Abstract
Free full text

Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression
Abstract
Gene-specific oligonucleotide probes are currently used in microarrays to avoid cross-hybridization of highly similar sequences. We developed an approach to determine the optimal number and length of gene-specific probes for accurate transcriptional profiling studies. The study surveyed probe lengths from 25 to 1000 nt. Long probes yield better signal intensity than short probes. The signal intensity of short probes can be improved by addition of spacers or using higher probe concentration for spotting. We also found that accurate gene expression measurement can be achieved with multiple probes per gene and fewer probes are needed if longer probes rather than shorter probes are used. Based on theoretical considerations that were confirmed experimentally, our results showed that 150mer is the optimal probe length for expression measurement. Gene-specific probes can be identified using a computational approach for 150mer probes and they can be treated like long cDNA probes in terms of the hybridization reaction for high sensitivity detection. Our experimental data also show that probes which do not generate good signal intensity give erroneous expression ratio measurement results. To use microarray probes without experimental validation, gene-specific probes ~150mer in length are necessary. However, shorter oligonucleotide probes also work well in gene expression analysis if the probes are validated by experimental selection or if multiple probes per gene are used for expression measurement.
INTRODUCTION
DNA microarrays are widely regarded as a powerful tool for large-scale gene expression measurements. The two main DNA microarray platforms are cDNA and oligonucleotide microarrays. cDNA microarrays are made with long double-stranded DNA molecules generated by enzymatic reactions such as PCR (1), while oligonucleotide microarrays employ oligonucleotide probes spotted by either robotic deposition or in situ synthesis on a solid substrate (2). It should be noted that, in this article, the immobilized DNA molecules are referred to as the probes and the labeled gene transcripts for hybridization as the targets, as suggested in Vol. 21, Supplement, Chipping Forecast, Nature Genetics 1999.
If the probes are not optimized for sequence specificity, both types of DNA microarrays can generate false-positive data due to non-specific cross-hybridization to highly similar sequences, gene families (3,4), or alternatively spliced variants (5). Cross-hybridization of one probe to several targets occurs more often with cDNA microarrays than with gene-specific oligonucleotide microarrays. In this article, 25–30mer probes are short oligonucleotide probes and 50–80mer probes are long oligonucleotide probes. Long DNA probes refer to probes of 100–150mer in length. cDNA probes are derived from cDNA clones and are ≥500 bases in length. Literature reports (3,4) have shown that, if the targets have >70–80% global sequence homology to the cDNA probe, they can hybridize indiscriminately to the cDNA probe. In addition, high local sequence similarity between different sequences also causes significant cross-hybridization (3). Long oligonucleotide probes are also prone to cross-hybridization. For instance, any non-target sequence showing 75% similarity to a 50mer oligonucleotide probe results in cross-hybridization (6), and the same is true for non-target sequences showing 70% similarity to a 60mer probe (7). These observations have suggested that the percentage sequence homology is a reasonable predictor of cross-hybridization (4). To overcome this cross-hybridization problem, a general practice adopted by several laboratories is to design oligonucleotide probes targeting regions of low sequence similarity (6–8).
However, the use of oligonucleotide probes to replace cDNA probes in microarrays for expression profiling has generated discussion about the discordant results obtained using these two types of probes (9,10), the optimal oligonucleotide probe length and the number of oligonucleotide probes needed to obtain reliable expression data for a gene (11). Literature data (7,11) indicate that longer oligonucleotides (e.g. 60–80mers) provide significantly better detection sensitivity than shorter probes (e.g. 25 or 30mers). However, these long oligonucleotide probe microarrays use only one probe per gene, despite the fact that oligonucleotide hybridization is highly sequence dependent (12). It has been reported that oligonucleotide probes binding to different regions of a gene yield different signal intensities (2,7,13), and it is difficult to predict whether an oligonucleotide probe will bind efficiently to its target sequence and yield a good hybridization signal on the basis of sequence information alone (14). Because of this, multiple probes per gene have been used in oligonucleotide array designs to obtain reliable quantitative information of gene expression (2,7,13). Early versions of in situ synthesized 20mer oligonucleotide arrays employed 20 probe pairs per gene to provide statistically reliable quantification (2). On the basis of accumulated experimental results, probes that do not yield good hybridization signals were excluded to reduce the number of probes per gene. Five probes per gene has been suggested to be a good number for 30mer probes (11). When the best single probe for a gene was selected experimentally from eight 60mer candidate probes, it successfully detected gene expression at low levels (7). An experimental study showed that >25% of the probes in a set of 15 357 80mer probes were incapable of producing a usable hybridization signal (http://www.clontech.com/archive/APR02UPD/BDAtlas.shtml). To ensure good hybridization signals, every oligonucleotide probe should be tested experimentally, but the large-scale screening process is extremely time-consuming and costly.
