Abstract
Free full text

Nicotinamide N-methyltransferase: a potential biomarker for worse prognosis in gastric carcinoma
Abstract
In clinical practice, cancer stage (or grade) and some biomarkers, such as carcinoembryonic antigen (CEA) and CA199, are widely used to predict the prognosis of gastric carcinoma patients. Due to the limited role of prognostic indicators for gastric carcinoma, this condition remains one of the most fatal human malignancies with a dismal prognosis. Nicotinamide N-methyltransferase (NNMT, EC.2.1.1.1), a metabolizing enzyme, is involved in the development and progression of various carcinomas. However, the prognostic and biological functions of NNMT in gastric carcinoma are not yet clear. In the present study, NNMT was found to be overexpressed at the mRNA and protein levels in gastric carcinoma tissues compared with adjacent tissues. Importantly, the survival analysis verified that NNMT expression is an independent prognostic factor for overall survival of gastric cancer patients. Moreover, NNMT expression was related to primary tumor size, lymph node metastasis, distant metastasis, and TNM (tumor, node, and metastasis) stage. We also demonstrated that knockdown of NNMT inhibits cellular proliferation, invasion and migration in vitro and in vivo. Overall, the results of this study suggest that NNMT is a promising prognostic predictor for gastric cancer patients and could be used as a new target for gastric cancer therapy.
Introduction
Gastric cancer is the second leading cause of cancer-related mortality and the fourth most common cancer globally after lung, breast and colorectal cancers [1]. In East Asia, especially China, gastric cancer accounts for approximately half of all cancers [2]. Delayed detection of the primary tumor, progressive growth, and late detection of metastases hinder the search for a cure for gastric cancer [3]. Currently, reliable predictors for gastric carcinoma are lacking. Douglas Hanahan and Robert A. Weinberg emphasized in their eminent review Hallmarks of cancer: the next generation that “Reprogramming energy metabolism could be an emerging hallmark of cancer” [4]. The relationship between metabolism-related enzymes and cancer has become a popular topic of cancer research.
Nicotinamide N-methyltransferase (NNMT), a phase II metabolizing enzyme, mainly catalyzes the methylation of nicotinamide and other pyridines into pyridinium ions [5]. Most studies of this metabolizing enzyme mainly examined its roles in metabolic diseases, such as diabetes [6] and obesity [7]. In 1984, Seifert, R was the first to prove that alteration of NNMT activity was involved in the development and progression of carcinoma in vivo [8]. Numerous subsequent studies demonstrated that NNMT was overexpressed in various cancers and was correlated with poor prognosis [9-16]. Studies have also indicated that NNMT influences the proliferative, migratory, invasive and differentiation profiles of various cancers [17-19]. For example, a previous study suggested that the phosphoinositide 3-kinase (PI3K)/Akt pathway plays a vital role in NNMT-dependent cellular invasion and matrix metallopeptidase 2 (MMP-2) activation in renal carcinoma [20]. Although NNMT expression was reported to be elevated in gastric carcinoma by a former study, the biological function and prognostic role of NNMT in gastric carcinoma still remain unclear [21].
The present study evaluated the role of NNMT as a prognostic determinant by analyzing NNMT expression in a large cohort of well-characterized gastric samples. To further explore the biological role of NNMT in gastric carcinoma, siRNA-mediated silencing was also performed in vitro and in vivo. Examining whether NNMT inhibition is a possible molecular approach for the treatment of gastric carcinoma has value.
Material and methods
Patient samples
For IHC analysis, 875 formalin-fixed paraffin-embedded (FFPE) tissue samples (35 chronic gastritis, 31 intestinal metaplasia, 32 low-grade intraepithelial neoplasia, 42 high-grade intraepithelial neoplasia, 641 carcinomas, and 94 adjacent tissue samples) from 781 patients were collected. These tissues were obtained from the Department of Pathology, Affiliated Hospital of Nantong University from 2003 to 2010. The details of these tissues and the IHC analysis are presented in Tables 1, ,22 and and3.3. In addition, 38 paired tissues (freshly frozen gastric carcinoma and adjacent tissues from Nanjing Medical University Affiliated Cancer Hospital, Second Affiliated Hospital of Nanjing Medical University and Nanjing Drum Tower Hospital) were also examined by qRT-PCR. Each patient provided written informed consent for the publication of this study, and the research protocol was approved by the Human Research Ethics Committee.
