Abstract
Free full text

Temporal regulation of EGF signaling networks by the scaffold protein Shc1
Abstract
Cell-surface receptors frequently employ scaffold proteins to recruit cytoplasmic targets, but the rationale for this is uncertain. Activated receptor tyrosine kinases, for example, engage scaffolds such as Shc1 that contain phosphotyrosine (pTyr) binding (PTB) domains. Using quantitative mass spectrometry, we find that Shc1 responds to epidermal growth factor (EGF) stimulation through multiple waves of distinct phosphorylation events and protein interactions. Following stimulation, Shc1 rapidly binds a group of proteins that activate pro-mitogenic/survival pathways dependent on recruitment of the Grb2 adaptor to Shc1 pTyr sites. Akt-mediated feedback phosphorylation of Shc1 Ser29 then recruits the Ptpn12 tyrosine phosphatase. This is followed by a sub-network of proteins involved in cytoskeletal reorganization, trafficking and signal termination that binds Shc1 with delayed kinetics, largely through the SgK269 pseudokinase/adaptor protein. Ptpn12 acts as a switch to convert Shc1 from pTyr/Grb2-based signaling to SgK269-mediated pathways that regulate cell invasion and morphogenesis. The Shc1 scaffold therefore directs the temporal flow of signaling information following EGF stimulation.
Many cell surface receptors associate with intracellular scaffold proteins that amplify signaling by providing docking sites for downstream effectors1. In the case of receptor tyrosine kinases (RTKs), autophosphorylated NXXY motifs recruit scaffolds with phosphotyrosine (pTyr)-binding (PTB) domains, such as members of the insulin-receptor substrate (IRS), Dok, FGF-receptor substrate 2 (FRS2) and Shc families2. Once associated with an RTK, the scaffold is itself phosphorylated at tyrosine motifs that recruit SH2 domain proteins, resulting in the activation of intracellular pathways3.
This begs the question as to why receptors use scaffolds for activities that might have been incorporated into the receptors themselves. We have investigated this issue using mammalian Shc1; the Shc1 gene encodes three proteins of 46, 52 and 66 kDa that share an N-terminal PTB domain and a C-terminal SH2 domain, flanking a central region (CH1) containing sites of phosphorylation at Tyr239/240 and Tyr3134,5. Modification of these tyrosines creates pYXN binding motifs for the SH2 domain of the Grb2 adaptor. Through its SH3 domains, Grb2 recruits proteins such as Sos1 and Gab1, that in turn activate the Ras-Erk MAP kinase and phosphatidylinositol 3′-kinase (PI3K)/Akt pathways6,7.
Shc1 is important for normal and oncogenic signaling by ErbB RTKs in mice8–10. In vivo, pTyr-binding by the PTB domain is required for all known function of Shc1, but downstream signals are transmitted through both pTyr-dependent and pTyr-independent mechanisms8,11. Indeed, some polypeptides are known to bind Shc1 in a pTyr-independent manner, including the endocytic adaptor α-adaptin and the tyrosine phosphatase Ptpn1212,13. Here, we demonstrate that Shc1 mediates a temporal switch in the signaling output of the EGFR.
Shc1 assembles an extensive EGF-regulated interactome
To map the dynamic properties of the EGF-regulated Shc1 signaling network, we generated Rat-2 cells stably expressing p52Shc1 doubly-tagged with FLAG and GFP (termed dt-Shc1) to a level comparable to that of the principal endogenous isoform, p52Shc1 (Supplementary Fig. 1a). Following EGF stimulation, we immunoprecipitated dt-Shc1 with anti-FLAG antibodies; using mass spectrometry (MS) we identified 41 binding partners involved in cellular functions such as protein phosphorylation, lipid metabolism, endocytosis, ubiquitination, and small GTPase regulation (Fig. 1). Several interactors either function in cytoskeletal rearrangement, consistent with the observation that Shc1−/− cells exhibit defects in focal contacts and actin stress fibres14, or potentially antagonize EGFR mitogenic signaling. For example, the Ras GTPase activating protein (GAP) Dab2ip is a tumour suppressor that controls both Ras and NF-κB activity15; the atypical kinase SgK269/PEAK1 modulates the cytoskeleton to control cell spreading and migration, and thus tumourigenesis16; the Arf GTPase activators Asap1 and Asap2 promote cancer cell invasiveness17,18; and the Rho guanine nucleotide exchange factor Arhgef5 is often overexpressed in breast cancers and helps form Src-induced podosomes19,20. We also mapped EGF-induced phosphorylation sites on Shc1 (S29, T214, Y239, Y240, Y313 and S335) and Shc1-associated proteins (Fig. 1 and Supplementary Table 1).
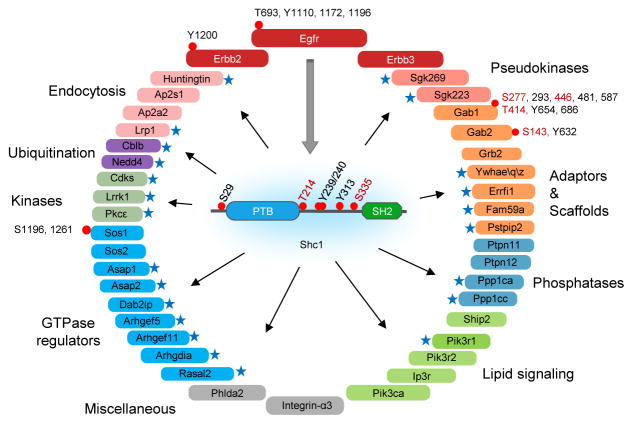
EGF-dependent Shc1 phosphorylation and protein interaction network identified by LC/MS in discovery mode. Novel Shc1-interacting proteins are marked by blue stars. Phosphorylation sites are shown by a red dot, with novel sites highlighted in red. Proteins are coloured according to functional groups.