357 80mer probes were incapable of producing a usable hybridization signal (http://www.clontech.com/archive/APR02UPD/BDAtlas.shtml). To ensure good hybridization signals, every oligonucleotide probe should be tested experimentally, but the large-scale screening process is extremely time-consuming and costly.
In view of the aforementioned facts, since the major difference between different microarray platforms is the immobilized probes, the differences in the expression measurement results obtained can be attributed to differences in the probes. The sequence and length of a probe are critical factors for DNA microarray performance. Different probe formats show different degrees of sequence-dependent hybridization variation, different hybridization sensitivities and specificities, and a different number of probes per gene. In this article, we present an approach that can be used to investigate these issues and its use in evaluating probe lengths and the number of probes per gene for optimal microarray analysis of gene expression.
MATERIALS AND METHODS
Probe design
The human UniGene database (build #153) was used to derive source sequences for probe design. The gene sequences were filtered to remove repetitive elements using the RepeatMasker program (A. F. A. Smit and P. Green, http://ftp.genome.washington.edu/RM/RepeatMasker.html), then aligned using the BLAST program (15) with source sequences compiled from the UniGene database and the TIGR gene indices. The probe selection strategy was mainly based on frequently used criteria (6–8) for setting a sequence similarity threshold for specificity and a range of GC content for uniform hybridization reaction. Any part of a gene having 75% local sequence similarity in a 50 base window with other genes was masked. The remaining unmasked sequences were considered unique sequences of the gene. Within these unique sequences 100 and 150mer probes were designed using the Primer3 program (Unix version 0.9, S. Rozen and H. J. Skaletsky, http://www-genome.wi.mit.edu/genome_software/other/primer3.html). The GC content was calculated for every oligonucleotide probe ranging from 25 to 70mer selected from within the unique sequences of the gene transcript using a one-base-shift tiling method. We selected the gene-specific probes with a GC content between 45 and 55% and used the Mfold program (16) to further screen for probes that bind to the target sequences with the maximal Gibbs free energy of secondary structure.
Array fabrication
The 25mer, 50mer and 70mer oligonucleotides and all PCR primer pairs were synthesized on a 1536-channel DNA synthesizer (17) constructed in-house. Unless otherwise specified, all the oligonucleotide probes were modified with spacer and 5′ amino-linker moieties. The spacer was positioned between the probe sequence and the linker to extend the probe sequence away from the surface for better access to the target molecules. The spacer was a single unit of hexa-ethyloxy-glycol and was added to the probe using DMT-hexa-ethyloxy-glycol CED phosphoramidite (ChemGenes) and standard phosphoramidite synthesis chemistry. The amino-linker was coupled to the spacer using the same phosphoramidite synthesis chemistry and was used to tether probes by their 5′ ends to the slide surface with covalent bonding. Probes 100mer or longer were generated by PCR using a sense primer with the same spacer and 5′ amino-linker modifications. The concentrations of the oligonucleotide probes and primers were measured on a UV spectrophotometer at 260 nm (SpectraMax Plus 384, Molecular Devices). All the unique cytochrome P450 gene probes were generated by PCR amplification of the 3′-untranslated regions (3′-UTRs) or exons in genomic DNA derived from human placenta tissue. The high-performance liquid chromatography (HPLC)-purified oligonucleotide probes and column-purified (QIAquick, Qiagen) long DNA probes were dissolved in 150 mM sodium phosphate buffer (pH 8.5). The probes were spotted in duplicate and covalently immobilized via the 5′ end to surface-activated slides (SurModics) using an in-house constructed arrayer. The subsequent processing of slides was performed according to the manufacturer's instructions. For long DNA probe arrays, the double-stranded probes generated by PCR were rendered single-stranded by heating in boiling water for 2 min. Quality assessment of array printing was performed using a fluorescent dye, SYTO 61 (Molecular Probes), to directly measure the amount of DNA retained on the slide surface (18).
Preparation of in vitro-transcribed polyadenylated RNA
Five plant genes (rbcl, rca, ga4, hat4 and atps) and two human genes (gapdh and β-actin) were PCR-amplified with gene-specific primers then cloned into the pCITE-4a(+) vector (Novagen). Thirty-one cytochrome P450 (CYP) family gene members were amplified by PCR using gene-specific primers (PCR amplicon size 800–1000 bp), with a T7 promoter sequence attached to the forward primer and (dT)25 attached to the reverse primer. In vitro-transcribed polyadenylated RNAs, generated using T7 RNA polymerase (Epicenter), were purified on an RNeasy column (Qiagen). All the in vitro-derived RNAs were quantified using a BioAnalyzer (Agilent).