Table 1
NNMT expression in gastric benign and malignant tissues
| Characteristic | NO. | NNMT | Pearson χ2 | P-value | |
|---|---|---|---|---|---|
|
| |||||
| High no. | % | ||||
| Gastric tissues | 36.490 | <0.001* | |||
| Chronic gastritis | 35 | 10 | 28.6 | ||
| Intestinal Metaplasia | 31 | 11 | 35.5 | ||
| Low-grade intraepithelial neoplasia | 32 | 12 | 37.5 | ||
| High-grade intraepithelial neoplasia | 42 | 17 | 40.5 | ||
| Carcinoma | 641 | 364 | 56.8 | ||
| Adjacent tissue | 94 | 30 | 31.9 | ||
Table 2
Correlation of high NNMT expression with clinical attributes in gastric cancer
| Groups | NO. | NNMT | Pearson χ2 | P-value | |
|---|---|---|---|---|---|
|
| |||||
| High no. | % | ||||
| Gender | 3.832 | 0.061 | |||
| Male | 464 | 246 | 53.0 | ||
| Female | 153 | 95 | 62.1 | ||
| Age | 1.978 | 0.161 | |||
| ≤60 years | 247 | 128 | 51.8 | ||
| >60 years | 370 | 213 | 57.6 | ||
| Histological type | 4.431 | 0.351 | |||
| Tubular | 512 | 300 | 57.6 | ||
| Hybrid | 7 | 3 | 42.9 | ||
| Mucinous | 28 | 11 | 39.3 | ||
| Signet ring cell | 21 | 11 | 52.4 | ||
| Othersa | 13 | 8 | 61.5 | ||
| Differentiation | 5.066 | 0.079 | |||
| High | 32 | 12 | 37.5 | ||
| Moderate | 157 | 87 | 55.4 | ||
| Low | 383 | 222 | 58 | ||
| Othersb | |||||
| Primary tumor size | 33.468 | <0.001* | |||
| Tis | 33 | 17 | 21.2 | ||
| T1 | 62 | 28 | 45.2 | ||
| T2 | 120 | 55 | 45.8 | ||
| T3 | 345 | 214 | 62 | ||
| T4 | 48 | 34 | 70.8 | ||
| Lymph node metastasis | 33.486 | <0.001* | |||
| N0 | 238 | 101 | 42.4 | ||
| N1 | 108 | 60 | 55.6 | ||
| N2 | 117 | 74 | 63.2 | ||
| N3 | 145 | 103 | 71 | ||
| Distant metastasis | 9.130 | 0.002* | |||
| M0 | 580 | 312 | 53.8 | ||
| M1 | 38 | 30 | 78.9 | ||
| TNM stage | 46.134 | <0.001* | |||
| 0 | 34 | 9 | 26.5 | ||
| I | 119 | 44 | 37.0 | ||
| II | 200 | 112 | 56.0 | ||
| III | 217 | 143 | 65.9 | ||
| IV | 38 | 30 | 78.9 | ||
| Preoperative CEA, ng/ml | 2.517 | 0.284 | |||
| ≤5 | 263 | 136 | 51.7 | ||
| >5 | 74 | 44 | 59.5 | ||
| Unknown | 281 | 162 | 57.7 | ||
| Preoperative CA199, U/ml | 2.590 | 0.274 | |||
| ≤37 | 276 | 143 | 51.8 | ||
| >37 | 50 | 30 | 60 | ||
| Unknown | 292 | 169 | 57.9 | ||
Table 3
Univariate and multivariate analyses of prognostic factors for overall survival in gastric cancer
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
| |||||
| HR | p>|z| | 95% CI | HR | p>|z| | 95% CI | |
| NNMT expression | ||||||
| High vs Low | 1.894 | <0.001* | 1.509-2.378 | 1.446 | 0.028* | 1.041-2.065 |
| Gender | ||||||
| Male vs Female | 0.930 | 0.559 | 0.730-1.185 | |||
| Age | ||||||
| >60 years vs ≤60 years | 1.463 | 0.001* | 1.169-1.831 | 0.434 | 0.878 | 0.633-1.217 |
| Histological type | ||||||
| Tubular vs Hybrid vs Mucinous vs Signet ring cell vs othersa | 0.929 | 0.257 | 0.817-1.055 | |||
| Differentiation | ||||||
| High vs moderate vs low vs othersb | 1.397 | <0.001* | 1.137-1.716 | 1.106 | 0.575 | 0.778-1.572 |
| Primary tumor size | ||||||
| T4 vs T3 vs T2 and T1 vs Tis | 2.017 | <0.001* | 1.747-2.329 | 1.436 | 0.014* | 1.075-1.918 |
| Lymph node metastasis | ||||||
| N3 vs N2 vs N1 vs N0 | 1.702 | <0.001* | 1.554-1.865 | 1.288 | 0.004* | 1.082-1.532 |
| Distant metastasis | ||||||
| M1 vs M0 | 3.745 | <0.001* | 2.641-5.312 | 3.195 | <0.001* | 1.697-6.013 |
| TNM stage | ||||||
| IV vs III vs II vs and I | 2.244 | <0.001* | 1.979-2.545 | 1.400 | 0.054 | 0.995-1.969 |
| Preoperative CEA, ng/ml | ||||||
| >5 versus ≤5 | 2.403 | <0.001* | 1.763-3.274 | 1.772 | 0.003* | 1.216-2.582 |
| Preoperative CA199, U/ml | ||||||
| >37 versus ≤37 | 2.570 | <0.001* | 1.806-3.649 | 1.524 | 0.051 | 0.998-2.326 |
TMA construction and IHC analysis
A tissue microarray system (Quick-Ray, UT06, UNITMA, and Korea) was used as described previously in the Department of Clinical Pathology, Nantong University Hospital, Jiangsu, China [22]. The immunohistochemical analysis was performed as previously described [23]. The IHC analysis to evaluate the protein expression of NNMT in gastric tissues was conducted using anti-NNMT mouse monoclonal antibody (3D8) [NBP2-00537].
NNMT immunostaining was scored by blinded observers according to the intensity and percentage of positive cells. The staining intensity was scored according to 4 grades: 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (intense staining). The product of the percentage of positive cells and the respective intensity scores was used as the final staining score, with a minimum value of 0 and a maximum value of 300. A cutoff value of 120 was found to be statistically significant using the X-tile software program (the Rimm Lab at Yale University; http://www.tissuearray.org/rimmlab/) [24].
