Abstract
Free full text

Inhibition of class I histone deacetylases blunts cardiac hypertrophy through TSC2-dependent mTOR repression
Abstract
Altering chromatin structure through histone posttranslational modifications has emerged as a key driver of transcriptional responses in cells. Modulation of these transcriptional responses by pharmacological inhibition of class I histone deacetylases (HDACs), a group of chromatin remodeling enzymes, has been successful in blocking the growth of some cancer cell types. These inhibitors also attenuate the pathogenesis of pathological cardiac remodeling by blunting and even reversing pathological hypertrophy. The mechanistic target of rapamycin (mTOR) is a critical sensor and regulator of cell growth that as part of mTOR complex I (mTORC1) drives changes in protein synthesis and metabolism in both pathological and physiological hypertrophy. Here, we demonstrated through pharmacological and genetic methods that inhibition of class I HDACs suppressed pathological cardiac hypertrophy through inhibition of mTOR activity. Mice genetically silenced for HDAC1 and HDAC2 had a reduced hypertrophic response to TAC and showed reduced mTOR activity. We determined that the abundance of tuberous sclerosis complex 2 (TSC2), an mTOR inhibitor, was increased through a transcriptional mechanism in cardiomyocytes when class I HDACs were inhibited. In neonatal rat cardiomyocytes, loss of TSC2 abolished HDAC-dependent inhibition of mTOR activity, and increased expression of TSC2 was sufficient to reduce hypertrophy in response to phenylephrine. These findings point to mTOR and TSC2-dependent control of mTOR as critical components of the mechanism by which HDAC inhibitors blunt pathological cardiac growth. These results also suggest a strategy to modulate mTOR activity and facilitate the translational exploitation of HDAC inhibitors in heart disease.
INTRODUCTION
Hypertrophic growth of the myocardium accompanies most forms of heart disease, the leading cause of death in industrialized nations (1). In various animal models, therapeutic targeting of this hypertrophic response affords benefit (2–4). Indeed, hypertrophic growth of the myocardium has been proposed as a therapeutic target in heart disease. However, translation of preclinical observations demonstrating benefit to specific therapies with clinical efficacy has remained elusive.
Reversible protein acetylation, controlled by enzymes which attach (histone acetyltransferases) or remove (histone deacetylases, HDACs) acetyl groups at lysine residues, is a major mechanism governing various cellular processes, both transcriptional and posttranscriptional. In the context of cardiac hypertrophy, small molecule HDAC inhibitors attenuate pathological cardiac remodeling, including hypertrophic growth, fibrosis, and declines in contractile function (2, 5, 6). Indeed, HDAC inhibition can reverse each of these maladaptive processes, even in the presence of persistent afterload stress and without provoking decreases in ventricular performance (6). These exciting data raise the prospect of therapeutic targeting of pathological cardiac growth.
Pharmacological suppression of HDACs has emerged with promise in oncology, and four HDAC inhibitors have received regulatory approval for use in patients. With respect to the myocardium, however, mechanisms whereby HDACs govern cardiomyocyte growth remain poorly defined. As elucidating mechanisms linking HDACs and cardiomyocyte growth is central to the path of translating this biology to the clinic, we set out to define underlying mechanisms.
RESULTS
Inhibition of class I HDACs attenuates cardiomyocyte growth through mTOR signaling
Class I HDACs are held to promote pathological cardiac growth, because pharmacological inhibition of their enzymatic activity blunts phenylephrine (PE)-induced hypertrophy in vitro (neonatal rat ventricular myocytes, NRVMs) and in vivo [thoracic aortic constriction (TAC)-induced pressure overload in mice] (2, 7). To begin to define mechanisms whereby inhibition of class I HDACs blunts cardiac growth, we exposed NRVMs to various growth cues: PE, an α-1 adrenergic receptor agonist; endothelin-1 (ET-1); insulin-like growth factor-1 (IGF-1), an inducer of physiological growth; and a 50% hypo-osmotic solution, a model of mechanical stretch. Cardiomyocyte growth was measured as tritiated leucine (3H-leucine) incorporation. As expected, each of these growth stimuli elicited robust increases in 3H-leucine incorporation at 48 hours (Fig. 1A). Also as expected (7), apicidin, a class I-specific HDAC inhibitor (fig. S1A), blunted the pathological growth triggered by PE (Fig. 1A). Trichostatin A (8), a broad-spectrum HDAC inhibitor (5), had similar effects (figs. S1B, S1C). Apicidin also suppressed hypertrophic growth triggered by ET-1 and by mechanical stretch, two additional cues of pathological growth. In each case, apicidin blunted 3H-leucine incorporation by ≈50% (Fig. 1A). Cell toxicity was minimal under these conditions, except for the longest exposures to the highest concentrations of TSA and apicidin (fig. S2A, S2B, S2C, S2D). Together, these data suggested that class I HDACs governed a cardiomyocyte growth mechanism.
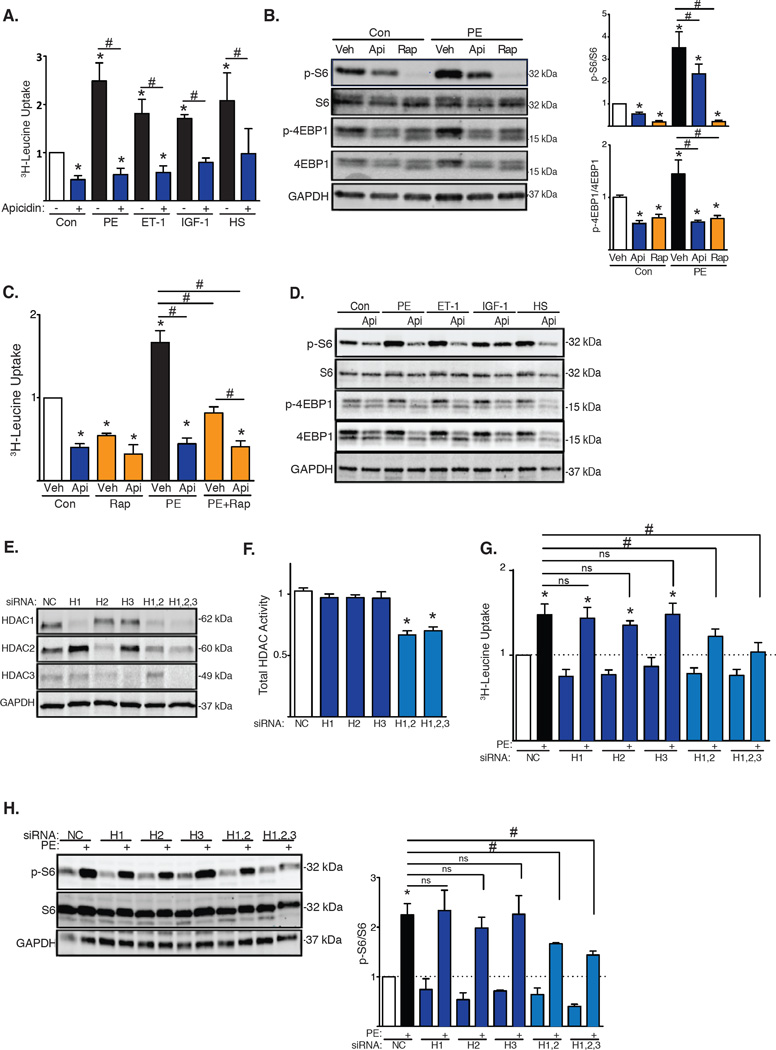
(A) 3H-Leucine incorporation in NRVMs stimulated with PE, ET-1, IGF1, or 50% hypo-osmotic solution (HS) and exposed to the HDAC inhibitor apicidin (Api) (n=4). Con, control. (B) Western blot of mTOR downstream targets in NRVMs treated with HDAC inhibitors or rapamycin for 6h with quantification of S6 and 4EBP1 phosphorylation (n=6). p, phospho. (C) 3H-Leucine incorporation in NRVMs exposed to HDAC inhibitors and/or rapamycin (n=3–6). (D) Western blot for mTOR activity in NRVMs treated with PE, ET-1, IGF-1, or 50% HS and apicidin (n=3). (E) Western blot analysis of HDAC1, HDAC2 and HDAC3 knockdowns in NRVMs (n=3). (F) Total HDAC activity of NRVMs depleted of HDAC1, HDAC2 and/or HDAC3 (n=4). H, HDAC. (G) 3H-Leucine incorporation in NRVMs depleted of HDAC1, HDAC2 and/or HDAC3 and stimulated with PE (n=6). (H) Western blot and quantification of S6 phosphorylation in NRVMs depleted of HDAC1, HDAC2 and/or HDAC3 (n=3). Values are mean ± SEM, analyzed by one-way ANOVA followed by Tukey's test; * denotes p<0.05 sample compared to control (white bar); # denotes p<0.05 between groups marked; n= independent experiments.