Sample labeling, hybridization and image analysis
For direct cDNA labeling, 2 μg of polyadenylated RNA sample was labeled by reverse transcription with 300 μM Cy3- or Cy5-dUTP (Amersham) at 42°C for 2 h in a 30 μl reaction mixture containing 1 μg of oligo(dT) (18–20mer, Invitrogen), 1.5 μg of random hexamer (Invitrogen), 200 μM dTTP, 500 μM dCTP, dATP and dGTP (Amersham), 400 U Superscript II Reverse Transcriptase (Invitrogen), 1 μM DTT and 1× reverse transcription buffer. After completion of the labeling reaction, 15 μl of 0.1 M NaOH/2 mM EDTA solution was added to stop the reaction and degrade the RNA. The reaction mixture was neutralized by the addition of 15 μl of 0.1 M HCl. The labeled cDNA was purified using size exclusion chromatography (YM-30 column, Millipore).
Hybridization of fluorescently labeled cDNA to the slide was performed at 42°C for 16–18 h in 10 μl of 5× SSC, 0.1% SDS and 50% formamide under an 18 × 18 mm cover-slip in a sealed chamber. After hybridization, the arrays were sequentially washed in 2× SSC, 0.1% SDS at 42°C for 5 min, in 0.2× SSC at room temperature for 1 min, and in 0.1× SSC at room temperature for 1 min. After drying by centrifugation, the arrays were scanned with a GenePix 4000B scanner (Axon Instruments). Array image acquisition and signal analysis were performed using GenePix Pro 4.0 software.
Measurement bias
To estimate the number of probes needed per gene to obtain statistically significant expression results, we randomly picked different numbers of probes from the available probes (population probes) for every gene. Chauvenet's criterion was used for rejection of outlying data points (19). Probes with an intensity deviation greater than a critical value, defined as the deviation divided by the sample standard deviation, were regarded as statistical outliers and excluded. The remaining probes were used for the calculation of the mean intensity. We used the average intensity (sample mean of signals) to represent the hybridization signal for a gene. The same approach was used to calculate the population signal of the population probes for a gene. The average expression measurement bias between the sample and population signal intensities (measurement bias hereafter) is defined as
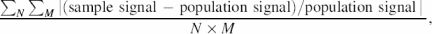
where M and N are, respectively, the number of iterative samplings and the number of genes used for the calculation.
The loss function
In order to identify the optimal probe length based on three factors, cross-hybridization (CH), measurement bias (MB) and the coefficient of variation (CV) of the signal intensity (SI), a loss function (20–23) was defined as L(CH, MB, SI) = w1·CH2 + w2·MB2 + (1 − w1 − w2)·SI2, where (0 ≤ w1, w2 ≤ 1) are the weights for CH, MB and SI. The weights, w1 and w2, for CH, MB and SI were set at 1/3 (equal weight) to balance these three components. The quadratic loss function was then simplified to L(CH, MB, SI) = CH2 + MB2 + SI2. This is a deterministic function, since no probability is involved.
RESULTS
Optimization of probe length: theoretical and empirical considerations
Effect of probe length on hybridization signal intensity variation
We analyzed two sets of literature data on genome-wide expression profiling, one for Escherichia coli (15 PM and MM pairs of 25mer probes per gene transcript, with ~80% of the genes having positive signals for at least 8 of the probe pairs) (13) and the other for Saccharomyces cerevisiae (8 60mer probes per gene) (7). The coefficient of variation, defined as the standard deviation divided by the mean of the hybridization signals for all the probes of each gene, was used as a measure of intensity variation. Based on statistical principles of sampling, the smaller the CV for the hybridization signals, the fewer the probes per gene needed for reliable gene expression measurement. The CV for the genome-wide average hybridization intensity was smaller for the 60mer probe set (0.55) than for the 25mer probe set (1.06).
To further explore the CV for signal intensity (hybridization efficiency) for longer probe lengths, we carried out experimental studies in which the same amount of seven Cy3-labeled cDNAs (five plant genes and two human genes) derived by in vitro-transcription of full-length gene clones (see Materials and Methods) were individually hybridized to an array of 336 probes. For each of the seven genes, there were six different probe lengths, with eight probes for each length. Probe selection was based on the algorithm described in the Materials and Methods. Figure Figure1A1A shows that the average CV for the hybridization intensity decreased monotonically with increasing probe length. These experimental results showed a reasonable agreement with the literature data of genome-wide expression measurements (7,13). The CV for hybridization intensity did not reach 0, but leveled off at a value of 0.1 for probes with 500 bases. This residual CV may result from spotting variation introduced by the arrayer, as the CV for the intensity of SYTO 61 staining of probes ranged from 7 to 12%.