RNA preparation, reverse transcription, and real-time quantitative PCR
Total RNA was isolated from tissue samples and cultured cells using Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. RNA was reverse transcribed into cDNA using the Prime Script RT reagent Kit (Takara, China). Quantitative real-time PCR was performed using the SYBR Prime-Script RT-PCR kit (Takara, Japan) and QuantStudioTM 6 Flex Real-Time PCR System. Relative NNMT expression was calculated and was normalized to β-actin using the 2-ΔΔCt method. The primers used in the quantitative real-time PCR analysis are presented in Table 4. All experiments were conducted at least three times.
Table 4
Primer series of NNMT and β-actin gene
| Gene | Forward sequence (5’-3’) | Reverse sequence (5’-3’) |
|---|---|---|
| NNMT | GAGCAGAAGTTCTCCAGCCT | ACCATTCGATTGTGTAGCCA |
| β-actin | TGGAGAAAATCTGGCACCAC | GAGGCGTACAGGGATAGCAC |
Cell culture and transfection
The gastric carcinoma cell lines MKN28, SGC7901, MGC803 and BGC823 were purchased from American Type Culture Collection (ATCC, Netherlands) and the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA) medium containing 10% fetal bovine serum (FBS, GIBCO) at 37°C in humidified air containing 5% CO2. Small interfering RNA (siRNA) transfection. NNMT siRNA (si1 and si2) and negative control siRNA were purchased from Gene Pharma (Shanghai, China). Cells (5×104 cells/well) were seeded into 6-well plates and were transfected with 10 nM siRNA in phosphate-buffered saline (PBS) using Lipofectamine-2000 (Invitrogen) according to the manufacturer’s instructions. The siRNA sequences are presented in Table 5.
Table 5
Sequences of siRNA for NNMT expression
| No. | Gene name | Sequence | |
|---|---|---|---|
|
| |||
| Sense (5’-3’) | Antisense (5’-3’) | ||
| 1 | siRNA-homo-791 | CCUCGGGAUUACCUAGAAATT | UUUCUAGGUAAUCCCGAGGTT |
| 2 | siRNA-homo-846 | GCCAGAUUCUUAAGCACCUTT | AGGUGCUUAAGAAUCUGGCTT |
| 3 | NC-siRNA | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
Western blot assay
The cells were harvested for protein extraction, and the extracted proteins were quantified as previously described using a 12% or 4~20% polyacrylamide gradient sodium dodecyl sulfate (SDS) gel [23]. After the proteins were separated in the gel and transferred to a polyvinylidene difluoride (PVDF) membrane, the membrane was blocked in 2% BSA in TBST for 1 h. Then, the membranes were incubated overnight (4°C) with anti-NNMT mouse monoclonal antibody (3D8) [NBP2-00537]. After incubation with Alexa Fluor 488-labeled Goat Anti-Mouse IgG (Beyotime Biotechnology, China) at room temperature for 2 h, the relative protein levels were calculated using GAPDH (Abcam, ab181602) as a loading control. Each assay was repeated three times.
Cell proliferation
The cell proliferation assay was performed using Cell Counting Kit-8 (CCK-8; Dojindo, Tokyo, Japan) at 48 h after transfection. Briefly, 2000 cells were plated per well in 96-well plates and were cultured for 24 h. Then, 10 μl of CCK-8 reagent was added to each well, and the plates were incubated at 37°C for 2 h. The absorbances at 450 nm (A450) and 650 nm (A650) were measured using a Synergy 2 microplate reader (BioTek, Winooski, VT, USA) and were detected in quadruplicate at various time points for 6 days. The final absorbance was calculated as A650 minus A450, and cell viability was normalized using the following formula: (final absorbance treated/final absorbance control) × 100%. All experiments were performed at least three times.
Flow cytometry
The cells were collected and washed three times with PBS and maintained in ethanol for at least 24 h at -20°C. Then, the cells were incubated in propidium iodide (PI) staining solution (RNase A 100 ug/ml and PI 500 ug/ml) for 30 min at 4°C. Next, the cells were analyzed using a FACScan flow cytometer (BD Biosciences), and the cell cycle distributions were calculated using the Cell Quest software package (BD Biosciences) according to the manufacturer’s protocol. The percentage of cells in G1, S, and G2 phase were counted and compared. Each assay was repeated three times.
Migration and invasion
The migration and invasion assays were performed using 24-well Transwell chambers (8 mm pores, Millipore, Billerica, MA). For the migration assay, tumor cells were resuspended in serum-free RPMI-1640 medium; 2×105 cells were seeded into the upper chambers, and 0.5 mL RPMI-1640 containing 10% FBS was added to the bottom chambers. Following a 24 h-incubation, the cells on the upper surface of the membrane were scrubbed off, and the migrated cells were fixed with 95% ethanol, stained with 0.1% crystal violet, and counted under a light microscope. The invasion assay protocol was similar to the migration assay protocol, except that the upper chambers were first covered with 1 mg/ml Matrigel. All experiments were conducted at least three times.
Xenograft experiment
MGC803 cells were transfected with sh-NNMT or negative control siRNAs (si1 and siNC, respectively) using Lipofectamine 2000 (Invitrogen). NNMT mRNA and protein levels were determined by qRT-PCR and Western blot, respectively. The cells were collected and injected into either side of the posterior flank of the same BALB/c nude mouse after 48 h. Tumor volume and weight were measured every 2 days. Tumor volume was measured as length × width2 × 0.5. At 25 days after injection, the mice were sacrificed, and the final tumor weights were measured. All experimental manipulations were undertaken in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Statistical analysis
All statistical analyses were performed using SPSS 21.0 statistical software (SPSS Inc. Chicago, IL) [25]. The relationships between NNMT expression and clinicopathological attributes were analyzed using the x2 test. Survival rates were calculated using the Kaplan-Meier method and were compared by the log rank test. The univariate and multivariate analyses followed the Cox proportional hazards regression model. A P-value of less than 0.05 was considered to indicate a statistically significant difference.