The mechanistic target of rapamycin (mTOR) kinase is a central regulator of cell growth in many contexts including both physiologic and pathologic cardiac hypertrophy (9). To determine whether mTOR activity is regulated downstream of HDACs, we treated NRVMs with an inhibitor of class I HDACs (apicidin) and/or an inhibitor of mTOR (rapamycin) under hypertrophic conditions. We quantified mTOR activity by evaluating by Western blot the phosphorylation of two downstream targets: ribosomal subunit S6 and 4EBP1. As expected, PE triggered significant activation of mTOR (Fig. 1B). Exposure to apicidin reduced mTOR activity ≈50% under both baseline and growth conditions (Fig. 1B). Additionally, apicidin and rapamycin each blunted 3H-leucine incorporation under both resting and PE-induced growth conditions (Fig. 1C). Because suppression of 3H-leucine incorporation was greater with apicidin as compared with rapamycin, HDACs may control mechanisms independently of mTOR to promote growth. Also, inhibition of mTOR (rapamycin) in the setting of HDAC inhibition did not confer additional suppression of 3H-leucine incorporation (Fig. 1C). Similarly, mTOR activity induced by other growth stimuli, including ET-1, IGF-1, or mechanical stretch, was suppressed by apicidin as evidenced by decreases in the phosphorylation of both S6 and 4EBP1 (Fig. 1D). These data support a model in which HDAC inhibition attenuates cardiac growth by suppressing mTOR signaling.
Class I HDACs function redundantly
To determine the role of specific HDAC isoforms, we performed siRNA knockdown of individual HDAC isoforms in NRVMs. Western blot analysis confirmed effective protein knockdown (Fig. 1E, fig. S3A, S3B). Knockdown of individual HDAC isoforms did not alter total cellular HDAC activity; however, knockdown of class I HDACs in combination resulted in reduced HDAC activity (Fig. 1F). Again, we initially focused on PE-induced cardiomyocyte hypertrophy. Silencing of HDAC1, HDAC2 or HDAC3 individually had no significant effect on PE-induced 3H-leucine incorporation. However, PE-induced cardiomyocyte growth was abrogated by knocking down HDAC1 and HDAC2, as well as all three HDAC isoforms simultaneously (Fig. 1G). The effects of HDAC isoform silencing on leucine incorporation were mirrored in the response of the fetal gene program, with PE-induced activation of ANF and BNP inhibited by HDAC1/2 siRNA treatment (fig. S3C). Similar to the hypertrophic response, PE-induced activation of mTOR, measured as S6 phosphorylation, was blunted when more than one class I HDAC isoform was silenced (Fig. 1H). Furthermore, simultaneous knockdown of HDAC1/2/3 in combination inhibited hypertrophy induced by various growth stimuli (fig. S3D). Together, these data uncover functional redundancy of class I HDAC isoforms in the regulation of mTOR and NRVM growth.
Inhibition of class I HDACs reduces TAC-induced mTOR activity
We next used surgical TAC (10) to trigger pressure overload-induced cardiac hypertrophy in vivo. Our lab has previously demonstrated that isoform-nonspecific inhibition of HDAC activity suppresses the hypertrophic response in this model (2). To determine the role of class I HDACs, we subjected vehicle or apicidin-treated wild-type mice to TAC for three days (fig. S4A), at which point histone acetylation was increased (fig. S4B). In vehicle-treated mice, markers of the hypertrophic response were increased at 3 days, a response that was attenuated in apicidin-treated animals (Fig. 2A). Treatment with apicidin did not affect body weight (fig. S4C). Activation of mTOR, measured as the phosphorylation of its downstream targets S6 and 4EBP1, was also increased in vehicle-treated TAC mice. Consistent with our observations in NRVMs, TAC-induced increases in mTOR activity were blunted in mice treated with apicidin (Fig. 2B).
