not been shown conclusively. We would like to ask you to modify the text accordingly. For instance, in the Abstract "We thereby demonstrate that even though there is substantial inter-annual variation between spring phytoplankton blooms, the accompanying succession of bacterial clades is governed by deterministic principles such as substrate-induced forcing" could be modified to something like: "We thereby hypothesize that even though…".
Reviewer #1:
The study reports the characterisation of planktonic blooms following the primary blooms of algae in the southern North Sea. Successions during spring phytoplankton blooms have been studied for four consecutive years. An enormous amount of data was collected by dense sampling and high-resolution taxonomic analyses. As I wrote in my first report, these data are of very high quality. In addition, metagenomic data were provided which revealed recurring patterns at the functional level, in particular concerning polysaccharide degradation.
The revised version of the manuscript was considerably improved. At least to me, many questions were well addressed. The manuscript is very well written, the outcome is very interesting. The methods used are state of the art. The discrepancy of data obtained by CARD-FISH and 16S rRNA tag sequencing was well discussed.
The authors added additional data such as the full set of physicochemical measurements and the data of the dominating algae taxa. The number of figures is extraordinary, but I agree with the authors, that is required to show the full data set in a reasonable manner. This is one of the reasons, why I expect the data being an extremely valuable source for further studies.
Reviewer #2:
We thank the authors for their response to our criticism. We appreciate their detailed and extended response to many of our technical suggestions, of which they responded to compellingly. The dataset and extensive analysis is indeed very impressive and valuable. However, we were surprised and disappointed that the authors made relatively few changes in their manuscript in response to our main concerns, so our view of the manuscript in terms of the novelty of general conclusions and quantitative assessment of "recurring patterns" has not significantly changed. We regret we cannot be more positive after reading their response.
We do now better understand what the authors mean by "recurring patterns,", i.e., that many clades appear at >5% abundance more than once following a spring bloom. However, we find this general observation itself to not be exceptionally novel. Note, for example, in the abstract as background for the study, the authors state that Flavobacteria, Gammaproteobacteria, and alphaproteobacteria are known associated with blooms (Abstract). Earlier published studies include reports where the same taxa are found in different years, including after blooms (admittedly with much less comprehensive overall system analysis than this study).
Specific suggestion: In the figures they should use arrows or other notation to point out the recurring clades and when they bloom.
Reviewer #3:
I feel that the revised manuscript is significant improved with respects to readability and articulation of the importance of the work.
Author response
[Editors’ note: the author responses to the first round of peer review follow.]
We think you have submitted a very interesting study and the reviewers were collectively enthusiastic about the efforts to collect the data. However, at the same time the reviewers also were concerned about the novelty yet provided compared to the Science paper of 2012. They felt that a more rigorous analysis will yield new and interesting insights into bacterial phytoplankton interactions. Furthermore, the reviewers indicated some experiments and also experimental and methodological improvements, which might lead to a study with deeper insight into the interaction of algae and bacteria, but this cannot be in the time schedule we expect for a major revision. Reviewer #1:
The manuscript describes the characterisation of planktonic blooms following the primary blooms of algae in the southern North Sea. Such successions during spring phytoplankton blooms were investigated for four consecutive years. An enormous amount of data was generated by dense sampling and high-resolution taxonomic analyses. These data are of very high quality. In addition, metagenomic data were provided which revealed recurring patterns at the functional level, in particular concerning polysaccharide degradation. The manuscript is very well written, the outcome is very interesting. The methods used are state of the art. I have two comments to be discussed:
1) The functional analysis is purely based on occurrence of e.g. CAZyme-encoding genes by metagenome sequencing. Of course, the findings are sound, i.e., a succession of bacteria with a distinct set of CAZyme-encoding genes was detected. I am not sure whether it would be possible to also carry out a proteome analysis to demonstrate the presence of these enzymes, many of them can be expected to be extracellular. I am not sure whether this is possible at all.