Bioinformatics
Level 3 TCGA data (TCGA_STAD_exp_HiSeq-2015-02-24) were used to evaluate the differential expression of NNMT between gastric carcinoma tissues (n=384) and normal tissues (n=37). Data were downloaded from the website of UCSC cancer browser: http://genome-cancer.ucsc.edu and were generated using an IlluminaHiSeq platform to examine pathologically diagnosed cancer tissue samples and relative normal tissue samples. The mRNA expression levels of various genes were measured and normalized. NNMT mRNA expression was obtained from the “genomic Matrix” file (using Editplus). Then, the data were grouped into a tumor tissue group and normal tissue group, and the Fisher’s t-test was used to compare the two groups.
The TCGA data was also used for KEGG Pathway enrichment analysis. The biological functions of the genes being examined were determined using the Database for Annotation, Visualization, and Integrated Discovery (DAVID). The pathway analysis using the KEGG Pathway database revealed that the differentially regulated genes were associated with critical biological functions. Analyses were performed to identify the biological functions and pathways of the identified differentially expressed genes (DEGs) based on the hypergeometric distribution algorithm. P<0.05 was chosen as the cut-off value for enriched functions and pathways.
Results
Expression of NNMT in gastric tissues
The Cancer Genome Atlas (TCGA) data analysis revealed that NNMT mRNA expression was upregulated in gastric carcinoma tissues compared with normal tissues (P<0.001) (Figure 1A). To verify those results, NNMT expression was analyzed in 38 paired primary cancerous and adjacent noncancerous tissues of gastric carcinoma by quantitative real-time polymerase chain reaction (qRT-PCR). NNMT mRNA was overexpressed in gastric carcinoma tissues, with an average upregulation of 1.833-fold (P=0.003) (Figure 1B), which is consistent with the results of the TCGA data analysis.
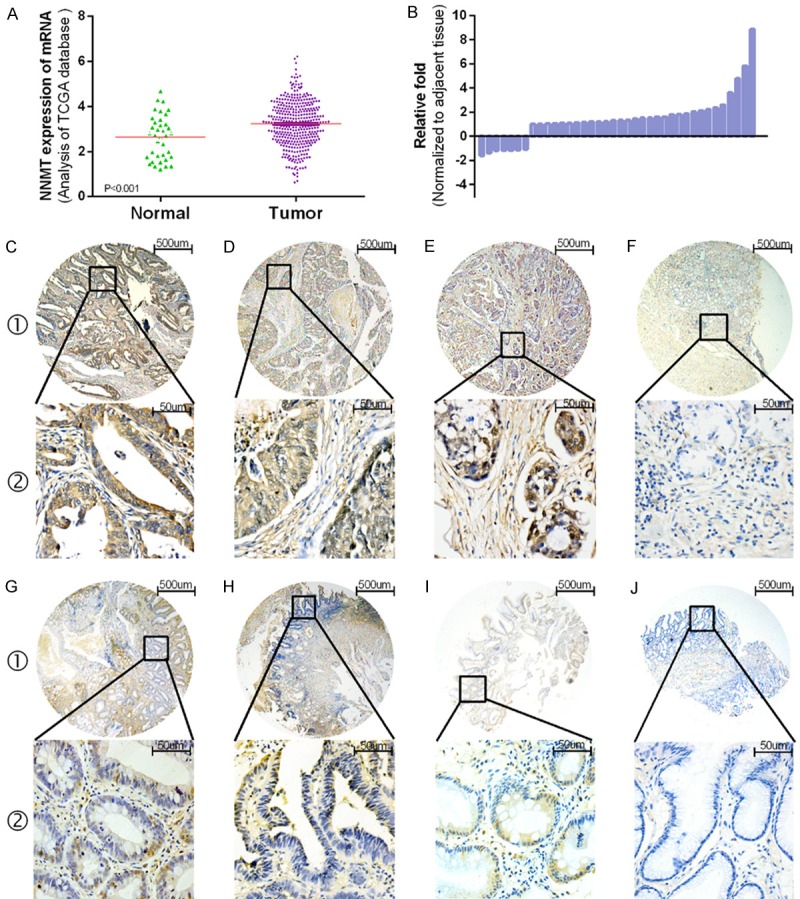
NNMT mRNA and protein expression levels in gastric tissues. (A) TCGA data analysis indicates that the NNMT mRNA level in 384 gastric carcinoma tissues is significantly increased compared with 37 non-cancerous tissues (P<0.001). (B) NNMT was detected in 38 pairs of gastric tissues by qRT-PCR. The level of NNMT in gastric carcinoma tissues was significantly increased compared with non-tumorous tissues (p=0.003). (C) Representative pattern of NNMT protein expression in gastric carcinoma tissues. Strongly positive tumor cytoplasmic immunohistochemical staining of NNMT in gastric carcinoma samples. (D, E) NNMT was expressed in both the cytoplasm and stroma, as indicated by strong IHC staining. (F) Representative non-cancerous tissue sample with low NNMT expression. (G, H, J) Precancerous lesions with low NNMT expression, such as low-grade intraepithelial neoplasia (G1 and G2), high-grade intraepithelial neoplasia (H1 and H2), chronic gastritis (I1 and I2), and intestinal metaplasia (J1 and J2).