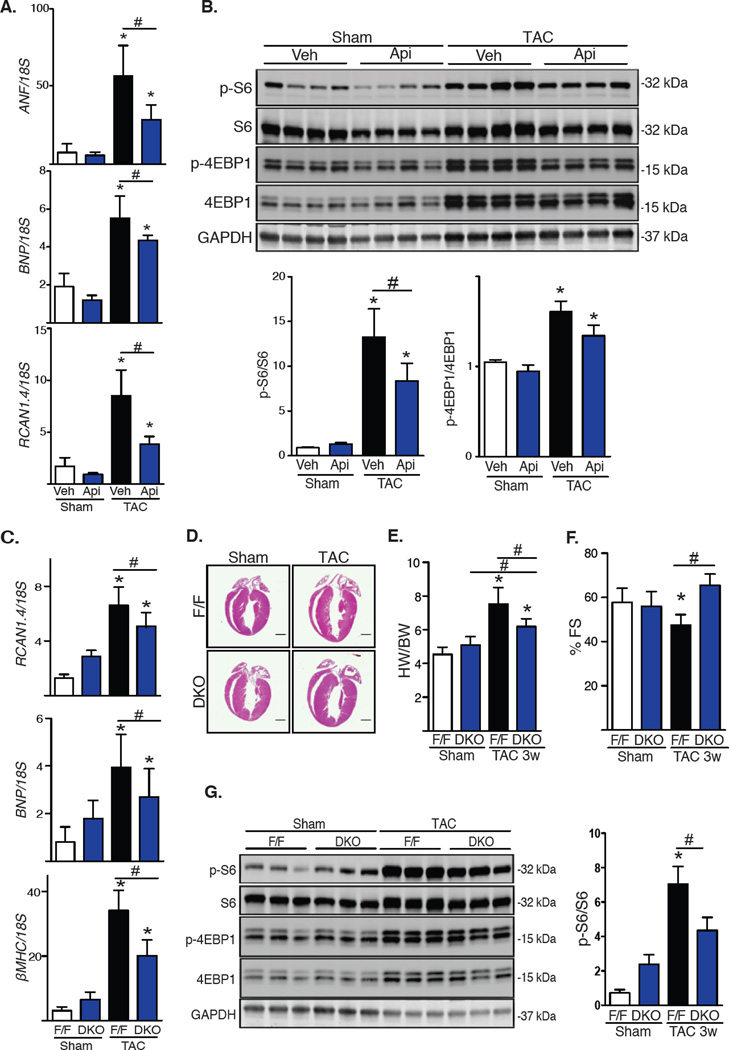
(A) mRNA expression of the hypertrophy markers ANF, BNP and RCAN1.4 from LV after 3 days of TAC and treatment with apicidin (Api, 2 mg/kg) or vehicle (n=4). Veh, vehicle. (B) Western blot analysis of mTOR activity in the left ventricle after TAC for 3 days with quantification (n=6). (C–G) DKO mice 3 weeks after TAC: (C) mRNA expression of hypertrophy markers ANF, BNP, and βMHC (n=4–5). (D) Hematoxylin and eosin staining of four-chamber heart sections after TAC (n=2). Scale bar: 2 mm. (E) Ventricular percent fractional shortening (FS) of control and DKO mice 3 weeks after TAC (n=8–9). (F) Heart weight (HW)/body weight (BW) ratios of control and DKO mice 3 weeks after TAC (n=8). (G) Western blot of S6 and 4EBP1 phosphorylation with quantification (n=4–5). Values are mean ± SEM, analyzed by one-way ANOVA followed by Tukey's test; * denotes p<0.05 sample compared to control (white bar); # denotes p<0.05 between groups marked; n=mice per genotype and treatment.
Cardiomyocyte-specific silencing of HDAC1 and HDAC2 reduces TAC-induced growth and mTOR activity
Our in vitro findings suggesting that HDAC1 and HDAC2, acting redundantly, promoted cardiac hypertrophy led us to determine whether HDAC1 and HDAC2 deficiency altered mTOR activity in vivo. Because cardiomyocyte-specific deficiency of the genes coding for HDAC1 and HDAC2 is lethal at P14 (11), we engineered conditional HDAC1 and HDAC2 double knockout (DKO) mice by crossing HDAC1ff/ff;HDAC2ff/ff mice with an αMHC-Mer-Cre-Mer-expressing line. Animals at 4–6 weeks of age were injected with tamoxifen to activate the Cre recombinase (fig. S4D). After treatment, mRNA measurements confirmed reduced expression of these genes (fig. S4E). DKO mice appeared normal and healthy and did not manifest changes in heart mass or contractile performance for at least 3 weeks after tamoxifen exposure (fig. S4F). Body weight was similar between control and DKO mice (fig. S4G).
HDAC1/2 DKO mice at 8–10 weeks of age were subjected to TAC for 3 weeks. Whereas control (F/F) mice manifested robust ventricular hypertrophy as measured by heart weight/body weight (HW/BW) ratios and expression of fetal gene markers, growth in DKO mice was significantly blunted (Fig. 2C, 2D, 2E). Ventricular performance was decreased in TAC-treated wild-type mice but not in DKO animals (Fig. 2F). Whereas mTOR activity was normal in DKO mice under resting conditions, TAC-induced increases in mTOR activity were blunted in DKO mice (Fig. 2G). Further, in addition to phosphorylation status, HDAC silencing in DKO mice in vivo decreased S6 and 4EBP1 protein abundance (Fig. 2G). These findings suggest that HDAC inhibition affects both the kinase activity of mTOR in vivo as well as the abundance of its substrates through a yet to be elucidated mechanism. Together, these data corroborated our in vitro findings, pointing to HDAC1/2 as critical regulators of stress-induced mTOR activation and cardiomyocyte growth.
Suppression of mTOR by HDAC inhibition is dependent on transcription
To define mechanisms linking HDAC activity and mTOR, we turned to our in vitro model of NRVM growth. Class I HDACs are largely localized to the nucleus, with a small pool residing in the cytosol (12). Because of this cytoplasmic pool of class I HDACs, we first tested whether mTOR was reversibly acetylated in NRVMs under conditions of hypertrophic growth. Immunoprecipitated mTOR was not detectably acetylated, and mTOR was not immunoprecipitated by an acetylated lysine antibody (fig. S5A). Thus, we concluded that HDAC-dependent control of mTOR did not involve mTOR as an HDAC substrate. The activity of AKT, an important modulator of mTOR activity, was assessed indirectly by Western blot. Our results show that HDAC inhibition did not affect the phosphorylation of AKT 6 hours after treatment, a point when S6 phosphorylation was reduced (fig. S1D).
We next examined the kinetics of mTOR inhibition by apicidin. NRVMs exposed to apicidin or trichostatin A (8) displayed a progressive decrease in mTOR activity over time (measured as S6 phosphorylation), requiring 6 hours to reach statistical significance (fig. S6A). On the other hand, rapamycin completely inhibited mTOR activity in less than 3 hours (fig. S6A). This 6 hour delay in mTOR repression hinted at transcriptional regulation as a possible mechanism. To test this notion, NRVMs were treated with cycloheximide or actinomycin D to block transcriptional or translational processes, respectively. In these experiments, we found that HDAC inhibition did not blunt mTOR activity in the presence of cycloheximide or actinomycin D (fig. S6B). Given that blocking transcription or translation robustly activates mTOR (13) (fig. S6B), these data suggested that DNA transcription was required for HDAC inhibitor-dependent repression of mTOR.
TSC2 mRNA abundance is increased by class I HDAC inhibition
mTOR is the main catalytic subunit of two distinct kinase complexes: mTOR Complex 1 (mTORC1) and mTOR Complex 2 (mTORC2). mTORC1 is composed of the kinase mTOR, regulatory-associated protein of mTOR (Raptor), mammalian lethal with Sec13 protein 8 (mLST8), proline-rich AKT substrate 40 kDa (PRAS40) and DEP-domain-containing mTOR-interacting protein (DEPTOR) (14). The mTORC1 and mTORC2 complexes share in common mTOR, mLST8 and DEPTOR. However, mTORC2 includes rapamycin-insensitive companion of mTOR (Rictor), mammalian stress-activated protein kinase interacting protein (mSIN1) and protein observed with Rictor-1 (Protor-1) (15). Because our findings pointed to a role for gene transcription in HDAC inhibitor-dependent mTOR repression, we measured the abundance of mRNAs encoding several direct binding components of both mTORC1 and mTORC2 in NRVMs treated with TSA or apicidin under resting and growth conditions (fig. S7A). PRAS40 expression was significantly reduced, but this reduction was also detected in NRVMs treated with rapamycin, suggesting that reduced abundance of PRAS40 was a secondary response to mTOR inhibition (fig. S7B). The abundance of the mRNA encoding the mTOR inhibitor Deptor (16) was increased after HDAC inhibition, a response not elicited by rapamycin (fig. S7B). However, siRNA knockdown in NRVMs of Deptor did not affect PE-induced mTOR activity (fig. S8C, S8D).