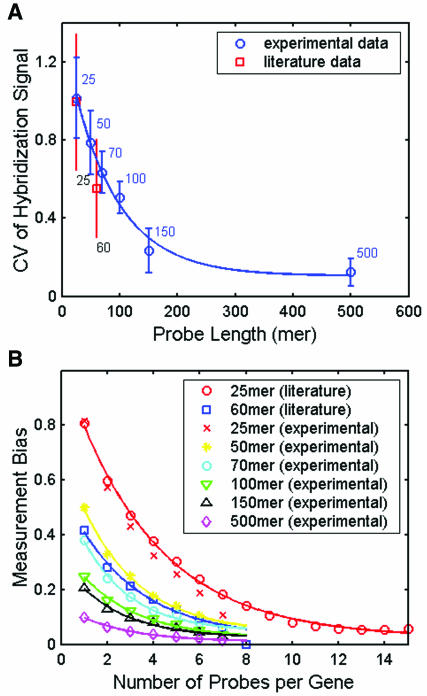
Effect of probe length and the number of probes per gene on expression measurement. (A) Effect of probe length on the variation (CV) in the hybridization signal using different length probes for the same genes. The experiments were repeated at least four times and the fluorescence signals were measured by Cy3 intensity. (B) Effect of the number of probes per gene on measurement bias.
Effect of the number of probes per gene on measurement bias
To evaluate measurement bias, we used the previous two literature datasets and the experimental data for the seven Cy3 labeled cDNAs. Figure Figure1B1B shows that the measurement bias (M = 100) decreased with an increase in the number of probes per gene. Fewer probes per gene were required for the longer probes to achieve the same bias as shorter probes. Although we used only seven genes in the experiments, the results corresponded quite well with the genome-wide literature data for E.coli and S.cerevisiae (7,13). These results show that a single 60–70mer probe per gene selected using a computational approach without experimental validation may not accurately measure the ‘true’ gene expression signal intensity and that multiple oligonucleotide probes per gene are required to obtain statistically reliable gene expression data.
Influence of probe length on cross-hybridization
Although long DNA or cDNA probes provide a low measurement bias and signal intensity variation, they often exhibit poor discrimination and hybridize to similar sequences, causing cross-hybridization problems. Cross-hybridization is also dependent on the stringency of the hybridization protocols. Several reports on the use of microarrays and commercial user manuals [(6,7,11,24), Amersham 30mer Uniset Bioarray, and MWG 50mer array protocol manuals] show that hybridization at 42°C in 30–50% formamide-based buffer works well for spotted DNA microarrays with probe lengths from 25mers to >1 kb (cDNA probes). Using fixed hybridization conditions in this study, the extent of cross-hybridization depends mainly on the sequence similarity between the sample targets and the microarray probes.
It is well known that the position of mismatched bases on short probes dramatically affects hybridization behavior (25). For instance, a single central mismatch in a 25mer probe completely eliminated the hybridization signal (13) and a single mismatch at 50 bases from the end of the probe attached to the slide surface of a 60mer microarray significantly reduced the hybridization signal (7). We therefore took these aforementioned mismatch positions on probes (25 and 60mer) into consideration in the cross-hybridization prediction below.
To compute the effect of probe length on cross-hybridization, we performed an in silico prediction of probe cross-hybridization to non-target human transcripts. We first randomly sampled 1000 genes from the UniGene database (build #153) and selected probes with lengths ranging from 25 to 1000 bases for each gene. Probe sequences of a particular length were chosen by a tiling method, one base at a time, along each gene. These probes were then aligned with the TIGR gene index (HGI version 9) using the BLAST program to analyze whether they would cross-hybridize to non-target sequences. A probe was considered prone to cross-hybridization if its sequence composition met the criteria for sequence similarity (see Materials and Methods) and mismatch positions described above.
By performing computations for over 106 probes for the 1000 genes, we found that among those prone to cross-hybridization probes, as predicted by the sequence similarity computation, only 3.2% of the 25mer probes and 3.5% of the 60mer probes had single mismatches at the center (25mer probes) or 10 bases from the 5′ end (60mer probes). These results show that sequence similarity is the dominant factor in genome-wide cross-hybridization computation. The percentage of the probes for the 1000 genes that would cross-hybridize versus probe length is shown in blue in Figure Figure2A.2A. The plot shows that randomly chosen 50mer probes give the minimum cross-hybridization.