EGF-induced dynamic phosphorylation of Shc1
To analyze the dynamics of Shc1-based signaling in EGF-stimulated cells, we developed an MS approach based on scheduled multiple reaction monitoring (sMRM) that was quantitative and linear over four orders of magnitude21–23 (Supplementary Fig. 1–4, Supplementary Tables 2–4 and Supplementary Text). Using sMRM we mapped Shc1 phosphorylation at 16 time points covering the first 90 minutes following EGF stimulation. Phosphorylation of both Tyr313 and Tyr239/240 peaked at 1–2 min, consistent with their being direct EGFR substrates, followed by dephosphorylation to baseline levels after 60 min (Supplementary Fig. 5a). In contrast Ser29, Thr214 and Ser335 were phosphorylated with distinct kinetics (Fig. 2a and Supplementary Fig. 5b). Phosphorylation of Ser29 started at ~40 seconds and peaked at 3 min, indicating that it is a substrate for a Ser/Thr kinase that is rapidly activated by the EGFR. Thr214 phosphorylation began 1 min after EGF addition and peaked at 5 min, implicating a kinase further downstream of the receptor. Phosphorylation of Ser335 reached a maximum only at 20 min. The totality of measured phosphorylation sites in the EGF-induced Shc1 signaling network showed a similar pattern; Tyr sites were rapidly phosphorylated followed by distinct waves of Ser/Thr phosphorylation (Fig. 2b), as revealed by principal component analysis (PCA) (Supplementary Fig. 6).
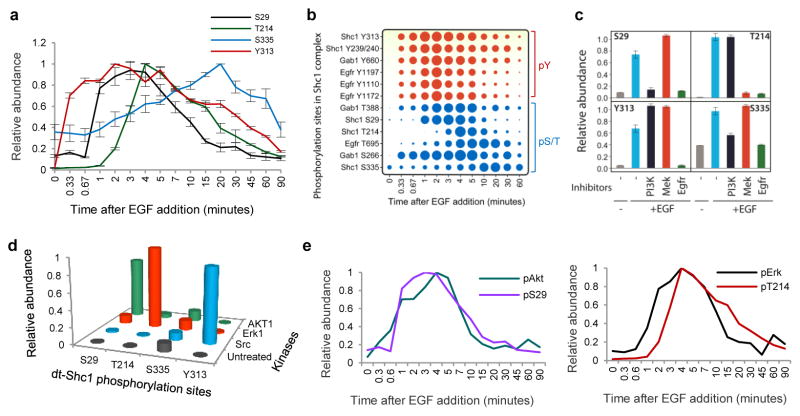
a, Temporal profiles of individual Shc1 phosphorylation sites following EGF stimulation. dt-Shc1 was affinity purified from EGF-stimulated fibroblasts at various time points. Relative abundance of dt-Shc1 phosphopeptides was quantified by sMRM and plotted using a quasi logarithm time scale to expand the early phase of phosphorylation. b, Temporal profiles of all analyzed phosphorylation sites in the EGF-induced Shc1 complex. The size of each dot is proportional to the relative abundance of the corresponding phosphopeptide. c, Differential inhibition of Shc1 phosphorylation by kinase inhibitors as quantified by sMRM. d, In vitro kinase/sMRM analysis. Affinity purified dt-Shc1 was incubated with recombinant kinases in vitro. Phosphorylation of dt-Shc1 sites was quantified by sMRM. e, Activation kinetics of Akt and Erk1 were measured by quantitative immunoblotting, and overlaid with the phosphorylation kinetics of Shc1 S29 and T214 from panel a, respectively. Inhibitors used: EGFR: AG1478; PI3K: LY294002; Mek: PD98059. Results are representative of three independent experiments. Error bars are s. d. from all transitions for a given protein/peptide from all technical repeats.
Shc1 feedback phosphorylation by Erk MAPK and Akt
EGFR phosphorylation of Shc1 stimulates the Ras-Erk MAPK and PI3K-Akt pathways, which may mediate Shc1 feedback phosphorylation at Ser/Thr sites. The EGFR inhibitor AG1478 abolished phosphorylation of all Shc1 Tyr and Ser/Thr sites (Figure 2c). Shc1 Ser29 lies in an RXXS/T substrate motif for AGC kinases, and its phosphorylation was diminished by the pan-PI3K inhibitor LY294002 and by a p110α PI3K-specific inhibitor (Fig. 2c and Supplementary Fig. 7, 8a)24. Treatment with an Akt kinase inhibitor (Akt inhibitor IV) specifically blocked Ser29 phosphorylation, and Ser29 was selectively phosphorylated by Akt in vitro (Supplementary Fig. 8b and Fig. 2d). In contrast, Thr214 is followed by a proline; its phosphorylation was blocked by the MEK inhibitor PD98059 and it was selectively phosphorylated by Erk MAPK in vitro (Fig. 2c and 2d). Thus EGF stimulation initially induces Shc1 tyrosine phosphorylation, followed very rapidly by Akt-mediated Ser29 phosphorylation, and with a slightly longer lag phosphorylation of Thr214 by Erk. The slower kinetics observed for Thr214 as compared to Ser29 phosphorylation is consistent with Erk being more distal to the EGFR than Akt, which is supported by direct analysis of Erk and Akt phosphorylation kinetics (Fig. 2e and Supplementary Fig. 9). The kinase responsible for Ser335 phosphorylation remains to be identified (Fig. 2c and d). Kinase feedback loops therefore phosphorylate Shc1 on distinct residues, and with different kinetics, potentially affecting the nature of signaling following EGF stimulation.