We found that the mRNA abundance of TSC2, which encodes an inhibitor of mTOR, increased upon inhibition of class I HDACs. TSC2 expression did not increase in NRVMs treated with rapamycin, suggesting that this effect was not secondary to the inhibition of mTOR (Fig. 3A). Consistently, TSC2 mRNA abundance also increased in NRVMs depleted of HDAC1, HDAC2 and HDAC3 (Fig. 3B). Of note, PE alone did not alter TSC2 mRNA abundance, suggesting that basal HDAC activity was sufficient to relieve the TSC2 “brake” on mTOR. The 2-fold increase in TSC2 mRNA when HDACs were inhibited was roughly consistent with the relative increases in TSC2 protein that we detected by Western blot (Fig. 3C). Similarly, mouse hearts subjected to TAC for 3 days and treated with apicidin manifested increased abundance of TSC2 transcript and protein (Fig. 3D and 3E). We also observed a trend toward an increase in TSC2 mRNA abundance in DKO mice at baseline and after TAC (Fig. 3F).
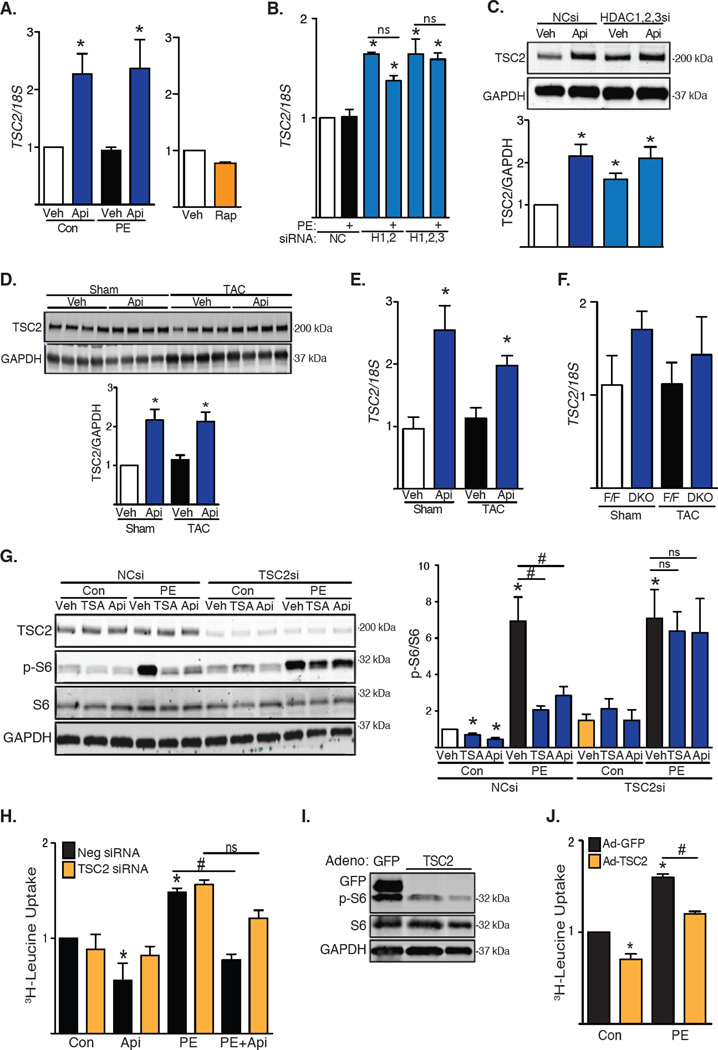
(A) RT-PCR analysis of NRVMs exposed to apicidin (Api) or rapamycin (Rap) under basal and PE-induced growth (n=6–7). Veh, vehicle; Con, control. (B) TSC2 mRNA abundance in NRVMs depleted of HDAC1, HDAC2 and/or HDAC3 (n=4). NC, Negative Control; H, HDAC. (C) Western blot of TSC2 in NRVMs depleted of class I HDACs and treated with apicidin (n=4). si, siRNA. (D) Western blot of TSC2 in WT mice subjected to TAC for 3 days in the presence of apicidin (n=4). (E) TSC2 mRNA abundance in apicidin-treated WT mice subjected to TAC for 3 days (n=4). (F) TSC2 mRNA abundance in DKO mice 3 weeks after TAC (n=4–5). (G) Western blot analysis of mTOR activity in NRVMs depleted of TSC2. Treatments with PE and HDAC inhibitors were performed for 24h. Quantification of S6 ribosomal subunit phosphorylation (n=3–5). (H) Leucine incorporation in NRVMs depleted of TSC2 under basal conditions and after PE treatment for 48h (n=4). (I) Western blot analysis of S6 phosphorylation in NRVMs over-expressing TSC2 (n=3). (J) 3H-leucine incorporation in NRVMs over-expressing TSC2 (n=3). Values are mean ± SEM, analyzed by one-way ANOVA followed by Tukey's test; * denotes p<0.05 sample compared to control (white bar); # denotes p<0.05 between groups marked; n=independent experiments for NRVMs or mice per group and treatment.
TSC2 is required for HDAC-dependent regulation of mTOR, and increased TSC2 abundance attenuates cardiomyocyte hypertrophic growth
We next determined whether TSC2 was required for the ability of apicidin to inhibit the mTOR pathway. Treatment of NRVMs with siRNA targeting TSC2 prevented hypertrophic stimulation from increasing mTOR activity, as measured by phosphorylation of S6 at baseline and with PE treatment (Fig. 3G and fig. S8A, S8B). This finding is contrary to that reported previously in other muscle cell types (17). Moreover, treating NRVMs with HDAC inhibitors failed to decrease mTOR activity in the absence of TSC2, both at baseline and with PE treatment, pointing to a requirement for TSC2 activity in HDAC-dependent control of mTOR activity (Fig. 3G). We next determined whether hypertrophic growth was also influenced by TSC2. Consistent with our findings regarding mTOR activation, knockdown of TSC2 did not induce growth at baseline, nor did it exacerbate PE-induced growth. However, silencing of TSC2 partially rescued the growth response after exposure to HDAC inhibitors (Fig. 3H), pointing to a requirement for TSC2 in HDAC inhibition-dependent suppression of mTOR activity. Finally, we employed a gain-of-function approach in which we expressed TSC2 at low amounts, comparable to the 2-fold increases triggered by the inhibition of class I HDACs. Expression of TSC2 reduced PE-induced S6 phosphorylation (Fig. 3I) and blunted PE-induced NRVM hypertrophy (Fig. 3J). These data point to a role for increased abundance of TSC2 in mediating the anti-hypertrophic effects of HDAC inhibition.