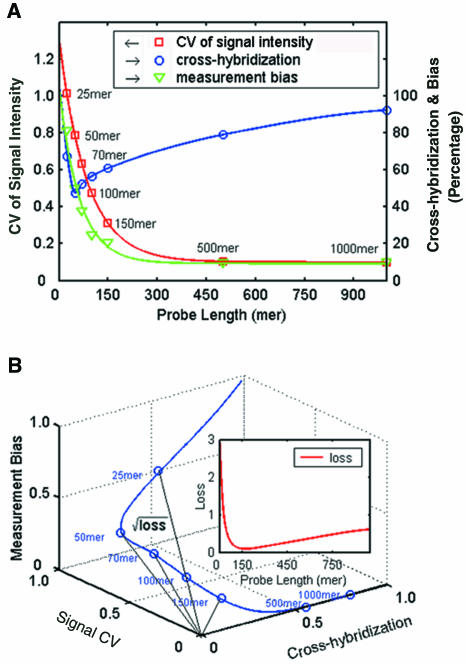
Determination of the optimal probe length. (A) Plots of cross-hybridization, measurement bias for one probe per gene, and the CV of the signal intensity versus probe length. All the solid lines were fitted by regression analysis of the data points. The scale for cross-hybridization and measurement bias is on the right of the figure. (B) The data from the three curves in (A) were plotted against each other in an XYZ plot, the minimum and maximum values for all parameters being normalized, respectively, to 0 and 1. The Euclidean distance between the curve and the origin represents the square root of the loss. The inset shows a plot of loss versus probe length to locate the minimum.
Probe length optimization
In addition to the cross-hybridization curve, Figure Figure2A2A combines the CV curve in Figure Figure1A1A with extrapolation to 1000 bases and the average measurement bias data extracted from the Figure Figure1B1B data for a single probe per gene. As shown in Figure Figure2A,2A, hybridization signal variation and measurement bias work in opposition to cross-hybridization for selecting an optimal probe length. Thus, the optimal probe length is a compromise length chosen to minimize these conflicting effects. We have developed a mathematical approach for the optimization. A statistical loss function (see Materials and Methods), defined as the square of the Euclidean distance to the origin (Figure (Figure2B),2B), was used to identify the optimal probe length, a smaller value of the loss function reflecting that the compound negative effects were closer to zero. The quadratic curve for loss function versus probe length up to 1000mer, displayed in the inset in Figure Figure2B,2B, showed that the minimum loss of 0.085 was seen with a 150–160mer probe. The loss function calculation provides a reference for optimal probe length selection. This conclusion was supported by the experimental validation below.
Optimization of probe length: experimental evaluation
Effects of probe length and probe concentration on hybridization sensitivity
In vitro-transcribed polyadenylated RNA for a plant gene, rbcl, was directly labeled with Cy3 dye by reverse transcription, then hybridized to an array containing rbcl probes of five different lengths ranging from 25 to 150 bases at various probe concentrations (0.2–100 μM). The probe sequence used for the indicated length was the best probe experimentally selected from the eight probes for each length using the same procedure for obtaining the data shown in Figure Figure1A.1A. A 500mer cDNA clone-derived probe at a concentration of 1 μM was spotted on the array for comparison. Figure Figure3A3A shows curves of the signal intensity versus probe concentration for different probe lengths. The experimental results showed that long DNA probes gave a more intense hybridization signal than the long oligonucleotide probes at every probe concentration tested, but that higher concentrations of long oligonucleotide probes gave the same signal intensity as lower concentrations of long DNA probes. For example, 50–70mer probes at a concentration of 20–40 μM gave a similar hybridization signal to 150 or 500mer probes at a concentration of 1 μM. These results show that long DNA probes, which extend farther away from the slide surface than oligonucleotide probes, are more accessible to free target molecules for hybridization. Although oligonucleotide probes are less accessible, a high surface density resulting from a high spotting concentration largely improves their poor hybridization signal intensity. These findings are in agreement with those in previous reports (11,12,26).