The nature of Shc1 interactome switches over time
sMRM-based quantitation revealed that various binding proteins associated with Shc1 with diverse kinetics. We assigned each Shc1-binding protein, except for Shcbp1, to one of three clusters, based on the time required for maximal binding (Fig. 3a and Supplementary Fig. 10–14). Following EGF stimulation, Cluster 1 proteins were maximally bound to Shc1 at ~1–2 min, largely through pTyr/Grb2-dependent interactions (discussed below). This group includes ErbB receptors that bind the Shc1 PTB domain, and effectors primarily involved in stimulating the Ras-Erk MAP kinase and PI3K pathways, including scaffold proteins (Gab1/2, Fam59a/GAREM), Ras/Rho GEFs (Sos1/2, Arhgef5), protein/lipid kinases and phosphatases (Pik3, Ptpn11, Lrrk1) and an E3 ubiquitin-protein ligase (Cblb)25,26. Several of these targets are novel Shc1-binders, such as Arhgef5, Lrrk1, and Fam59a, potentially with positive roles in proliferation and migration27. Members of this group showed distinct dissociation kinetics. For example, Sos1 and Sos2 were more transiently associated with Shc1 than Grb2, potentially due to the disassembly of the Grb2/Sos complex caused by feedback phosphorylation of Sos28. We subdivided this cluster into two groups, 1a and 1b; Cluster 1b proteins bound a few seconds more rapidly to Shc1 following EGF stimulation compared with Cluster 1a, and remained associated for longer, suggesting a more prolonged involvement in Shc1 signaling.
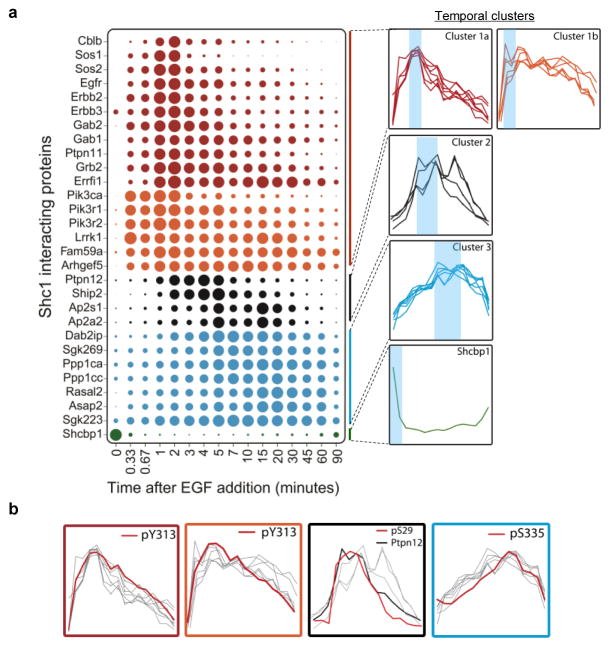
a, dt-Shc1 associated proteins were quantified as a function of time following EGF stimulation. The size of each dot is proportional to the relative abundance of the associated protein. Proteins were divided into three clusters based on the similarity of their association rates with dt-Shc1 and were colour-coded accordingly. Shcbp1 has a unique binding profile. At right: individual binding curves from each cluster were overlaid, with blue shading over the regions with maximal protein binding. b, Overlays of each temporal cluster with kinetic profiles of Shc1 phosphorylation sites (pY313 vs. Cluster1a and 1b; pS29 vs. Cluster 2; pS335 vs. Cluster 3).
The tyrosine phosphatase Ptpn12, the endocytic adaptor protein Ap2 and the lipid phosphatase Ship2 comprise Cluster 2, and bound maximally to Shc1 for between 2–5 min, followed by a rather rapid disassociation. Cluster 3 proteins associate more slowly with Shc1, with binding peaking between 15–20 min. Several proteins in this cluster have been implicated in cytoskeletal reorganization, including SgK269/PEAK1, Sgk223/pragmin, and Asap229; others, including the tumour suppressors RasGAP Dab2ip and the Ppp1ca/Ppp1cc Ser/Thr protein phosphatases, suppress Ras/MAPK signaling15,30, and can therefore downregulate signaling induced by Cluster 1 proteins. Cluster 3 proteins share similar association/disassociation kinetics, and could therefore coordinately control cell morphology, movement and proliferation. Finally, Shcbp1 is associated with Shc1 prior to stimulation but is displaced following EGF treatment, suggesting an inhibitory activity; indeed Shcbp1 and pTyr bind competitively to the Shc1 SH2 domain31.
We saw a similar pattern of EGF-induced Shc1 phosphorylation and interacting proteins using primary embryonic fibroblasts from mice in which FLAG-tagged Shc1 had been knocked into the endogenous Shc1 locus8, indicating that the results obtained with dt-Shc1 are physiologically relevant (Supplementary Fig. 15 and 16). This coordinated assembly and disassembly of Shc1 complexes suggests that Shc1 signaling properties switch during the course of growth factor stimulation from activation of the Erk/PI3K pathways to the control of cytoskeletal architecture, cell movement, and signal reversal.
Grb2-independent late phase Shc1 complex assembly
Work on Shc1 signaling has focused on the pYXN-mediated recruitment of Grb232. However, the delayed binding of Cluster 2 and 3 proteins (Supplementary Fig. 17) argues that Shc1 has Grb2-independent functions. To test this notion, we stably expressed dt-Shc1 in MEFs in which the Grb2 gene can be inducibly deleted (Supplementary Fig. 18). Binding of Cluster 1 proteins to Shc1, except for upstream RTKs, was almost completely abolished following Grb2 deletion (Fig. 4a and Supplementary Fig. 19), indicating that Grb2 couples Shc1 pTyr sites to these targets during early EGF signaling. PCA confirmed that Shc1 tyrosine phosphorylation correlates with the binding of Cluster 1 proteins (Fig. 4b). In contrast, association of Cluster 2 and 3 proteins with Shc1 upon EGF stimulation was retained in Grb2-deficient cells (Fig. 4a). The delayed wave of Shc1-binders is therefore Grb2-independent, but correlates with Ser29 and Ser335 phosphorylation (Figure 4b).