To determine whether TSC2 could be regulated by direct acetylation, we tested immunoprecipitation of endogenous TSC2 by probing against acetylated lysine. We did not detect any acetylation of TSC2 by this method (fig. S5B). To further explore potential mechanisms by which HDAC inhibitors promote TSC2 expression, we tested the acetylation of the TSC2 promoter with chromatin immunoprecipitation studies. We analyzed 2000 base pairs upstream and 500 base pairs downstream of the TSC2 transcription start site. We tested NRVMs as well as MEFs. Our results suggest changes in histone acetylation contribute to the increased expression of TSC2 (fig. S10A, S10B).
HDAC inhibition-dependent suppression of mTOR activity in human cells
To determine the effect of HDAC inhibitors in human cells, we tested H9 embryonic stem cells differentiated into cardiomyocyte-like cells (H9-CM) (18). Upon differentiation, H9-CM cells contract spontaneously, develop sarcomeric structures, and express the cardiogenic markers troponin I and α-actinin (Fig, 4A). Consistent with our findings in rodent models, treatment with the HDAC inhibitor apicidin reduced mTOR activity and resulted in a 2-fold increase in TSC2 mRNA abundance (Fig. 4B, 4C). Exposure to apicidin did not decrease the abundance of α-actinin or alter the cardiomyocyte-like phenotype of these cells (Fig. 4D). We also tested wild-type and TSC2+/− MEFs. On exposure to apicidin, wild-type and TSC2-heterozygous cells each showed a 2-fold increase in TSC2 steady-state abundance (Fig. 4E). Whereas apicidin inhibited mTOR activity in wild-type cells, it did not reduce mTOR activity in TSC2-heterozygous or TSC2-knockout MEFs (Fig. 4F). Additionally, cell growth in wild-type MEFs as measured by 3H-leucine incorporation was reduced by HDAC inhibition, an effect that was lost in the knockout cells (Fig. 4G).
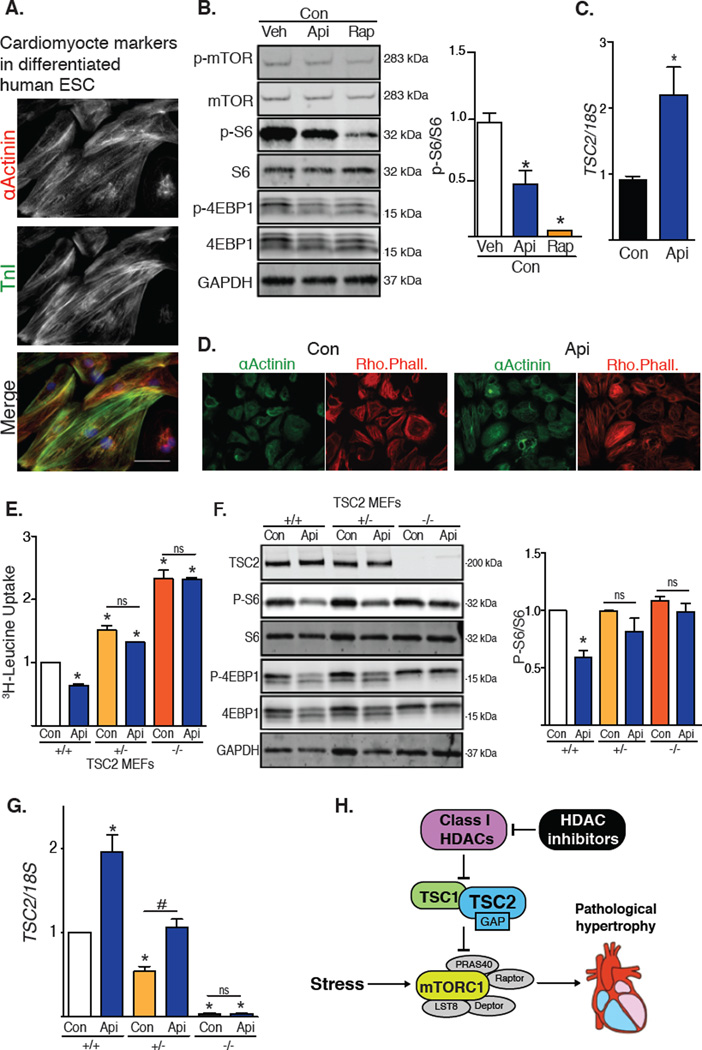
(A–C) H9-CM cells: (A) Immunostaining for α-actinin and troponin I (n=2); scale bar: 40 µm. (B) Western blot of mTOR downstream targets from H9-CM cells treated with apicidin or rapamycin for 24h (n=3). (C) TSC2 expression in H9-CM exposed to apicidin for 24h (n=3). (D) Representative images of H9-CM cells exposed to apicidin for 24h and probed for α-actinin and rhodamine phalloidin (Rho. Phall.) (n=3). (E–G) MEFs: (E) Tritiated leucine incorporation in MEFs depleted of TSC2 treated with apicidin for 48h (n=6). (F) Western blot of mTOR activity in WT, TSC2 heterozygous, or TSC2 homozygous MEFs treated with apicidin for 6h. Quantification of S6 phosphorylation (n=4). (G) TSC2 expression in MEFs exposed to HDAC inhibitor (n=4). (H) Working model: Inhibition of class I HDACs promotes expression of TSC2, which leads to inhibition of mTORC1 in pathological cardiac hypertrophy. Values are mean ± SEM, analyzed by one-way ANOVA followed by Tukey's test; * denotes p<0.05 sample compared to control (white bar); # denotes p<0.05 between groups marked; n=independent experiments.
Small molecule inhibitors of HDAC activity have been successful in impeding the proliferation of some cancer cells in vitro and in vivo (19), possibly through reducing mTOR activity (20). Treatment of HeLa cells with apicidin increased, rather than decreased, the phosphorylation of S6 and did not alter TSC2 mRNA abundance (fig. S9A). Finally, we also tested additional cell types derived from embryonic rat heart (H9c2), rat skeletal muscle (L6), or mouse skeletal muscle (C2C12). Similar to NRVMs, HDAC inhibition induced increases in TSC2 mRNA and suppressed mTOR activity in H9c2 (fig. S9B) and L6 cells (fig. S9C) but not in mouse skeletal C2C12 myoblasts (fig. S9D).