Then, we used immunohistochemical staining to examine NNMT protein expression in gastric carcinoma tissues, benign gastric disease tissues and corresponding noncancerous tissues. As presented in Table 1, NNMT protein expression was increased in gastric carcinoma tissues compared with benign disease tissues and noncancerous tissues (Pearson χ2=36.490, P<0.001). Also, the progression of precancerous lesions was positively correlated with NNMT expression. Figure 1C-J demonstrates that NNMT expression was localized to the tumor cytoplasm in a multitude of immunohistochemistry (IHC) specimens (Figure 1C), whereas NNMT was expressed in both the cytoplasm and stroma in the other groups (Figure 1D, ,1E1E).
Association of NNMT expression with clinicopathological characteristics in gastric cancers
The relationship between NNMT expression and the clinicopathological attributes of gastric cancer are presented in Table 2. High NNMT positive staining within the cytoplasm and interstitium was significantly associated with primary tumor size (Pearson χ2=33.468, P<0.001), lymph node metastasis (Pearson χ2=33.486, P<0.001), distant metastasis (Pearson χ2=9.130, P=0.002) and TNM stage (Pearson χ2=46.134, P<0.001). NNMT expression was not associated with any other clinical parameters, including gender, age, histological type, differentiation, carcinoembryonic antigen (CEA) status and CA199 status.
Prognostic value of NNMT protein expression in gastric cancer
We examined possible prognostic factors for gastric cancer using both univariate and multivariate Cox regression analyses. The univariate analyses showed that increased expression of NNMT, age, differentiation, primary tumor size, lymph node metastasis, distant metastasis, TNM stage, CEA status and CA199 status were associated with the prognoses of gastric carcinoma patients in terms of overall survival rate. The prognostic indicators identified in the univariate analyses were included in the multivariate analyses, which further indicated that NNMT expression was associated with poor overall survival (hazard ratio (HR), 1.446; P=0.028; 95% confidence interval (CI), 1.041-2.065) as was primary tumor size, lymph node metastasis, distant metastasis and CEA status. The detailed findings are presented in Table 3. Kaplan-Meier survival curves revealed that low expression levels of NNMT and CEA and early cancer stage were associated with an enhanced rate of survival and that patients positive for the two markers NNMT and CEA had a significantly worse prognosis (Figure 2). These Kaplan-Meier survival curves will help us analyze the five-year survival rate of patients with gastric cancer (Figure 2A, ,2B,2B, ,2F2F).
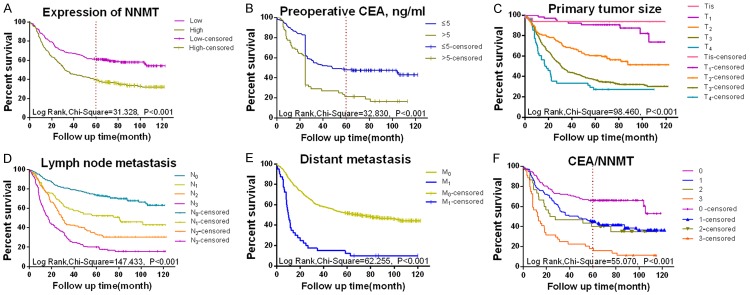
Survival curves of gastric carcinoma generated by the Kaplan-Meier method and the log-rank test. A. Overall survival curves of NNMT+ (green line, High) and NNMT- tissue samples (purple line, Low). B. Overall survival curves by primary tumor size: Tis (pink line, Tis), T1 (purple line, T1), T2 (orange line, T2), T3 (green line, T3) and T4 (blue line, T4). C. Overall survival curves by stage of lymph node metastasis: node-negative (blue line, N0), L1 (green line, L1), L2 (orange line, L2), and L3 (purple line, L3). D. Overall survival curves by distant metastasis: negative (green line, M0) and positive (light blue line, M1). E. Overall survival curves by preoperative CEA status: high (green line, ≤5) and low (blue line, >5). F. Overall survival curves by CEA and NNMT status: CEA-/NNMT- (purple line, 0), CEA-/NNMT+ (blue line, 2), CEA+/NNMT- (green line, 3) and CEA+/NNMT+ (orange line, 4).
Knockdown of NNMT inhibits cellular proliferation, invasion and migration in vitro
To better demonstrate the role of NNMT in gastric carcinoma, we first measured the expression of NNMT in four gastric cancer cell lines (MKN28, SGC7901, MGC803 and BGC823). The MGC803 and BGC823 cell lines exhibited increased NNMT expression levels compared with the other two cell lines (Figure 3A), and these cell lines were used for NNMT silencing experiments conducted using two siRNA molecules (Table 5). The NNMT mRNA and protein levels were analyzed to determine the specific effects of siRNA treatment on NNMT expression. Compared with the negative control, both NNMT mRNA and protein levels were significantly reduced in the siRNA groups. NNMT mRNA expression was approximately 4-fold reduced in the siRNA groups compared with the negative control group (Figure 3B).