We did carry out ten metaproteome analyses of soluble fractions of bacterioplankton cells that correspond to the 10 new metagenomes of 2010 to 2012 that we present in our study. The smallest metaproteome dataset of the 2010 to 2012 blooms comprises only 1,058 and the largest 3,994 proteins (25,832 proteins for all 10 time points combined). Since the blooms of 2010 to 2012 were also less intense than the bloom of 2009, these metaproteomes yielded mostly conventional highly expressed proteins such as chaperonins, ribosomal proteins and transporters. While the proteomes were not deep enough to detect many expressed CAZymes, we could however, as in 2009, detect remarkably high levels of TonB-dependent outer membrane receptors/transporters among the proteins with the highest expression levels. We will use this expression of TonB-dependent receptors as an indirect measure for the expression of CAZyme-encoding polysaccharide utilization loci (PULs) in a future study. In order to achieve this, we have systematically isolated and sequenced close to 50 genomes of North Sea Flavobacteriia. We are working on a systematic investigation of the co-localization of TonB-dependent receptors (TBDRs) and CAZymes in PULs of these isolates and predict polysaccharide substrate specificities. We will subsequently use the metaproteome data in conjunction with these (meta-)genomic data to use TBDR expression as a proxy for expression of CAZymes. This is a much more laborious, but also targeted approach, which has the potential for additional insights on relevant polysaccharide substrates, which we plan to publish in a separate paper.
2) I think the manuscript contains sufficient novelty compared to the Science paper of 2012, where some of the authors showed that substrate-controlled succession of marine bacterioplankton populations is induced by a phytoplankton bloom. In the current manuscript a far more comprehensive analysis was provided. In general, I think the manuscript reports very interesting data, and also functional relationships. Also, I expect the data being an extremely valuable source for further studies. We would like to thank reviewer #1 for this statement. The dataset we present is indeed unusually large (e.g. 10x more metagenomic information than in the 2012 Science paper), which prevents analysis of every possible aspect. Instead, it is our intention to explain the dataset and to focus the analyses of biodiversity and gene function repertoires on selected relevant key aspects in continuation of our initial 2012 Science publication.
We agree with reviewer #1 that the dataset will allow us and others to explore additional aspects, for example the diversity and genomic potential of bacterioplankton groups that were not in our focus, such as for example members of the alphaproteobacterial Roseobacter or the betaproteobacterial OM43 clades, or to focus on other relevant gene functions besides those involved in the concerted decomposition of polysaccharides.
In order to make the dataset more accessible, we now include the full set of physicochemical measurements and the data of the dominating algae taxa as supplementary material (new Supplementary files 1 and 2).
Reviewer #2:
This is a significant contribution to the understanding of how bacteria respond to spring phytoplankton blooms. It appears technically sound in design and data reporting, and provides great detail that will inform our understanding of this important phenomenon. My main concern: There are clearly consistencies in the bacterial responses yearly, however the primary conclusion that there are "recurring patterns" of bacterioplankton dynamics is not statistically supported, and frankly vague. A clear quantitative (and justified) definition is needed, otherwise one can focus on what parts repeat and say there is a repeating pattern, or focus on differences and say there isn't.
We are communicating a very densely sampled large dataset and for reasons of practicality we focused on the major clades. Within these clades, there is a clear pattern of recurrence, not in the sense that the order of clades is identical each year (which is not expected due to variations in phytoplankton dynamics), but in the sense that these clades recur from year to year, which we prefer to show as original data.
We used a quantitative criterion of equal or larger than 5% relative abundance in at least two of the analyzed four for the most abundant clades, and some of the clades (defined mostly on the level of genus) that we describe responded with even much higher peaks. We made this more clear in the revised manuscript (subsection “bacterioplankton - diversity and bloom characteristics”, fourth paragraph). Annual recurrence of these clades is independent of any stochastic occurrence of other clades, and this means that we did not hand-pick results in order to make up recurrence as this reviewer implies. We focused our study further to clades from Bacteroidetes and Gammaproteobacteria, two clades known for their potential to degradepolysaccharides and demonstrated in the 2012 Teeling paper to respond to phytoplankton blooms. We only touch upon other abundant groups such as SAR11, Roseobacter or OM43 that lack a specific potential for polysaccharide degradation.
The succession details in their earlier Science paper (Teeling et al., 2012) differ here. If one accepts a broad qualitative definition of "recurring patterns," the results are confirmatory of previous work by the authors and others, cited by recent reviews (Buchan et al., Nature Reviews Microbiology, for example), showing that many taxa, including those reported here, have repeated seasonality or bloom responses. Admittedly few prior studies covered the fine temporal scale measured here, and that is a considerable strength.
The purpose of this study was to test our hypothesis that spring phytoplankton blooms trigger rapid successions of flavobacterial and gammaproteobacterial clades specialized for polysaccharide degradation and to investigate this succession in a very comprehensive dataset collected in four consecutive years.