DISCUSSION
Pharmacotherapy targeting epigenetic mechanisms is clinically efficacious in oncology (21), and considerable preclinical evidence suggests that this strategy holds promise in cardiovascular disease (22). Arguably the strategy closest to clinical translation involves pharmacological suppression of HDACs (23). Here, we set out to delineate mechanisms whereby HDAC inhibitors blunt pathological cardiac hypertrophy. We employed in vitro and in vivo models of pathological cardiac growth, finding that class I HDACs redundantly regulated cardiac hypertrophy induced by various growth cues. We found that HDAC inhibitors suppressed the activity of mTOR, a regulator of cell growth. We went on to delineate mechanisms of HDAC-dependent control of mTOR, which we found involved governance of expression of TSC2, which encodes an mTOR inhibitor. Due to the robust translational potential of pharmacological HDAC inhibitors (24–26), we explored this biology in human embryonic stem cell-derived cardiomyocytes, finding that HDAC inhibitors also increased TSC2 abundance and inhibited mTOR activity in these cells. In aggregate, these data define a mechanism of therapeutic manipulation of pathological cardiac hypertrophy involving class I HDACs, TSC2, and mTOR (Fig. 4H). Furthermore, our data uncover a means of therapeutically modulating mTOR activity.
HDAC-dependent control of mTOR activity
We and others have shown that broad-spectrum HDAC inhibitors have beneficial effects in models of pathologic cardiac hypertrophy by blunting increases in ventricular size and mass, improving contractile performance, and reducing fibrosis (2, 5). These compounds also reduce the pathological remodeling observed during ischemia/reperfusion injury in both rodents and rabbits, specifically targeting the reperfusion phase of injury (27). Therefore, we set out here to decipher mechanisms whereby HDAC inhibitors elicit benefits to the heart. In so doing, we have delineated critical elements of the biology of HDAC-dependent governance of cardiomyocyte growth and remodeling. Our starting point was the established notion that broad-spectrum HDAC inhibitors blunt mTOR activity in some cancer models (8); however, underlying mechanisms had not previously been defined. Further, the role specifically of class I HDACs in mTOR activity in the context of cardiac growth had not been elucidated.
The family of HDACs is divided into four classes: class I HDACs, comprising HDAC1, HDAC2, HDAC3 and HDAC8, are considered to be pro-growth, contributing to hypertrophic remodeling of disease-stressed cardiomyocytes (11, 28). In this study, we showed that inhibition of class I HDACs repressed the activity of the central growth kinase mTOR. This is particularly relevant in the context of pathological cardiac growth. While inhibition of mTOR complex I by genetic and pharmacological means leads to diminished cardiac hypertrophy (29, 30), HDAC inhibitors are even more effective in diminishing the growth response, even though inhibition of mTOR by HDAC inhibitors is not as complete as with rapamycin. This observation suggests that additional mechanisms exist downstream of HDAC inhibitors to regulate cardiac hypertrophy independent of mTOR. Finally, in a model of cardiac ischemia/reperfusion, inhibition of mTOR at the time of reperfusion reduces infarct size (31), findings similar to our observations in the context of HDAC inhibition.
Small molecule inhibitors specific for class I HDACs blunt pathological cardiac hypertrophy similar to pan-HDAC inhibitors (7). Cardiomyocyte-specific loss of either HDAC1 or HDAC2 has no impact on the response to hypertrophic stimuli (11). We report here that combinatorial silencing of both HDAC1 and HDAC2 blunts stress-induced hypertrophic growth of the myocardium, consistent with our model of functional redundancy of these two enzymes. Of note, mTOR inhibition and hypertrophy suppression in DKO were diminished relative to that seen in wild-type mice exposed to HDAC inhibitors. This is possibly due to the fact that we measure only a partial decrease (~50%) of HDAC1 and HDAC2 RNA abundance in the DKO mice, coupled with the effects of untargeted HDAC3.
mTOR regulates cellular autophagy by suppressing autophagic flux (32). We have reported previously that chronic administration of HDAC inhibitors decreases autophagic activity in the heart under hypertrophic stress (6). We have also reported that under conditions of reperfusion after an ischemic event, acute treatment of the heart with HDAC inhibitors results in the stimulation of autophagic flux (27). These seemingly contradictory findings can be explained by examining a time-course of HDAC treatment. Evidence from our lab shows that treating NRVMs with HDAC inhibitors promotes autophagy at early time points (6 hours), consistent with the inhibition of mTOR reported here; this is followed by a second phase in which autophagy is inhibited (27). This inhibition appears to be independent of mTOR, because mTOR is still inhibited even after 24 hours of HDAC inhibition. Rapamycin, on the other hand, continues to promote autophagic flux at these late time points.
We cannot exclude the possibility that inhibiting HDACs may alter other signaling nodes that regulate autophagy at later time points. The mechanism by which HDAC inhibitors regulate autophagy at longer exposure times remains to be determined. However, it is possible that simultaneous inhibition of mTOR and autophagy with HDAC inhibitors may elicit a stronger negative effect on hypertrophy compared to that seen with inhibiting mTOR alone (Fig. 1C). Further, it is possible that inhibiting HDACs may alter other signaling nodes regulating cardiac growth, including calcium handling, contractility, and other kinases, such as ERK. Finally, class I HDAC inhibition blunted mTOR activity in the setting of physiological cardiomyocyte growth. This finding suggests that eventual clinical application of these agents must be approached with caution, as untoward effects on catabolic balance, protein synthesis, and normal cell growth could arise.
HDAC-dependent regulation of TSC2 expression
mTORC1 includes Rheb, which is active when bound to GTP. Upstream of mTORC1 is the tuberin (TSC1)/hamartin (TSC2) complex, which has GAP activity towards Rheb and hence functions as an inhibitor of Rheb and mTOR activity (33). Deletion of TSC1 in heart and muscle driven by smooth muscle protein 22 promoter leads to cardiac hypertrophy, which can be rescued by mTOR inhibition with rapamycin, suggesting an important role of the TSC1/TSC2 complex in the regulation of mTOR during cardiac hypertrophy (34). We report here that class I HDACs regulate TSC2 mRNA. Defective activity of the TSC1/TSC2 complex in the heart triggers development of rhabdomyosarcomas at embryonic stages that halt or regress after birth (35). In neonatal cardiomyocytes, TSC2 deficiency does not promote hypertrophy on its own, but rather leads to increased DNA synthesis (36). In mice, silencing TSC1 in adult heart triggers robust cardiac hypertrophy (34). The TSC1/TSC2 complex is often inhibited under growth conditions to facilitate mTOR activation (37). There are a few scenarios in which the TSC1/2 complex is endogenously active, such as in starvation stress. We report here that HDAC inhibitors regulate the activity of the TSC1/TSC2 complex by promoting expression of TSC2, which encodes the catalytic component.
Our evidence points to HDAC-dependent control of mTOR involving transcriptional events. A previous study in smooth muscle-like cells from a patient with tuberous sclerosis showed that epigenetic regulation of the TSC2 promoter through methylation represses TSC2 expression (38). Whether increased TSC2 mRNA abundance reported here are due to direct changes in histone acetylation or derive from indirect mechanisms requires further investigation.
In summary, an emerging literature points to pharmacological suppression of HDAC activity as a means of targeting pathological cardiac hypertrophy (2), ischemia/reperfusion injury (27), and modulating autophagy (6). This report is a first step at defining underlying events, uncovering a mechanism of HDAC-dependent repression of TSC2 expression, resulting in the release of active mTOR. When HDAC activity is suppressed, either pharmacologically or through genetic ablation, TSC2 abundance increases, thereby inhibiting mTOR activity. This suppression of mTOR, in turn, has yet-to-be-defined effects on protein synthesis and autophagy. Together, this study delineates a molecular cascade – involving TSC2 expression and mTOR activity – whereby HDAC inhibitors have beneficial effects on pathological cardiac remodeling.