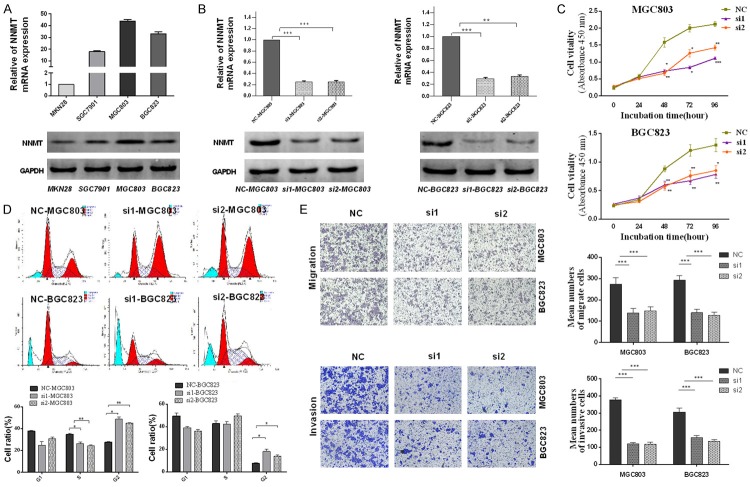
Knockdown of NNMT inhibited the proliferation, migration and invasion of gastric carcinoma cell lines in vitro. A. NNMT expression in MKN28, SGC7901, MGC803, and BGC823 cell lines. β-actin was used as the internal control in qRT-PCR, and GAPDH was used as the internal control in Western blot. B. In MGC803 and BGC823 cells, NNMT mRNA and protein levels were significantly reduced after transfection with NNMT siRNA (si1 and si2). C. NNMT promotes proliferation of gastric carcinoma cells. Cell proliferation assay using the CCK-8 kit: compared with negative control (NC), siRNA-mediated silencing of NNMT significantly inhibited cell proliferation in MGC803 and BGC823 cell lines. D. NNMT alters the cell cycle of gastric carcinoma cell lines. MGC803 and BGC823 cells transfected with NC (siRNA negative control) or NNMT siRNA (si1 and si2). Thirty-six hours after transfection, cells were stained and analyzed by flow cytometry. The cell cycle was arrested at the G2 phase. E. NNMT promotes migration of gastric carcinoma cell lines (MGC803 and BGC823). Compared with negative control (NC), cells transfected with NNMT siRNA (si1 and si2) showed weaker migratory and invasive abilities.
Induction of cell cycle arrest is an important mechanism for controlling tumor growth. We assessed the role of NNMT in regulating gastric carcinoma cell cycle progression to determine the effect of NNMT on gastric carcinoma cell growth. In MGC803 cells, NNMT downregulation caused a significant increase in the percentage of G2 phase cells and a concomitant decrease in the percentage of S phase cells, thus indicating the occurrence of G2 cell cycle arrest (Figure 3D). CCK-8 assays in MGC803 and BGC823 cell lines revealed that knockdown of NNMT reduced cell proliferation compared with the negative controls (Figure 3C).
To investigate the effect of NNMT on migration and invasion, two specific small interference RNAs, si1 and si2, and a negative control (si-NC) were transfected into MGC803 and BGC823 cells. Transwell migration assays indicated that NNMT knockdown led to a significant decrease in cell migration in MGC803 and BGC823 cell lines. To further assess the role of NNMT in the pathogenesis of gastric carcinoma, we also assessed whether NNMT knockdown inhibits invasion in MGC803 and BGC823 cell lines. Consistent with the previous results, knockdown of NNMT inhibited invasion in both cell lines (Figure 3E).
Knockdown of NNMT inhibited cell proliferation in vivo
To test whether NNMT regulates cell proliferation in vivo, we established xenograft tumor models in nude mice using MGC803 cells transfected with sh-NC or sh-NNMT. All nude mice developed xenograft tumors at the injection site, and the xenograft tumors were harvested 7 days after injection. The average tumor volume and weight in the sh-NNMT group was significantly reduced compared with the negative control group (Figure 4D, ,4E).4E). And the IHC staining was performed on xenograft tumors, and the Ki-67 staining signal was weaker in the sh-NNMT group compared with the sh-NC group (Figure 4C). These data demonstrate that NNMT may inhibit tumor growth in vivo.
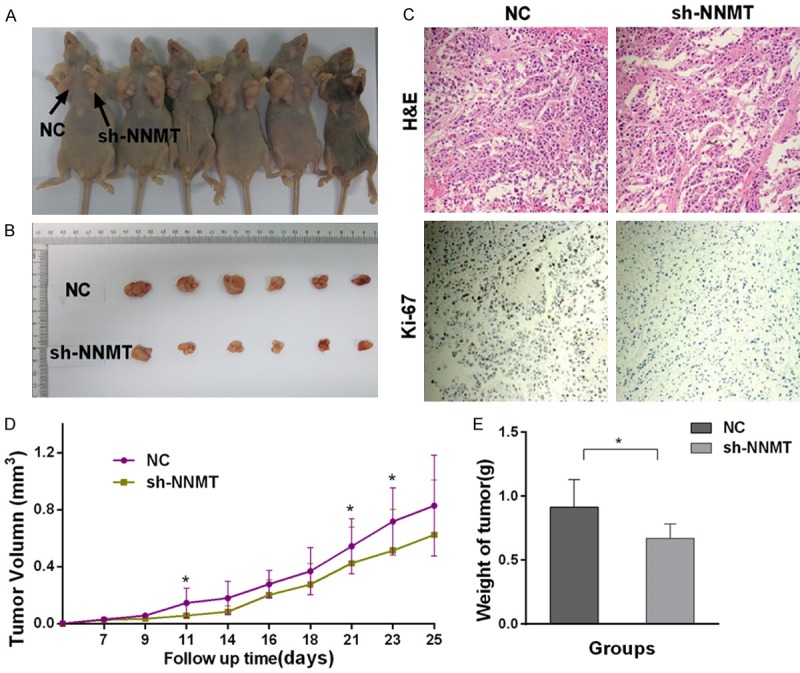
Knockdown of NNMT inhibited the proliferation of gastric carcinoma cell line in vivo. A, B. MGC803 cells were transfected with either Si-NNMT or sh-NC.The injection sites are indicated by arrows. D, E. The xenograft tumor weight and volume in the sh-NNMT group were significantly reduced compared with the sh-NC group. C. IHC staining was performed on xenograft tumors, and the Ki-67 staining signal was weaker in the sh-NNMT group compared with the sh-NC group. (Single asterisk indicates P<0.05; double asterisks indicate P<0.01; triple asterisks indicate p<0.001; error bars indicate means ± SEM).