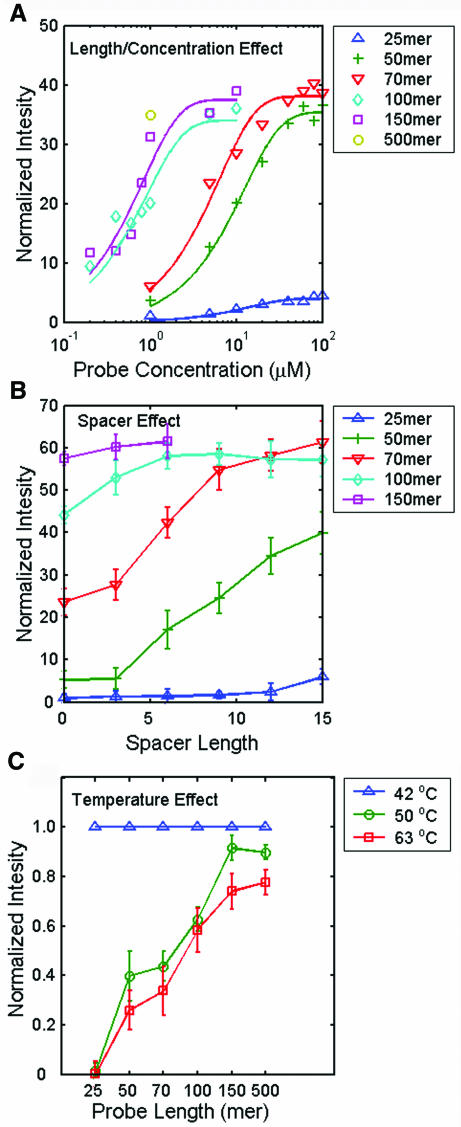
Assessment of discrepancies using different lengths of probes for hybridization. (A) Hybridization intensities at six probe lengths as a function of spotted probe concentrations. All the solid lines were fitted by regression analysis of the data points. Intensities were normalized to the intensity observed using the 25mer at a concentration of 1 μM. (B) Effect of spacer length on the mean hybridization intensity at five probe lengths. The basic single spacer unit had a length equivalent to a (dT)3 oligonucleotide, so the scale is based on the equivalent number of nucleotides. Intensities were normalized to the intensity obtained using the 25mer probe with no added spacer. (C) Mean hybridization intensities at three hybridization/washing temperatures as a function of probe length. Intensities measured at a given probe length were normalized to that at 42°C. The above experiments were performed three times for signal averaging.
Spacer effect
Figure Figure3B3B shows that increasing the probe length by addition of spacers of longer length enhanced the hybridization intensity. The spotting concentrations of 25–70mer oligonucleotides and 100–150mer PCR-derived single-stranded probes were 10 and 1 μM, respectively, as suggested by the slide manufacturer. The spacer effect was greater for 50–70mer probes, but negligible for long DNA probes (100–150mer); for the 25mer shown, much longer spacers were required to show a significant effect (data not shown). Previous reports have indicated that the addition of a spacer has a large effect on the hybridization signal intensity for 15–30mer oligonucleotides, but that the signal decreases with spacer length after an optimal length is reached (26–28). The hybridization intensities for the 100 and 150mer probes eventually reached the same plateau level (Figure (Figure33B).
Temperature effect
The above experimental results indicated that 150mer or longer probes did not benefit from spacer addition, were long enough to overcome steric and diffusion limitation and could be treated like long cDNA probes in the hybridization reaction. This was verified by the experimental results shown in Figure Figure3C,3C, in which hybrid stability was analyzed by the hybridization and post-hybridization washing temperature. We observed a reduction in hybridization intensity at higher hybridization/washing temperature for all of the six different probe lengths, but the effect started to reach a plateau at longer probe lengths. The data showed that 150 and 500mer probes gave similar results.
Array performance comparison of 70 and 150mer probes
Long oligonucleotide probes are becoming widely employed in commercial arrays for gene expression profiling (6,7,29). We therefore performed a series of experiments using the cytochrome P450 (CYP450) gene family to compare the array performance of 70 and 150mer probes. This gene family consists of many members with >80% sequence homology. The same criteria for avoiding cross-hybridization mentioned above were used to design gene-specific probes for the members of the gene family. Figure Figure4A4A shows the compiled results of 31 individual hybridization reactions. Each column shows the results of one Cy3-labeled, in vitro-transcribed CYP450 polyadenylated RNAs hybridized to an array containing the 31 150mer CYP450 gene probes spotted at 1 μM. No significant cross-hybridization was detected. These data demonstrate that gene-specific 150mer probes able to identify unique regions of a gene can be obtained using the computation approach.