To the best of our knowledge so far nobody has performed a polyphasic study of this scope including FISH cell counts, diversity assessment by tag sequencing, and metagenomic profiling for the same study site over a multiple year time span. To cite the 2014 review of Alison Buchan et al. that this reviewer brings forward:
"However, such studies often differ with respect to bloom location, duration and intensity of sampling, phytoplankton species succession, geochemistry and temperature, which prevents quantitative generalizations of bacterial composition and responses."
Here we address exactly this problem by presenting a high temporal resolution study from one and the same location that spans four consecutive years.
To quantitatively show dynamics are recurring, statistics should be applied using each year as a sort of replicate to determine which aspects of the microbial successions repeat, including relationships to algal taxa.
As stated above, the recurrence of abundant taxa can visually be seen in the data we show. We actually selected eLife because it is an electronic journal able to accommodate significantly more of our high-resolution original data than a printed journal. Any statistical approach with n=4, i.e. four years as replicates, has severe limitations, yet we show recurrence of specific clades in 3-4 out of 4 years. Likewise, we could further extend the analysis that we already did with respect to the relationships of specific bacterial genera to algal taxa. However, we would like to make this reviewer aware of the fact that our study deals with free-living bacteria in the size fraction 0.2 – 3 µm and not with bacteria that are directly algae-associated such as phycosphere bacteria. In any case, the statistical analyses provided show that there is no simple one to one relationship between occurrence of dedicated algae taxa and dedicated clades of free-living bacteria. This is actually a major new result of this study, consistent with our hypothesis that the availability of substrates commonly occurring in microalgae such as laminarin are a major cause for the bacterial succession, and more important than one-to-one interactions of particular phytoplankton and bacterioplankton species (see Discussion, eighth paragraph). This is now also stated in the final sentence of the concluding remarks (see end of Discussion).
The spring "blooms" are quite variable in their dynamics, as acknowledged by the authors, and the response differs, as expected. If bottom up controls are the dominant factor controlling the bacterial composition as suggested, the why are dynamics not similar within a year when there is a secondary peak of chlorophyll after the first bloom (see specific comments)? Could it be top-down controls? We designed the study in a way that it is strictly focused on the first spring bloom phase, since at the end of the winter period top-down factors should beminimal. Hence, we rephrased as follows:
“Dominance of bottom-up over top-down control is assumed to be characteristic for the initial phases of spring phytoplankton blooms. […] Hence, top-down control by predation sets in only during later bloom phases. This situation is distinct from summer and fall phytoplankton blooms.”
By no means we stated that bottom-up control is the dominant factor for all taxa for the entire year, but bottom-up control likely dominates during early bloom stages when predators and viruses are not yet abundant. Of course top-down control by grazing or virus mortality cannot be excluded, in particular during later bloom phases in which successions would be much more difficult to predict.
Regarding associations (or lack thereof) with phytoplankton, I think the assessment of the phytoplankton dynamics is not universal enough to allow full conclusions. For instance, only one member an important group of phytoplankton, the haptophytes, is reported (Phaeocystis) – are there no others?
In fact, we have very detailed phytoplankton data for all four years, and besides Phaeocystis, we discuss Chaetoceros debilis, Chaetoceros minimus, Mediopyxis helysia, Rhizosolenia styliformis and Thalassiosira nordenskioeldii, Chattonella spp., and Phaeocystis spp. as dominant groups.Likewise, all of these groups and the major phytoplankton clades were statistically tested for association. We added the respective primary data as new Supplementary file 2.
Additionally, among the most numerically abundant taxa were the Green Algae, which have many significantly different species, but here they are "lumped" together.
Green algae simply did not play a major role during the North Sea spring phytoplankton blooms that we analyzed, which in most cases are dominated by diatoms. This is why we did not include any information on this paraphyletic group in the abundance plots (Figure 3) and treated them as one group in statistical analyses. We included the primary algae count data (new Supplementary file 2) for reference that shows that the numbers of green algae were minute compared to the major algae taxa. We do have high-resolution data of many more algae taxa at hand (more than 200 algae taxa are continuously identified at Helgoland in the framework of the Helgoland Roads time series since 1962), and these data are available on request. (see: https://www.awi.de/forschung/biowissenschaft/oekologie-der-kuesten/arbeitsgruppen/ag-langzeitoekologieforschung-old-version/long-term-observations/helgoland-roads.html)
It is unlikely that particular microbial interactions would occur with whole families or orders of phytoplankton, but instead be rather very specific (e.g. see Amin et al. 2015 and references therein). Unfortunately, the manuscript only reports a relatively small number of phytoplankton species – and it would be good to see calculations of the fraction of total phytoplankton these represent (e.g. biomass totals to compare with chlorophyll-based totals to see what% may be missing. Some chlorophyll increases were not accompanied on a daily basis with counts).