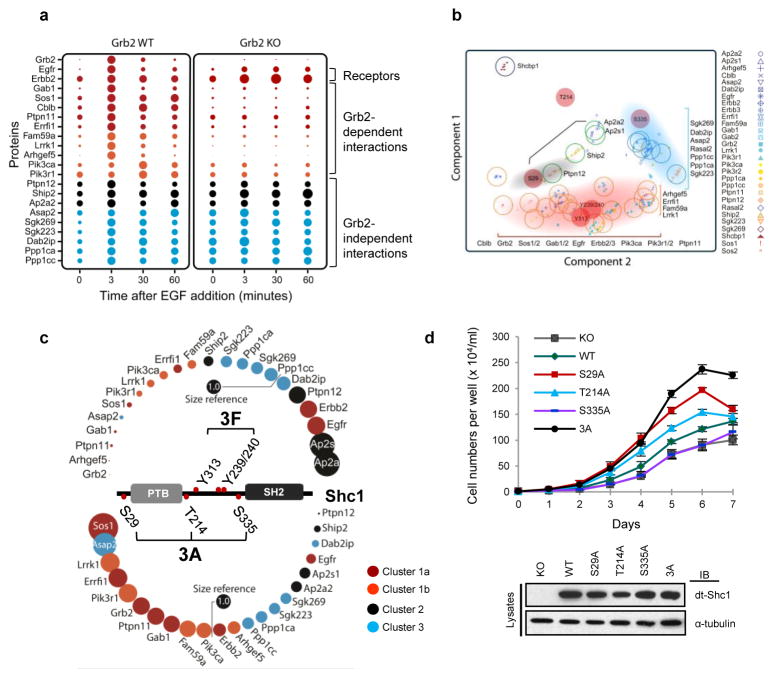
a, EGF-induced dt-Shc1 protein interactions were quantified by sMRM in Grb2flox/flox MEFs, with (WT) or without (KO) functional Grb2. Dots are coloured according to the temporal clusters defined in Fig. 3a. b, Correlations between Shc1 phosphorylation and protein binding revealed by PCA. The center of each open circle marks the mean PCA value for each protein. The open circles are coloured according to the protein’s cluster assignment. The red-filled circles are Shc1 phosphorylation sites. Shaded areas indicate co-modulations between specific Shc1 phosphorylation sites and binding clusters. c, Shc1 complex assembly in Shc1-deficient MEFs stably expressing WT dt-Shc1 compared to phosphosite mutants (3F: Y239/240/313F; 3A: S29/335/T214A) at 5 minutes post-EGF stimulation. The relative abundance of each protein from the WT dt-Shc1 complex was set at 1.0 (Size reference). Changes in protein-binding to Shc1 mutants are represented as the fold change over WT. d, Proliferation of Shc1-deficient MEFs expressing WT or mutant dt-Shc1 as quantified by cell counting. Lower panel: expression levels of dt-Shc1 variants. Error bars are +/− s.d. from three technical replicates. All results represent a minimum of three independent experiments.
In the absence of Grb2 there was increased and sustained phosphorylation of Y313 and Y239/240, especially during the later phase of EGFR signaling (>10-fold increase at 60 min) (Supplementary Fig. 20a). At the same time, Ser29 and Thr214 phosphorylation decreased by 47% and 56% (at 3 min), respectively. This argues that feedback phosphorylation of Shc1 Ser29 and Thr214 relies on pathways activated by Grb2-mediated signaling. Loss of Grb2 also caused an increased and prolonged tyrosine phosphorylation on the EGFR (Supplementary Fig. 20b), presumably due to failed recruitment of a tyrosine phosphatase.
To investigate whether Shc1 Ser/Thr phosphorylation sites are involved in Grb2-independent protein interactions, we quantified the Shc1 protein-interaction network in Shc1-null MEFs stably expressing either wild-type dt-Shc1, or dt-Shc1 mutants lacking all three phospho-Ser/Thr sites (3A), or all three pTyr sites (3F). The Shc1 3F mutant selectively lost association with Cluster 1 proteins, consistent with lack of Grb2 recruitment. Conversely, binding to Cluster 2 and 3 proteins, except for Asap2, was reduced or abolished in the Shc1 3A mutant (Fig. 4c and Supplementary Fig. 21). Analysis of Shc1 mutants lacking individual Ser/Thr sites indicated that this effect was largely due to loss of the Ser29 site (Supplementary Fig. 22–24). Replacing Ser29 with alanine suppressed binding of Shc1 to the tyrosine phosphatase Ptpn12, consistent with the temporal correlation between Ser29 phosphorylation and Ptpn12-binding (Fig. 3b and and4b).4b). Previous work has indicated that an atypical NPLH motif on Ptpn12 is recognized by the Shc1 PTB domain33, but that stable association also requires phosphorylation of Ser29 N-terminal to the PTB domain13. The depletion of Ptpn12 from the Shc1 complex observed with the 3A and S29A mutants correlated with enhanced binding of the pro-mitogenic Cluster 1 proteins (Fig. 4c and Supplementary Fig. 21–22), likely due to increased Shc1 tyrosine phosphorylation and Grb2-binding in the absence of the Ptpn12 phosphatase, but attenuated binding of inhibitory Cluster 2 and 3 proteins. The pSer29-dependent recruitment of Ptpn12 (a Cluster 2 protein) may therefore switch Shc1 from pro-mitogenic/survival signaling mediated by Grb2-associated Cluster 1 proteins to a form that attenuates signaling and stimulates cytoskeletal reorganization through Cluster 3 proteins. This is consistent with Ptpn12 acting as a tumour suppressor in human breast cancer34.
PCA also showed that Shc1 Ser335 phosphorylation is co-modulated with the binding of Cluster 3 proteins. However, substitution of Shc1 Ser335 significantly decreased binding of the majority of Shc1 partners (Supplementary Fig. 23), suggesting that this site may stabilize Shc1 signaling complexes. The timing of Thr214 phosphorylation did not correlate with any of the Shc1 protein-protein interactions (Fig. 4b), and substitution of Thr214 only marginally affected the Shc1 interactome (Supplementary Fig. 24).