MATERIALS AND METHODS
Bioethics
All studies conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication, 8th Edition, 2011) and were approved by the Institutional Ethics Review Committees of the University of Texas Southwestern Medical Center.
Neonatal rat ventricular myocytes (NRVMs) and siRNA knockdowns
Cardiomyocytes were isolated from the left ventricle of 1–2-day-old Sprague-Dawley rats. Lysates were digested with collagenase, and the resulting cell suspension was pre-plated to remove fibroblasts. Myocytes were then plated at a density of 1250 cells/mm in DMEM 4.5 g/L glucose medium containing 10% FBS and 100 µmol/L bromodeoxyuridine. For siRNA knockdown experiments, NRVMs were transfected 36 hours after plating with siRNA constructs (Sigma) using Lipofectamine RNAiMax (Invitrogen) in Optimem (6h). Experiments were launched 24h after knockdown. For hypertrophy studies, cells (48h after plating) were exposed to serum-free DMEM containing: phenylephrine (PE) [50 µM], endothelin-1 (ET-1) [100 nM], insulin growth factor-1 (IGF1) [10 nM], or 50% hypo-osmotic solution. HDAC inhibitors were employed as follows: apicidin (Api) at 0.2 µM and 1 µM; trichostatin A at 60 nM.(8)
Western blot analysis
Whole cell protein lysates were obtained from NRVMs or from tissue with M-PER mammalian extraction buffer (Thermo Scientific) containing protease and phosphatase inhibitors (Roche). Tissue protein extracts were passed through glass wool to remove DNA. Cell lysates were separated by SDS/PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis. Antibodies were used that targeted: mouse S6 ribosomal protein, rabbit phospho-S6 ribosomal protein (Ser235/236), rabbit 4EBP1, rabbit phospho-4EBP1 (Thr37/46), rabbit S6K, mouse phospho-S6K (Thr389), rabbit TSC2, mouse HDAC1, rabbit HDAC3, rabbit acetyl lysine, mouse mTOR, and rabbit phospho-mTOR (Ser2448) all from Cell Signaling; mouse GAPDH (Fitzgerald Industries Int.); mouse α-tubulin (Sigma); rabbit HDAC2 (Invitrogen). Blots were scanned, and bands were quantified using an Odyssey Licor (version 3.0) imaging system.
Real-time PCR
Total RNA was harvested from NRVMs or left ventricles using TRIzol (Invitrogen) according to the manufacturer's protocol. cDNA was prepared from RNA using a high capacity cDNA reverse transcription kit (Applied Biosystems). Real-time PCR was performed using SYBR green on an ABI 7000 Prism Sequence Detection System (Applied Biosystems). To confirm amplification specificity, PCR products were routinely subjected to melting curve analysis. Negative controls containing water instead of cDNA were run concomitantly. Data for each transcript were normalized to reactions performed with 18S rRNA primers, and fold change was determined using the comparative threshold method (Supplemental Table 1).
3H-Leucine incorporation
NRVMs were cultured with 3H-leucine (1 mCi/mL, Perkin Elmer) at the time of treatment. Cells were washed (3×) with ice-cold PBS and incubated with 10% trichloroacetic acid (30 min, 4°C) followed by three washes with ice-cold 95% ethanol. Samples were incubated in 0.5N NaOH (6h, 37°C) with gentle agitation, then neutralized with 0.5N HCl, and subjected to scintillation counting (Beckman).
HDAC activity
Total cellular HDAC activity was measured with the Fluor-de-Lys HDAC Fluorometric Cellular activity assay kit (Enzo Life Sciences, BML-AK503-0001). In brief, NRVMs plated in 96 wells were treated with HDAC inhibitor or HDAC RNAi and subsequently incubated with 200 µM Fluor-de-lys substrate (4h). The cell-permeable acetylated substrate fluoresces when deacetylated. Fluor-de-lys lysis buffer, trichostatin A, and developer solution were added 15 min before the end of the experiment, and fluorescence was measured in a microplate reader (Ex 360 nm, Em 460 nM).
In vivo cardiac hypertrophy and histology
Male C57/BL6 mice (8–10 weeks old) were subjected to TAC (10) for 3 weeks as previously described (2). Control animals underwent sham operations. HDAC1 and HDAC2 conditional cardiomyocyte-specific combinatorial knockout (DKO) mice were generated by crossing HDAC1 and HDAC2 floxed mice (11) with C57BL6 Mer-Cre-Mer mice. To drive Cre expression, 5 daily doses of tamoxifen (20mg/kg) were delivered intraperitoneally at 4–6 weeks of age. For HDAC1/2 DKO mice, cardiac function was determined after tamoxifen exposure, as well as pre- and post-surgery. For histology, animals hearts were fixed (4% paraformaldehyde, room temperature with agitation) followed by routine paraffin processing. Hematoxylin and eosin and Masson’s trichrome staining were performed according to routine procedures.
Echocardiography
Echocardiograms were performed on conscious, gently restrained mice using a Vevo 2100 system and an 18 MHz linear probe. A short axis view of the left ventricle at the level of the papillary muscles was obtained, and M-mode recordings were performed from this view. Measurements of interventricular septal thickness (IVS), left ventricular internal diameter (LVID), and left ventricular posterior wall thickness (LVPW) were obtained from 2D parasternal short axis views in diastole. Left ventricular mass was calculated by the cubed method as 1.05×((IVS + LVID + LVPW)3 – LVID3) (mg) (39), which we have independently verified (40). Left ventricular internal diameter at end-diastole (LVIDd) and end-systole (LVIDs) were measured from M-mode recordings. Fractional shortening was calculated as (LVIDd − LVIDs) / LVIDd (%).
Embryonic stem cell differentiation into cardiomyocytes
Embryonic stem cells (H9 cell line, from “WiCell”) were maintained on matrigel in mTeSR culture medium. Cardiomyocyte differentiation was performed as described (41), with modifications (10). When cells reached 85–90% confluency, the medium was switched to RPMI 1640 (Life Technologies, 11875-093) with B27 Supplement minus insulin (Life Technologies, A1895601) (days 0–8) supplemented with small molecule inhibitors: 6 µM CHIR99021 (Selleckchem, S2924) on day 0 for 48 hours and 5 µM IWR-1 (Sigma) on day 4 for 48 hours. Medium was changed to RPMI 1640 with B27 Supplement, serum free (Life Technologies, 17504-044) on day 8. Cells were metabolically selected for 10 days with medium lacking glucose, replated, and used for experimentation after day 30.