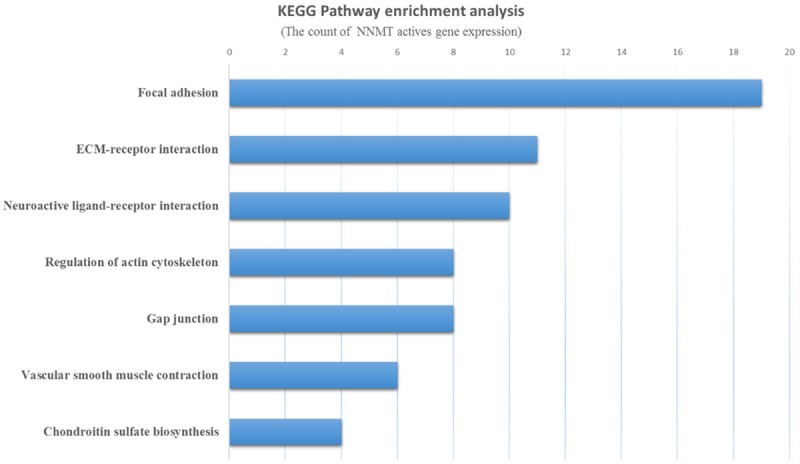
KEGG pathway enrichment analysis
A Kyoto Encyclopedia of Genes and Genome (KEGG) Pathway enrichment analysis of the resulting gene signature was performed. The most significant five KEGG terms for NNMT are presented in Figure 5 and include focal adhesion, ECM-receptor interaction, neuroactive ligand-receptor interaction, regulation of actin cytoskeleton, gap junction, vascular smooth muscle contraction and chondroitin sulfate biosynthesis.
Discussion
“Altered energy metabolism is proving to be as widespread in cancer cells as many of the other cancer-associated traits that have been accepted as hallmarks of cancer” [4]. NNMT, a metabolic enzyme, has attracted great attention. NNMT is normally expressed in adipose and liver tissue and participates in biotransformation of many drugs and xenobiotic compounds [7]. Recent studies have reported that NNMT is involved in the progress of cancers as an oncogene. Among the literature we reviewed, one of the most thoroughly explored relationships was that between NNMT and colorectal cancer. Researchers have reported that NNMT plays an important role in colorectal cell proliferation, differentiation, and apoptosis and that the diagnostic value of NNMT appears to be at least comparable to that of the typical biomarker CEA [5,26]. Both gastric and colorectal carcinoma originate from the digestive system, and they share many similar characteristics, such as a similar histological structure and route of metastasis. Based on these similarities, similar diagnostic and even treatment strategies, such as CEA, may be utilized for these two cancer types.
To develop rational and specific approaches for early diagnosis and treatment, this study explored the relationship between NNMT and gastric carcinoma. Cancer stage (or grade) and some biomarkers, such as CEA and CA199, are used to predict gastric carcinoma prognosis clinically [27]. To facilitate prognostic studies, we previously collected a considerable amount of specimens with relevant clinical parameters (including survival parameters) [28].
In our study, similar to a previous report [21], NNMT mRNA and protein levels were both upregulated in gastric carcinoma compared with normal tissues, and the TCGA database indicated a similar tendency. Previous studies reported that NNMT was overexpressed in several cancer types. Using serum tests, the sensitivity of NNMT was increased compared with CEA (ROC=0.84 vs. ROC=0.79) in colorectal carcinoma patients [26]. A recent study also found that NNMT was elevated in urine [29]. Consistent with the aforementioned results, we not only found that NNMT was overexpressed in gastric carcinoma tissues but also that increased NNMT level was associated with the aggravation of precancerous lesions. This finding has rarely been reported previously. Considering our relatively large sample size, this finding may help clinicians make better treatment decisions and eventually improve patient prognosis.
Similar to CEA, to a certain extent, the prognostic value of NNMT was very significant. In recent decades, studies have suggested that NNMT overexpression predicts poor prognosis in various carcinomas. For example, a previous study reported that NNMT mRNA upregulation was correlated with worse OS in bladder carcinoma [30]. Another study reported that NNMT protein expression was a prognostic factor in renal carcinoma, but the sample size of that study was relatively small (only 46) [11]. In our study, gastric carcinoma patients with increased NNMT expression had worse survival in both univariate and multivariate analyses, indicating that NNMT is a possible prognostic indicator for gastric carcinoma. A more specific analysis revealed that patients positive for two markers, NNMT and CEA, had a significantly worse prognosis than those who were positive for one or neither of the two markers. The 5-year survival rates for patients with different NNMT and CEA expression levels ranged from 75% (NNMT+/CEA+) to 15% (NNMT-/CEA-), suggesting that a more accurate clinical diagnosis can be obtained by combining these two markers.