The initial wave of pTyr-dependent binding proteins is therefore followed by a second network of binding partners enriched for regulators of cytoskeletal organization and cell migration, trafficking and inhibitors of mitogenic signaling. Consistent with the biochemical data, Shc1-null MEFs expressing the dt-Shc1 3A mutant lacking all three Ser/Thr phosphorylation sites proliferated more rapidly than cells expressing wild-type dt-Shc1 (Fig. 4d), and cells expressing the S29A Shc1 single-site mutant showed a large increase in mitotic activity compared with the T214A or S335A mutants.
These data argue that feedback phosphorylation of Shc1 Ser29 is particularly important for the binding of Cluster 2 and 3 proteins, including targets that negatively regulate cell growth. Erk-mediated phosphorylation of Thr214 also restricts cell proliferation, but through a device other than recruitment of proteins analyzed here.
Late phase Shc1 complex assembly by SgK269
In EGF-stimulated cells, Cluster 3 proteins bind Shc1 with similar kinetics, suggesting that their recruitment to Shc1 might be coordinated by an adaptor other than Grb2. When we used the Ppp1ca, Ppp1cb or Ppp1cc isoforms as baits for affinity purification and LC/MS analysis, we identified SgK269 as a prominent binding partner for Ppp1ca and Ppp1cc, but not Ppp1cb (Supplementary Fig. 25a). This recalls the selective binding of Shc1 to SgK269 and Ppp1ca/Ppp1cc, which are among the Cluster 3 proteins. Ppp1ca/Ppp1cc did not associate with the closely related Sgk223 (Supplementary Fig. 25b). These observations suggest that Ppp1ca and Ppp1cc might be recruited to Shc1 through SgK269, a large protein with a C-terminal pseudokinase domain and an N-terminal region with numerous potential sites of phosphorylation and predicted interaction motifs (Supplementary Fig. 26). Lentiviral-mediated shRNA knock-down of SgK269 expression in HeLa cells reduced Ppp1ca/Ppp1cc binding to Shc1 (Supplementary Fig. 27), suggesting that SgK269 acts as a bridge between Ppp1ca/Ppp1cc and Shc1. Surprisingly, knocking down SgK269 also suppressed the association of other Cluster 3 proteins with Shc1, including Dab2ip, Asap2 and Sgk223 (Fig. 5a; Rasal2 was not detectable in HeLa cells). This effect was specific to SgK269, as shRNA-induced silencing of Dab2ip expression only negatively affected the binding of Asap2 (Supplementary Fig. 28). These data argue that SgK269 is a scaffold that enables Shc1 to switch from Grb2-dependent mitogenic activity to Grb2-independent functions. We confirmed that EGF induced binding of SgK269 to endogenous Shc1, and that this interaction increased during the late phase of EGFR signaling (Supplementary Fig. 29a).
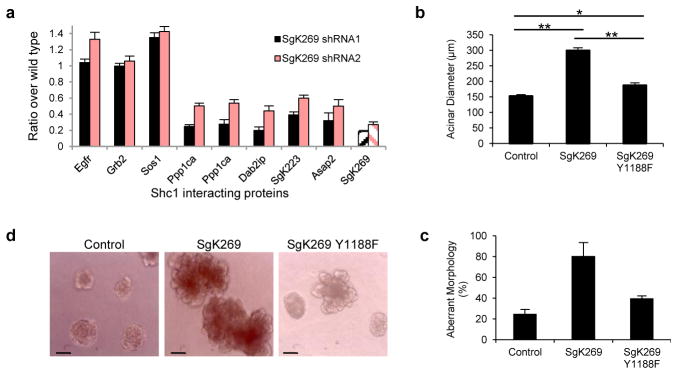
a, Cells stably infected with shRNAs for SgK269 or luciferase were stimulated with EGF. Association of Shc1 with Cluster 3 proteins and representative Cluster 1 proteins was quantified by sMRM. Error bars are +/− s. d. from all transitions for a given protein/peptide from all technical repeats. b–c, Effects of the SgK269 Y1188F mutation on acini size and morphology were analyzed 12 days after inoculation into Matrigel. The diameters of ~100 acini were measured (+/− s.e.m., ** p<0.0001, * p<0.001). Data in a–c are representative of two independent experiments. d, Representative images of acini.
As judged by mutagenesis experiments, phosphorylation of SgK269 Y1188, which lies in a PTB-binding NPXY motif, is necessary for binding to Shc1 (Supplementary Fig. 29b, c), suggesting that tyrosine phosphorylation prompts SgK269 recognition by the Shc1 PTB domain. Using a matrigel assay, we showed that over-expressing wild-type SgK269 in MCF-10A human epithelial breast cells generated acini that had a two-fold increased diameter, a multi-lobular morphology and non-cleared lumens, while control cells formed rounded, hollow acini. In contrast, a Y1188F SgK269 mutant that fails to bind Shc1 was relatively inactive in this assay (Fig. 5b–d and Supplementary Fig. 30). These observations support a role for the late-forming Shc1-SgK269 complex in regulating acinar morphogenesis.
Conclusion
RTKs frequently employ PTB-containing scaffolds to recruit their targets. Here we define a dynamic signaling network surrounding the Shc1 scaffold for ErbB RTKs (Fig. 6). Following EGF stimulation, tyrosine phosphorylated Shc1 rapidly binds Grb2 and Grb2-associated proteins that stimulate mitogenic and survival pathways. There is subsequently a switch in the Shc1 interactome to non-SH2 domain proteins involved in cytoskeletal reorganization, trafficking and downregulation of pro-mitogenic pathways. These latter complexes are linked to Shc1 serine/threonine phosphorylation, which may be a mechanism by which Shc1 monitors the state of pathway activation following growth factor stimulation, and directs a switch in its own output accordingly.