We concur that associations of algae and phycosphere bacteria can be quite specific, but those bacteria were not subject of our study in which we examined the 0.2 – 3 µm fraction. We did include species level assignments of phytoplankton that was dominant in terms of biomass (Figure 3A-D bold lines) except for the raphidophyte Chattonella that dominated in 2012. In this context we would like to point out to this reviewer that the 2012 bloom was distinct from the usually diatom-dominated spring blooms at Helgoland, and still we found the same bacterioplankton clades responding. This is a clear indication against the proposed strong effect of algal identity on the sampled 0.2 – 3 µm bacterioplankton fraction. It is actually among the main outcomes of this study that indeed families and orders of phytoplankton and not particular species are important during spring blooms in the Southern North Sea, i.e. all diatoms and several other microalgae have laminarin as the main storage polysaccharide.
We are sorry that within this multi-year study we were not able to provide daily measurements for all parameters. As we explained in the manuscript, there is no strict one-to-one correlation between algae biomass and chlorophyll a. Reliable biomass data is very hard to obtain, and this reviewer certainly agrees that sizes of algae depend heavily on the physiological state. There are of course published values for all of the major taxa that occurred during the blooms we investigated, but usage of such values would be very rough estimations with a considerable margin of error, which is why we did abstain from including such error-prone biomass data in statistical analyses.
Furthermore, the main way the authors have looked for correlation between phytoplankton and bacteria is via Spearman's correlation for apparently all of the years together. By visual examination, many of the "responding" bacteria seem to have a delayed reaction to the increases in chlorophyll, so the authors should also look for time-delayed correlations, in addition to year-by-year studies.
We agree that a delayed reaction of the responding bacteria is obvious. For a brief analysis of such delays, lagged correlation approaches (LSA, localsimilarity analysis) could be applied generally. However, for these analyses, consistent and coherent datasets are required without any missing values. Due to inherent limitations of field sampling, such datasets are scarce and only few studies fulfilling the LSA requirements were published until now. In the context of our study consistency was hard to achieve due to the unpredictability of the exact onset of the spring phytoplankton blooms.
We did in fact compute forward regressions for each year that yielded no significant correlations. Only a lagged co-occurrence could be proven since the interrelation-mediating metabolites (e.g. polysaccharides) were not analyzed, since this is technically extremely demanding. Future studies will need to include quantitative analyses of structurally resolved polysaccharides.
CARD-FISH identified only about 70% of the bacteria, a shortcoming to understanding the full community response, addressable with their less frequent 16S observations. The broad groups appear to have little consistent response to the 'blooms.' Why no direct comparisons between CARD-FISH and 16S sequences?
CARD-FISH does have the decided advantage over 16S rRNA tag sequencing that it provides reliable cell counts. The disadvantage is that it is a laborious technique when done for a very large number of samples, which is why regrettably few studies include such data. In contrast to many other high-profile microbiome studies we actually provide exact numbers for those clades expected to recur. In order to counteract the disadvantages that we only count what we are probing for by CARD-FISH, we have investigated all samples by in-depth 16S rRNA tag sequencing to also identify novel clades. The tag data that we provide represent a dense series for 2010 to 2012 and all of these samples were deeply sequenced (rarefaction analyses not included in the manuscript show that we sequenced multiple times beyond exhaustion of any increase in novel taxa).
As for a direct comparison of CARD-FISH and tag data, we would like to remind this reviewer that perfect consensus of both methods is not possible for fundamental reasons, because the taxon frequency obtained by tag sequencing are biased among other factors by differential PCR amplification. Also CARD-FISH probe specificities and taxonomic affiliations based on tag sequencing do not match perfectly. This is well known and inherent to the way both technologies work. We advise to use the probe match tool implemented in the Silva database for an exact evaluation of the groups that the CARD-FISH probes that we used target. All of these probes are published (see the references that we provided), including information on their specificities. A technical discussion of multiple reasons why CARD-FISH numbers and tag frequencies might differ is beyond the scope of this manuscript.