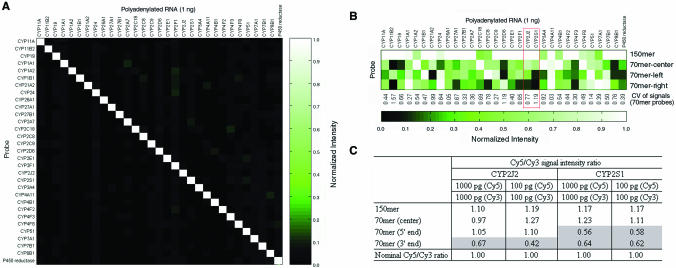
Array performance comparison of 70 and 150mer probes. (A) High specificity was seen using 31 CYP450 gene-specific 150mer probes. Each column represents the signal intensities of a CYP450 gene member hybridized to each of the 31 CYP450 probes. All signal intensities in each column were normalized to the brightest. Only the correct probe/target pairs yielded detectable signals. (B) Hybridization intensity variation using three 70mer CYP450 probes selected from within the corresponding 150mer gene-specific probe. (C) Probes with low hybridization sensitivity do not accurately measure the differential expression ratio. Lower signal intensity probes deviate greater from the nominal differential expression ratio of 1. All experiments were performed in triplicate for signal averaging. The Cy5/Cy3 intensity ratio was normalized by setting the arithmetic mean of the ratios of every spot on the array to 1.
In Figure Figure4B,4B, the same set of 31 in vitro-transcribed CYP450 cDNAs was individually hybridized to arrays containing probes for the 31 CYP450 genes with 4 probes per gene. The 4 probes were the 150mer probe and 3 70mer probes, selected by the aforementioned probe design algorithm from the center, 5′- or 3′ end of the 150mer gene probe. All four probes had a GC content between 45 and 55% and a similar melting temperature. Similar to that described in Figure Figure3A,3A, the 70 and 150mer probes were spotted on the P450 array at the optimal concentrations of 40 and 1 μM, respectively. In general, the 70mer CYP450 probes gave a poorer signal intensity than the corresponding 150mer probes. The average hybridization signal variation (CV of 0.6) calculated from the 31 sets of 3 70mer probes was the same as that in Figure Figure1B.1B. In total, ~23% (21/93) of the 70mer CYP450 probes gave a hybridization signal <20% of the strongest signal for that gene.
The current microarray experimental protocol employs two-color fluorescence detection for differential expression ratio measurement. It has been reported that, using the same set of probes, the variation in the differential expression ratio is lower than that in the absolute hybridization signal (7), suggesting that multiple probes per gene may not be needed in most applications using differential expression ratio measurement. Our experimental results showed that this holds provided that an experimental pre-screening process is employed to select the best single probe with a good signal intensity for a gene. For instance, Figure Figure4C4C shows that the probes giving a good signal intensity for CYP2J2 and CYP2S1 (framed in Figure Figure4B),4B), but not those giving a poor signal intensity, can provide accurate differential expression ratios.
DISCUSSION
In addition to sequence composition and probe length, target length is an important parameter in hybridization studies. Because of the poor hybridization efficiency of short probes, a protocol for RNA amplification using in vitro transcription (IVT) to generate labeled cRNA (30) is used to increase the amount of target molecules for hybridization. The process is followed by a fragmentation step to break down possible secondary structures in labeled cRNA target molecules and to increase the diffusion rate of the target molecules for microarray hybridization reactions with 20–30mer probes [(2,11), Amersham 30mer Uniset Bioarray protocol manual). For microarray experiments using long oligonucleotide probes, IVT is not used routinely and is only necessary when the amount of RNA is very low, e.g. RNA derived from tissue specimens.
The present study used a single hybridization protocol and probes of different lengths to examine their effects on gene expression measurement. To minimize experimental bias introduced by target length, a reverse transcription and labeling protocol using oligo(dT) and random primers (31) was employed. The protocol generated cDNA fragments ranging in length from 150 ± 60 to ~1000 bases, as revealed by slab gel electrophoresis (data not shown). The short target length facilitates the interaction of labeled cDNA with the probes during hybridization.
Previous literature reports on the analysis of oligonucleotide probe length have focused mainly on specificity and sensitivity (11,26) and, as far as we are aware of, this is the first study to examine the effect of probe length on hybridization intensity variation tested with different probes for a given gene and on measurement bias in gene expression profiling. We believe that all of these factors contribute to the discordant expression results observed using different microarray platforms. Since oligonucleotide hybridization efficiency is highly sequence dependent and cDNA probes are too long to avoid non-specific cross-hybridization, these two microarray platforms may not give concordant results. For instance, Tan et al. (32) evaluated three commercial microarrays, Agilent cDNA microarray, Affymetrix GeneChip and Amersham CodeLink Uniset Bioarray and found substantial differences using these three platforms. On the other hand, Barczak et al. (29) reported a good correlation (r = 0.8–0.9) between relative gene expression levels measured using a long oligonucleotide array (a single 70mer probe per gene) and Affymetrix GeneChip. On the basis of the reasonable assumption that the experimental protocols for these commercial microarrays have been optimized for gene expression profiling and that the additional RNA amplification (IVT) step employed in the short oligonucleotide probe microarray experiments does not introduce bias, a plausible explanation for the contradictory findings is sequence-dependent hybridization variation and measurement bias.