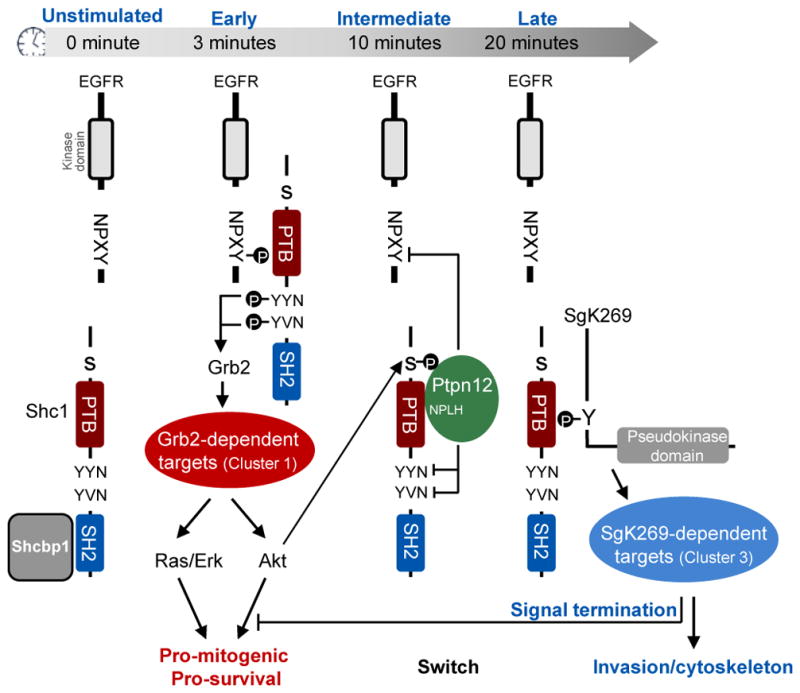
The figure depicts the different phosphorylation events and protein interactions involving Shc1 as a funtion of time following EGF stimulation. See text for details.
The dynamic properties of Shc1 can be appreciated by following the successive partners for its pTyr recognition domains. Shc1 is initially associated with Shcbp1, which may inhibit the precocious association of Shc1 with other targets31. Following EGF stimulation, Shcbp1 is displaced as Shc1 is recruited to pTyr motifs on autophosphorylated ErbB RTKs, notably through binding of the PTB domain to NXXpY motifs; this facilitates the phosphorylation of Shc1 YXN sites, resulting in their binding to the Grb2 SH2 domain and the recruitment of Cluster 1 proteins that stimulate the Ras-Erk MAP kinase and PI3K-Akt pathways, favouring cell proliferation and survival. The subsequent feedback phosphorylation of Shc1 on S29 by Akt recruits the tyrosine phosphatase Ptpn12 (a Cluster 2 protein), which also occupies the Shc1 PTB domain through an NPLH motif33. Ptpn12 antagonizes pro-mitogenic EGFR signaling, potentially by both displacing Shc1 from the EGFR and by dephosphorylating its Grb2-binding YXN motifs. Grb2 is then replaced by the SgK269 pseudokinase/scaffold that promotes the invasiveness of breast epithelial cells35; SgK269 binds the Shc1 PTB domain through a phosphorylated NPXY site, and brings in Ppp1c serine/threonine phosphatases and other Cluster 3 proteins. Taken together, we propose that Shc1 is a hub that determines the timing with which EGF-induced signaling switches between distinct states. This may be a more general property of scaffolds required for signaling from distinct types of cell surface receptors.
METHODS
Antibodies and reagents
We used anti-FLAG™ M2 agarose (Sigma) for dt-Shc1 immunoprecipitation and anti-Shc1 (BD) for immunoprecipitation of endogenous Shc1. We also used the following antibodies: anti-EGFR pY1073, anti-EGFR pY1068, anti-EGFR (Lifespan LS-C6640), anti-pAKT S473, anti-AKT, anti-MAPK p44/42, anti-Grb2 (CST). The MEK inhibitor PD 98059 and the PI3K inhibitor LY294001 were purchased from Cell Signaling Technology. The isoform specific PI3K inhibitors (PIK-90 and TGX221) were generous gifts from Kevan Shokat (UCSF, USA). We also used the following recombinant proteins: AKT1 (GST-tagged, Cell Sciences, CRA004B), Src (Cell Sciences, CRS155A), MAPK1 (Millipore, 14-439), EGFR (Abnova, H00001956-P02), and PI3K (p110a/85a) (SignalChem, P27-18H-10). The phosphatase inhibitor cocktail set II (524625) and IV (524628) were purchased from Calbiochem.
Plasmid constructs and site-directed mutagenesis
The cDNA expressing p52Shc1 with N-terminal FLAG and eGFP tags was cloned into the pCAGGS expression vector (at HindIII and EcoRI restriction sites). To generate Shc1 phosphorylation site mutants, we used the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene) and the following primers (forward):
S29A: GGACCAGACACGGGGCCTTTGTCAATAAGCC;
T214A: ACCGAAGCTGGTCGCCCCCCATGACAG;
S335A: GCTGGGCCCCCAAATCCTGCTCTTAATGGCAGTGCACCC;
3F (Y313/239/240F): CCCCCTGACCATCAGTTCTTCAATGACTTTCCAGGG and TCTTCGATGACCCCTCCTTTGTCAACATCCAGAAT.
Generating stable eGFP and FLAG-tagged p52Shc1 (dt-Shc1)-expressing Rat-2 and mouse embryonic fibroblast (MEF) cell lines
Wild type Rat-2 fibroblasts and Shc1−/− MEFs were transfected with plasmid constructs expressing dt-Shc1 or its mutants using polyethylenimine (PEI). Cells were sorted in three rounds using fluorescence activated cell sorting (FACS) cycles, and divided into three pools based on the signal intensity of the eGFP fluorescence. The dt-Shc1 expression levels of the three pools of cells were examined by immunoblotting and compared with the level of endogenous p52Shc. A pool of cells expressing dt-Shc1 levels comparable to that of endogenous p52Shc1 were selected (Supplementary Fig. 1).