Some groups explored with the 16S not found in CARD-FISH are generally ignored, but CARD-FISH could represent as little as about 40% during blooms. Notable such bacteria include Verrucomicrobia (increases in May 2010, 2011,2012). Also, chloroplast data should be separated from figures of 16S tag sequencing results because they are not bacteria or archaea. This is a hypothesis-driven study in which we indeed monitored particular bacterial clades using of CARD-FISH. The reviewer is thus correct that we did not include all abundant bacterial groups in the Discussion. Since the intention of this paper was not the general assessment of all aspects of bacterial diversity we in particular “neglected” groups such as SAR11, OM43 or Roseobacter, which are known to lack significant polysaccharide degradationpotential. A complete discussion of all data retrieved is simply impossible for a dataset of such size and hence we focused on key aspects that we think are prominent and of major importance. However, based on the reviewer’s remark we included a short comment on Verrucomicrobia (subsection “bacterioplankton - diversity and bloom characteristics”, third paragraph), which are for sure a very interesting group that responded to the investigated spring phytoplankton blooms with decreasing maximum abundances of 7.7% (2010), 5.0% (2011) and 2.9% (2012). This decrease correlates with decreasing bloom intensities, which agrees with a proposed role of Verrucomicrobia in polysaccharide decomposition (e.g. Manuel Martinez-Garcia et al., 2012).
As suggested we removed the chloroplast data and all tags ascribed as Eukaryota from the calculations of tag frequencies and recalculated all graphsin the respective figure (now Figure 4) accordingly.
Important: Is the variability observed largely temporal (local successions) or is it a mixture of spatial and temporal variability (most likely)? Are the blooms very patchy or regional? Satellite imagery would help, even occasional. Minimally – deserves discussion.
Based on the reviewer’s remarks we added satellite data (new Figure 2), which we already had analyzed, yet did not include. These data indicate that the variability is largely temporal, but there are of course also spatial influences. We discussed this in detail for the Helgoland sampling site in our Science publication of the 2009 spring bloom using satellite images and particle drift simulations. Particle drift simulation data could be computed and made available on request, but would further inflate the dataset.
[Editors’ note: the author responses to the re-review follow.]
The manuscript has been improved but there are some remaining issues that need to be addressed before acceptance, as outlined below: Although we acknowledge that the revised version was considerably improved we still felt that some points made by the reviewers were valid and simply disagreed by the authors. On the other side, we also see that the dataset is very valuable and that the authors had made more new, convincing, and quantitative conclusions. There are several points which need to be addressed. In addition to those outlined by the reviewers. We ask you to tone down the language associated with the conclusions, especially those related to the presumptive underlying drivers of the observed patterns ("substrate-induced forcing"). This conclusion is based on metagenomic analysis and functions that have not been shown conclusively. We would like to ask you to modify the text accordingly. For instance, in the Abstract "We thereby demonstrate that even though there is substantial inter-annual variation between spring phytoplankton blooms, the accompanying succession of bacterial clades is governed by deterministic principles such as substrate-induced forcing" could be modified to something like: "We thereby hypothesize that even though…".
We agree that our data does not provide a direct proof of deterministic effects such as substrate-induced forcing, but rather provides indirect indications that such effects might play a major role during spring bloom successions. We hence toned down the language of our conclusions throughout the manuscript as requested. The above-mentioned part of the Abstract was rephrased to:
"We therefore hypothesize that even though there is substantial inter-annual variation between spring phytoplankton blooms, the accompanying succession of bacterial clades is largely governed by deterministic principles such as substrate-induced forcing."
In the Results section, we rephrased as follows:
"Metagenome taxonomic classification provided sufficient data for analysis of CAZyme repertoires of the flavobacterial NS5 marine group, NS3a marine group, Formosa, Polaribacter, and Cryomorphaceae, and the gammaproteobacterial Alteromonadales and Reinekea clades (Supplementary file 10). For most of these clades, the analyses revealed fingerprint-like patterns, which corroborates the hypothesis that these clades have distinct glycan niches that are relatively stable across years."
In the Discussion, we rephrased as follows:
"The existence of recurrent key players during North Sea spring phytoplankton blooms suggests that the bacterioplankton community composition during and after such blooms is governed by deterministic effects."