Immunofluorescence staining
For immunofluorescence staining, cells were fixed with 4% formaldehyde, permeabilized with 0.1% triton X-100 in PBS, blocked with 3% FBS, incubated overnight with primary antibody (mouse α-actinin, Sigma, 7811) and troponin-I (rabbit, Santa Cruz Biotechnology, sc-15368) at 4°C diluted 1:400 in primary antibody, washed with PBS, followed by incubation with appropriate secondary antibody and counter-stained with DAPI.
Chromatin Immunoprecipitation
MEFs and NRVMs were treated overnight with 200 nM apicidin. The cells were subjected to ChIP assay using a commercially available Kit (Abcam ab500). Sheared chromatin was incubated overnight (with rotation, 4°C) with anti-acetyl lysine ChIP grade antibody (Abcam), anti-histone 3 ChIP grade antibody (Abcam) or negative control rabbit IgG. DNA pulldown and purification were performed according to the manufacturer’s protocol (Abcam ab500). DNA samples were subjected to real-time PCR with SYBR green using species-specific primers that span 2,000 base pairs upstream and 500 base pairs downstream of the TSC2 transcription tart site.
Statistics
Data are presented as mean ± SEM. The unpaired Student's t test was used for comparison between two independent groups and ANOVA followed by Tukey post-hoc test for pair-wise comparisons. For all statistical comparisons, p < 0.05 was considered significant. All statistical analyses were performed using GraphPad Prism (version 6.01) software.
Acknowledgments
We thank members of the Hill lab for constructive comments, and we thank Julia Kim for her contribution toward understanding of the effect of HDAC inhibitors in HeLa cells.
Funding: This work was supported by grants from the NIH (HL-120732; HL-100401 to J.A.H.), AHA (14SFRN20740000 to J.A.H.), CPRIT (RP110486P3 to J.A.H.), and the Leducq Foundation (11CVD04 to J.A.H.), CONICYT, Chile: FONDAP 15130011 (to S.L.), NIH (HL-097768; HL-072016 to B.A.R.) and AHA (11POST7950051 to D.R.).
Footnotes
SUPPLEMENTARY MATERIAL
Fig. 1S. Effect of class I HDAC inhibition on NRVM hypertrophy and AKT
Fig. 2S. Characterization of HDAC inhibition in NRVMs
Fig. 3S. Effect of siRNAs targeting class I HDACs on NRVM hypertrophy
Fig. 4S. In vivo experimental model
Fig. 5S. mTOR and TSC2 are not acetylated in NRVMs treated with HDAC inhibitors
Fig. 6S. Inhibition of mTOR by HDAC inhibitors likely depends on transcription
Fig. 7S. RT-PCR screen of components of mTORC1, mTORC2, and the TSC1/TSC2 complex
Fig. 8S. Deptor is not required for HDAC-dependent inhibition of mTOR
Fig. 9S. HDAC inhibitor-dependent suppression of mTOR is cell type-specific
Fig. 10S. ChIP analysis of the TSC2 promoter
Author contributions: C.R.M.: study design, data collection, data analysis, and manuscript preparation; D.L.L.: data collection, data analysis, and manuscript preparation; Z.P.: data collection, and data analysis; H.I.M.: data collection, and data analysis; N.J.: data collection, and data analysis; V.K.: data collection, and data analysis; G.C.: data collection, and data analysis; D.R.: data collection, and data analysis; B.A.R.: study design, data collection, data analysis, and manuscript preparation. J.W.S.: study design, data collection, data analysis, and manuscript preparation; S.L.: study design, data collection, data analysis, and manuscript preparation; T.H.G.; study design, data collection, data analysis, and manuscript preparation; J.A.H.: study design, data collection, data analysis, and manuscript preparation.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
Full text links
Read article at publisher's site: https://doi.org/10.1126/scisignal.aad5736
Read article for free, from open access legal sources, via Unpaywall:
https://stke.sciencemag.org/content/sigtrans/9/422/ra34.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
β-hydroxybutyrate recapitulates the beneficial effects of ketogenic metabolic therapy in polycystic kidney disease.
iScience, 27(9):110773, 20 Aug 2024
Cited by: 1 article | PMID: 39314240 | PMCID: PMC11418134
Therapeutic Potential of Natural Compounds Acting through Epigenetic Mechanisms in Cardiovascular Diseases: Current Findings and Future Directions.
Nutrients, 16(15):2399, 24 Jul 2024
Cited by: 0 articles | PMID: 39125279 | PMCID: PMC11314203
Review Free full text in Europe PMC
Forward genetic screen using a gene-breaking trap approach identifies a novel role of <i>grin2bb</i>-associated RNA transcript (<i>grin2bbART</i>) in zebrafish heart function.
Front Cell Dev Biol, 12:1339292, 08 Mar 2024
Cited by: 1 article | PMID: 38533084 | PMCID: PMC10964321
The MYPT2-regulated striated muscle-specific myosin light chain phosphatase limits cardiac myosin phosphorylation in vivo.
J Biol Chem, 300(2):105652, 13 Jan 2024
Cited by: 1 article | PMID: 38224947 | PMCID: PMC10851227
Go to all (55) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Signal-dependent repression of DUSP5 by class I HDACs controls nuclear ERK activity and cardiomyocyte hypertrophy.
Proc Natl Acad Sci U S A, 110(24):9806-9811, 29 May 2013
Cited by: 62 articles | PMID: 23720316 | PMCID: PMC3683796
Enhanced expression of glucose transporter-1 in vascular smooth muscle cells via the Akt/tuberous sclerosis complex subunit 2 (TSC2)/mammalian target of rapamycin (mTOR)/ribosomal S6 protein kinase (S6K) pathway in experimental renal failure.
J Vasc Surg, 57(2):475-485, 21 Dec 2012
Cited by: 24 articles | PMID: 23265586
Casein kinase-2α1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart.
Circulation, 123(21):2392-2403, 16 May 2011
Cited by: 73 articles | PMID: 21576649
Chromatin modifications remodel cardiac gene expression.
Cardiovasc Res, 103(1):7-16, 08 May 2014
Cited by: 34 articles | PMID: 24812277
Review
Funding
Funders who supported this work.
AHA (2)
Grant ID: 14SDG18440002
Grant ID: 11POST7950051
AHA (American Heart Association) (1)
Grant ID: 14SFRN20740000
CONICYT (Comision Nacional de Investigacion Cientifica y Tecnologica), Chile (1)
Grant ID: FONDAP 15130011
CPRIT (Cancer Prevention and Research Institute of Texas) (3)
Grant ID: RP110486P3
Grant ID: RP110486-AC
Grant ID: RP110486-P5
Leducq Foundation (1)
Grant ID: 11CVD04
NHLBI NIH HHS (10)
Grant ID: R01 HL126012
Grant ID: HL-072016
Grant ID: HL-100401
Grant ID: R01 HL120732
Grant ID: HL-097768
Grant ID: U01 HL100401
Grant ID: HL-126012
Grant ID: HL-120732
Grant ID: R01 HL072016
Grant ID: R01 HL097768
NIH (5)
Grant ID: HL-100401
Grant ID: HL-126012
Grant ID: HL-072016
Grant ID: HL-097768
Grant ID: HL-120732