In vitro kinase assay
MEFs were serum-starved for 2 hours. dt-Shc1 was affinity purified using anti-FLAG M2-conjugated agarose followed by washing 4 times with lysis buffer. The beads were equally divided into four aliquots and washed twice with relevant kinase buffers (Buffer A (for Erk and AKT): 25 mM Tris-HCl (pH 7.5), 5 mM beta-glycerophosphate, 0.5 mM DTT, 0.1 mM Na3VO4, 10 mM MgCl2, 1 mM EGTA, 50 nM calyculin A, 20 μM ATP; Buffer B (for Src), 5 mM MnCl2 was added into Buffer A). Purified recombinant human Src (100ng), Erk (100ng), AKT1 (100ng), or kinase buffer was added to the beads and incubated for 30 minutes at 37°C. The reactions were stopped by washing the beads once with 50 mM EDTA and twice with 50 mM ammonium bicarbonate. The beads were then digested with 0.4 μg trypsin overnight at 37°C and the phosphorylation sites on Shc1 were quantified by sMRM.
Proliferation assay
Shc1−/− MEFs reconstituted with dt-Shc1 or its mutants were seeded in quadruplicate at 0.5×104 cells/well into 24-well plates. Cells were trypsinized and the cell number was counted using a hemocytometer every 24 hours over a 7-day period.
Inducible deletion of Grb2 in MEFs
The Grb2 conditional allele was generated by flanking exon 2 of Grb2 with floxP sites to introduce a frameshift (Supplementary Fig. 18). MEFs were expanded from E13.5 Grb2flox/flox embryos and immortalized by the 3T3 protocol as previously described36. The immortalized Grb2flox/flox 3T3 MEF line was then infected with pMSCV-CreER retrovirus, and selected with 5μg/ml Blasticidin in DMEM supplemented with 10% FBS to generate a stable pool of CreER-expressing Grb2flox/flox 3T3 MEFs. For inducible deletion of Grb2, 4-OH tamoxifen (1μg/mL) was added into the culture medium for 48 hours. The deletion of Grb2 expression was confirmed by immunoblotting.
Lentivirus-mediated gene knockdown
Lentiviral short hairpin RNAs (shRNA) specifically targeting human SgK269 and Dab2ip genes were kindly provided by Dr. Jason Moffat (University of Toronto). shRNA sequences used are as follows:
SgK269 (Accession: NP_079052): shRNA1: CCACAAGTGTAATAAGCCATA; shRNA2: GAAGATCTCTTCCAGACTTTC;
Dab2ip (Accession: Q5VWQ8): GACTCCAAACAGAAGATCATT.
Lentiviruses were produced as previously described37 and used to infect HeLa cells for 24 hours, followed by puromycin (2ug/ml)-mediated drug selection for 5 days.
Knockdown efficiency was judged by quantifying the relative amount of targeted protein in the Shc1 complex after EGF stimulation in pooled colonies using sMRM.
Matrigel assay
MCF10A cells over-expressing SgK269 WT, SgK269 Y1188F or the control plasmid were grown in matrigel for 12 days, as previously described38. The resulting acini were photographed and the diameters of ~100 acini were measured using Image J (+/− S.E.M., ** p<0.0001, * p<0.001). Acinar morphology was determined by visual inspection (+/− SD, n=2).
Shc1 immunoprecipitation and on-bead tryptic digestion
Cell lines expressing dt-Shc1 were seeded at 1 × 107/15 cm dish (Nunc) in DMEM + 10% FBS. The following day, the indicated treatments were applied to the cells and they were immediately washed three times with ice-cold PBS to quench cell signaling, then lysed in NP40 lysis buffer (50 mM HEPES-NaOH (pH 8), 150 mM NaCl, 1 mM EGTA, 0.5% NP40, 100mM NaF, 2.5mM MgCl2, 10mM Na4P2O7, 1 mM dithiothreitol (DTT), 10% glycerol) supplemented with protease and phosphatase inhibitors (50 mM β-glycerolphosphate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM Na3VO4, 100nM calyculin A, 1 mM PMSF). The total cell lysates were centrifuged at 20,800 × g for 30 minutes to pellet the nuclei and insoluble material. Nuclear-free lysates were pre-cleared by one-hour incubation with protein A sepharose and normalized for total protein concentration using the Bio-Rad protein assay. dt-Shc1 was immunoprecipitated by incubating lysates with 5 μl (bed volume) anti-FLAG M2 antibody-conjugated agarose for 4 hours at 4°C. The beads were washed 4 times with lysis buffer and twice with 50 mM ABC before resuspending in 20 μl ABC (50 mM). Tryptic digestion was performed by directly adding trypsin (enzyme: substrate~1:50) to the beads and incubating at 37°C overnight. The digestion was stopped by adding 3 % formic acid to the reaction and the supernatant was transferred into a clean tube and dried.
Mass spectrometry analysis of the Shc1 interactome
Dried tryptic samples were reconstituted with 3% formic acid in HPLC grade water. Samples were loaded on to a 75 μm ID/360 μm OD pulled tip packed with 3 μm ReproSil C18 and analyzed on an TripleTOF 5600 mass spectrometer (AB SCIEX) or a QSTAR Elite mass spectrometer (AB SCIEX), each coupled to an Eksigent nanoLC Ultra 1D plus pump with a flow rate of 200 nl/min and a gradient of 2% to 35% acetonitrile over 90 min. The mass spectrometers were operated in information-dependent acquisition mode. For the TripleTOF 5600, a cycle time of 1.3 seconds was employed using a survey TOF scan of 250 millisecond (msec) at ~ 30,000 resolution followed by selection of the top 20 most intense peptides for MS/MS for 50 msec each with high sensitivity (at ~ 18,000 resolution). Only peptides with a charge state above +1 were selected for MS/MS and dynamic exclusion was set to 15 seconds for all ions within 20 ppm. For the QSTAR Elite, a cycle time of 5.25 seconds was employed using a survey TOF scan of 250 msec at ~ 10,000 resolution followed by selection of the top 5 most intense peptides for MS/MS using a Fragment Intensity Multiplier of 8 and a maximum accumulation time of 1 sec for each candidate. Enhance All was used for all MS/MS scans. Only peptides with a charge state above +1 were selected for MS/MS and dynamic exclusion was set to 20 seconds for all ions within 50 ppm. Q1 was set to Unit resolution on all MS/MS scans for both the TripleTOF 5600 and the QSTAR Elite.