Barczak et al. (29) also discovered that two different collections of 70mer probes sometimes yielded dramatic signal differences for the two probes for the same gene. Our experimental results, shown in Figure Figure4B4B and C, agree with this finding and strongly suggest that a large-scale hybridization screening for the single best oligonucleotide probe per gene or multiple probes per gene is needed for making microarrays that yield reliable gene expression measurements. Large-scale probe screening was used in a recent study by Lucito et al. (33), who tested ~700 000 unique 70mer probes to find the probes giving the strongest hybridization signals. Our present study also demonstrated that a single 70mer or longer oligonucleotide probe for a gene could be sufficient for accurate expression measurement if the probe is validated experimentally.
000 unique 70mer probes to find the probes giving the strongest hybridization signals. Our present study also demonstrated that a single 70mer or longer oligonucleotide probe for a gene could be sufficient for accurate expression measurement if the probe is validated experimentally.
The present study takes into account three factors to evaluate the performance of DNA microarrays of different probe lengths, investigates sequence-dependent effects and extends the study of the effect of probe length to beyond 500 bases. The experimental results suggest that gene-specific 150mer probes selected by a probe design computer program can minimize signal intensity variation and measurement bias to such an extent that prior experimental screening is not necessary, i.e. the hybridization properties of 150mer probes are similar to those of long cDNA probes and 150mer probes can be used for gene-specific expression measurements.
In summary, nucleic acid hybridization involves a multitude of variables not yet predictable by computation alone. A good probe design strategy is essential for accurate expression measurement. This study focused on characterizing the effect of probe length and the number of probes per gene on accurate microarray measurement of gene expression. Our data suggest that probes ~150mer in length are optimal for this purpose. Short or long oligonucleotide probes can also work well if the probes are validated by experimental hybridization selection or if multiple probes per gene are used. We believe that concordant results can be obtained using different microarray platforms if all the negative effects are minimized.
ACKNOWLEDGEMENTS
We thank Hung-Hui Chen, Ping-Ann Chen, Li-Jen Huang and Bi-Yun Lee for preparing the oligonucleotides and the gene-specific probes. We also thank Dr Tom Barkas for reviewing the manuscript for us. This project was supported by grants from the National Research Program for Genome Medicine (NSC 92-3112-B-001-013; NSC 93-3112-B-001-013-Y) of the National Science Council and a fellowship to C-.C.C. from the Medical Biotechnology Program of the Academia Sinica.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gnh099
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/32/12/e99/7038014/gnh099.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/100112588
Article citations
Weak-cooperative binding of a long single-stranded DNA chain on a surface.
Nucleic Acids Res, 52(15):8661-8674, 01 Aug 2024
Cited by: 0 articles | PMID: 38989620 | PMCID: PMC11347152
Experimental MicroRNA Detection Methods.
Methods Mol Biol, 2257:33-55, 01 Jan 2022
Cited by: 6 articles | PMID: 34432272
Review
Cell type-specific analysis by single-cell profiling identifies a stable mammalian tRNA-mRNA interface and increased translation efficiency in neurons.
Genome Res, 32(1):97-110, 02 Dec 2021
Cited by: 12 articles | PMID: 34857654 | PMCID: PMC8744671
Ribo-Pop: simple, cost-effective, and widely applicable ribosomal RNA depletion.
RNA, 26(11):1731-1742, 05 Aug 2020
Cited by: 6 articles | PMID: 32759389 | PMCID: PMC7566562
Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation.
Nat Commun, 11(1):4104, 14 Aug 2020
Cited by: 72 articles | PMID: 32796835 | PMCID: PMC7428014
Go to all (93) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A novel design of whole-genome microarray probes for Saccharomyces cerevisiae which minimizes cross-hybridization.
BMC Genomics, 4(1):38, 22 Sep 2003
Cited by: 21 articles | PMID: 14499002 | PMCID: PMC239980
Design and fabrication of spotted long oligonucleotide microarrays for gene expression analysis.
Methods Mol Biol, 381:213-225, 01 Jan 2007
Cited by: 0 articles | PMID: 17984521
Microarray analysis using disiloxyl 70mer oligonucleotides.
Nucleic Acids Res, 36(4):1334-1342, 10 Jan 2008
Cited by: 0 articles | PMID: 18187508 | PMCID: PMC2275082
A model of molecular interactions on short oligonucleotide microarrays.
Nat Biotechnol, 21(7):818-821, 08 Jun 2003
Cited by: 187 articles | PMID: 12794640