Our tissue microarray (TMA) data showed that elevated NNMT expression was associated with primary tumor size, lymph node metastasis, and distant metastasis and TNM stage. However, due to the specificity of the tumors, NNMT expression and the clinical profiles of the gastric cancer patients examined in this study differed from those of other cancers. For example, NNMT expression is associated with cancer cell differentiation but not with clinical stage in colorectal carcinoma. Also, an oral tongue squamous cell carcinoma study indicated that overexpression of NNMT was associated with smaller tumor size and lower TNM stage [31].
To further understand the biological role of NNMT in gastric carcinoma, we also performed NNMT knockdown studies and found that NNMT knockdown inhibits cell proliferation, invasion and migration both in vitro and in vivo.
NNMT knockdown caused cell cycle arrest at the G2-phase. NNMT may promote proliferation of gastric carcinoma cells by altering the cell cycle. Other studies also reported that NNMT expression was correlated with cancer cell proliferation [16,32]. However, not all previous research findings are consistent with our data. For example, a previous study found that overexpression of NNMT reduced the proportion of cells in the G1 phase and simultaneously increased the proportion of cells in the G2 and S phases [5]. We found that NNMT depletion can also inhibit cell migration and invasion. We also observed abnormal NNMT expression in mesenchymal tissues, indicating that NNMT may be involved in the epithelial-mesenchymal transition (EMT). NNMT was closely associated with other important members of molecular pathways, such as SHH, PI3k/Akt and stat3 [33-36]. Moreover, KEGG pathway analysis indicated that the genes that are co-expressed with NNMT are mainly involved in focal adhesion and extracellular matrix (ECM)-receptor interaction pathways, which are important for mechanical strength and participate in EMT [37]. Based on those findings, we conclude that NNMT plays a vital role in gastric cancer progression.
NNMT overexpression is associated with chemotherapy and radiotherapy resistance. Previous studies showed that NNMT may serve as a drug-resistance gene for Taxol, Erlotinib, Everolimus and Dasatinib chemotherapies [38-40]. Thus, targeting this drug-resistant gene will help obtain a better chemotherapy outcome, and this technique serves as a novel chemotherapy approach. Previous studies previously demonstrated that anti-NNMT agents potentially have therapeutic effects in cancer [15]. However, given that NNMT is not a membrane-bound protein, the therapeutic effect of anti-NNMT antibodies is limited. However, with the development of siRNA in vivo transfection, NNMT may have a great impact on cancer therapy in the future.
Overall, NNMT is a promising molecular biomarker that may predict the progression of precancerous lesions and patient survival in gastric carcinoma. Furthermore, NNMT may serve as a therapeutic target in the future. Further experiments arc necessary to confirm whether NNMT does indeed act as tumor suppressor in gastric carcinoma. Studies of the mechanisms of the involvement of NNMT in gastric carcinoma will need to be conducted.
Acknowledgements
This work was supported by the Major Scientific and Technological Special Project for “significant new drugs creation” (No. 2012ZX09103301-040), the Innovative Team of Jiangsu Province (No. LJ201126) and the National Natural Science Foundation (No. 81572390).
Disclosure of conflict of interest
None.
References
Articles from American Journal of Cancer Research are provided here courtesy of e-Century Publishing Corporation
Citations & impact
Impact metrics
Citations of article over time
Article citations
Uncovering Metabolic Alterations in HCT-116 Colon Cancer Cells upon Exposure to Bamboo Leaf Extract Obtained from Guadua incana Londoño.
Molecules, 29(13):2985, 23 Jun 2024
Cited by: 1 article | PMID: 38998936
Knockdown of nicotinamide N-methyltransferase suppresses proliferation, migration, and chemoresistance of Merkel cell carcinoma cells in vitro.
Hum Cell, 37(3):729-738, 19 Mar 2024
Cited by: 1 article | PMID: 38504052 | PMCID: PMC11016511
NNMT overexpression is an adverse prognostic factor in uterine leiomyosarcoma.
Turk J Med Sci, 54(4):804-810, 16 Apr 2024
Cited by: 0 articles | PMID: 39295619 | PMCID: PMC11407357
Gastric Cancer in the Era of Epigenetics.
Int J Mol Sci, 25(6):3381, 16 Mar 2024
Cited by: 3 articles | PMID: 38542354 | PMCID: PMC10970362
Review Free full text in Europe PMC
Inhibitors of NAD+ Production in Cancer Treatment: State of the Art and Perspectives.
Int J Mol Sci, 25(4):2092, 08 Feb 2024
Cited by: 2 articles | PMID: 38396769 | PMCID: PMC10889166
Review Free full text in Europe PMC
Go to all (43) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Identification of Biological Functions and Prognostic Value of NNMT in Oral Squamous Cell Carcinoma.
Biomolecules, 12(10):1487, 15 Oct 2022
Cited by: 1 article | PMID: 36291696 | PMCID: PMC9599733
[Expression of nicotinamide-N-methyltransferase in gastric cancer and its biological and clinicopathological significance].
Nan Fang Yi Ke Da Xue Xue Bao, 41(6):828-838, 01 Jun 2021
Cited by: 0 articles | PMID: 34238734 | PMCID: PMC8267982
Overexpression of Nicotinamide N-methyltransferase mainly covers stroma of colorectal cancer and correlates with unfavorable survival by its product 1-MNA.
J Cancer, 12(20):6170-6181, 26 Aug 2021
Cited by: 4 articles | PMID: 34539890 | PMCID: PMC8425209
Nicotinamide N-Methyltransferase in Head and Neck Tumors: A Comprehensive Review.
Biomolecules, 11(11):1594, 28 Oct 2021
Cited by: 21 articles | PMID: 34827592 | PMCID: PMC8615955
Review Free full text in Europe PMC