And finally in the concluding remarks, we rephrased the respective statement as follows:
"Our results suggest that besides stochastic also deterministic effects influence phytoplankton-bacterioplankton coupling during blooms."
Reviewer #2:
We thank the authors for their response to our criticism. We appreciate their detailed and extended response to many of our technical suggestions, of which they responded to compellingly. The dataset and extensive analysis is indeed very impressive and valuable. However, we were surprised and disappointed that the authors made relatively few changes in their manuscript in response to our main concerns, so our view of the manuscript in terms of the novelty of general conclusions and quantitative assessment of "recurring patterns" has not significantly changed. We regret we cannot be more positive after reading their response. We do now better understand what the authors mean by "recurring patterns,", i.e., that many clades appear at >5% abundance more than once following a spring bloom. However, we find this general observation itself to not be exceptionally novel. Note, for example, in the abstract as background for the study, the authors state that Flavobacteria, Gammaproteobacteria, and alphaproteobacteria are known associated with blooms (Abstract). Earlier published studies include reports where the same taxa are found in different years, including after blooms (admittedly with much less comprehensive overall system analysis than this study).
We again evaluated the critique of reviewer #2 thoroughly, but continue to disagree with respect to the "quantitative assessment of 'recurring patterns'". We did explain the extent of recurrence and what we deem are patterns. We for sure stand by our interpretation of the data we present. As for novelty, we do not claim that we are the first who found blooms of Flavobacteriia, Gammaproteobacteria and Roseobacter clade Alphaproteobacteria during and after phytoplankton blooms. However, our analysis extends far beyond these high-level taxa and we present high-time resolution data from four consecutive blooms in order to show that many taxa are recurring on the level of genus. To the best of our knowledge, this has not been shown before.
Specific suggestion: In the figures they should use arrows or other notation to point out the recurring clades and when they bloom.
All taxa are coded by identical colors throughout years. We tested inclusion of arrows and found the result not convincing. We will work with the eLife production team towards the best possible way of visualizing this dataset.
Articles from eLife are provided here courtesy of eLife Sciences Publications, Ltd
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Molecular insights into the hydrolysis and transglycosylation of a deep-sea <i>Planctomycetota</i>-derived GH16 family laminarinase.
Appl Environ Microbiol, 90(10):e0094224, 17 Sep 2024
Cited by: 0 articles | PMID: 39287396
Recovery of 240 metagenome-assembled genomes from coastal mariculture environments in South Korea.
Sci Data, 11(1):902, 20 Aug 2024
Cited by: 1 article | PMID: 39164259 | PMCID: PMC11336123
Genes for laminarin degradation are dispersed in the genomes of particle-associated Maribacter species.
Front Microbiol, 15:1393588, 12 Aug 2024
Cited by: 0 articles | PMID: 39188312 | PMCID: PMC11345257
Phaeocystis globosa and diatom blooms promote distinct bacterial communities and associations in a coastal ecosystem.
Environ Microbiol Rep, 16(4):e13313, 01 Aug 2024
Cited by: 1 article | PMID: 38988030 | PMCID: PMC11236930
Alpha-glucans from bacterial necromass indicate an intra-population loop within the marine carbon cycle.
Nat Commun, 15(1):4048, 14 May 2024
Cited by: 1 article | PMID: 38744821 | PMCID: PMC11093988
Go to all (176) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - SRA278189
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Annual dynamics of North Sea bacterioplankton: seasonal variability superimposes short-term variation.
FEMS Microbiol Ecol, 91(9):fiv099, 21 Aug 2015
Cited by: 18 articles | PMID: 26298013
Dissolved storage glycans shaped the community composition of abundant bacterioplankton clades during a North Sea spring phytoplankton bloom.
Microbiome, 11(1):77, 17 Apr 2023
Cited by: 15 articles | PMID: 37069671 | PMCID: PMC10108472
Dilution cultivation of marine heterotrophic bacteria abundant after a spring phytoplankton bloom in the North Sea.
Environ Microbiol, 17(10):3515-3526, 05 May 2014
Cited by: 37 articles | PMID: 24725270
A multiomics approach to study the microbiome response to phytoplankton blooms.
Appl Microbiol Biotechnol, 101(12):4863-4870, 19 May 2017
Cited by: 2 articles | PMID: 28526980
Review
Funding
Funders who supported this work.