All acquired raw files were converted to mgf format and searched against Ensembl databases (rat and mouse - release 44) using Mascot version 2.1 or ProteinPilot version 2.0.1 (AB SCIEX). The following parameters were used for the database searches: precursor mass accuracy: 30 ppm; MS/MS mass accuracy 0.1 Da for Elite and 0.05 Da for Triple TOF 5600, and modifications were the following: phospho-S/T/Y (variable), methionine oxidation (variable), and NQ deamidation (variable); one missed trypsin cleavage was accepted. All peptides with a Mascot score over 25 were selected for sMRM method building.
sMRM assay construction and optimization
The sMRM assay employs specific peptides and their fragments (termed transitions) as unique discriminators of individual proteins. Based on the fragmentation information acquired from the initial MS/MS scans, we built the sMRM assay to specifically quantify the Shc1 interactome. The MS/MS information from previous information-dependent acquisition (IDA) experiments were imported by the MRMpilot 1.1 (AB SCIEX) software to generate a list of potential sMRM transitions which was then manually filtered to contain only fragments ions greater than the precursor that were generated from doubly- or triply-charged precursor peptides with no methionine or tryptophan residues, and no N-terminal glutamine residues in general. However, for certain peptides, such as phosphopeptides, containing chemically unstable but biologically significant residues that could not be excluded from the list, the dominant forms of the peptide transition species were measured.
For confident peptide identification and quantification, at least three peptides per protein and a minimum of two transitions per peptide were targeted for sMRM analysis. The initial sMRM transition list then underwent multiple rounds of validation and optimization using non-scheduled MRM analysis of Shc1 immunoprecipitates. Transitions that showed low sMRM detection sensitivity were removed. Proteins that failed to produce a minimum of two unique peptides with sufficient detection sensitivity were not examined further. The peptide retention times were determined by MS/MS from multiple MRM Initiated Detection and Sequencing (MIDAS) runs using MultiQuant (version 2.1) software39. The final sMRM method consists of 381 transitions from 171 unique peptides, corresponding to 30 proteins (including Shc1) from the Shc1 interactome.
For data normalization, we chose 5 Shc1 peptides that were unlikely to either undergo post-translational modifications, other than oxidation on Met, Trp and His residues, or produce analytical artifacts (such as isobaric interference) (Supplementary Fig. 3a). These peptides showed linear responses over the concentration range of the samples (Supplementary Fig. 3b). Each biological experiment was measured with two technical repeats and a logistic regression analysis (LRA) showed low variation between the two measurements (Supplementary Fig. 4a, b). We also spiked 100fmol of digested α-casein into all samples as an analytical standard to monitor the LC-MS performance (Supplementary Table 4).
sMRM quantification
sMRM analysis was performed on hybrid triple quadrupole/ion trap mass spectrometers (4000QTrap and 5500QTrap; AB SCIEX). Chromatographic separations of peptides were carried out on a nano-LC system (Eksigent, Dublin CA) coupled to a 100 μm i.d. fused silica column packed with 5 μm ReproSil C18 as a trap column and a 75 μm i.d. fused silica column packed with 3 μm ReproSil C18 as the separation column. A micro Tee was used to connect both columns and a micro-union was used to connect the columns to an emitter. Peptides were separated with a linear gradient from 2–30% acetonitrile in 90 min at a flow rate of 300 nl/min. The MIDAS workflow was employed for sMRM transition confirmation. For all runs, the MS instrument was operated in the positive mode. LC-MS conditions, for all experiments, were evaluated using a 30 min gradient run of a mixture of 30 fmol of BSA and 60 fmol of α-casein (72 MRM transitions) before each sample run. In order to reduce the carry over, at least one clean-up run and one BSA run were performed between samples. Each sMRM run was scheduled using previously determined LC retention times with a 5-minute MRM detection window and a 3-second scan time with both Q1 and Q3 settings at unit resolution. We injected each biological sample at least twice for increased quantification confidence. In general, the technical replicates showed little variations (Supplementary Fig. 4).
sMRM data processing
The MS/MS spectra acquired by MIDAS were first searched against relevant Ensembl databases using Mascot to confirm the identities of peptides. The raw data was then imported into MultiQuant v2.1 (AB SCIEX) for automatic MRM transition detection followed by manual inspection by the investigators to increase confidence. Subsequently, the eXtracted Ion Chromatogram (XIC; the peak area) of each transition was calculated. The XIC of each transition is proportional to the real quantity (abundance) of the corresponding peptide. The data were visualized and exported as excel spreadsheets using MarkerView 1.2.1 (AB SCIEX) into a custom developed SQL based data-storage and management system (CoreFlow) for further analysis.
Statistical analysis
We used the R statistical package40,41 for generating the pseudo-3D view (dot blots) of the temporal and spatial profiles of Shc1 interactome and for the PCA analysis.
Relative abundance (RA)
To calculate the relative abundance of each protein or phosphopeptide in Shc1 immunoprecipitates, the mean XIC of a given sMRM transition from technical replicates was normalized to the Shc1 protein level. The normalized value was then converted to a percentage, using the highest value of that transition in a given experiment as 100. Percentages from all of the transitions representing the same protein or phosphopeptide were averaged and presented as relative abundance (RA) +/−standard deviations (s.d.). RA values for Shc1-interacting proteins from wild-type cells as compared to cells expressing mutated Shc1 or Grb2 genes were calculated slightly differently. Here, the highest XIC value of a given transition measured in the wild-type cells was set to 100. Dot blots were generated from the RA values using a mixture of the R Lattice package functions xyplot, bwplot and levelplot.
Principal component analysis (PCA)
PCA was applied to the RA values using the “princomp” function in R with parameters “corr = TRUE” and “scale=T”. We selected the first 2 or 3 main components from the PCA, which accounted for more than 80% of the variation of the data intercorrelation.
Acknowledgments
We thank J. Moffat (U Toronto) for shRNA lentiviruses and K. Shokat (UC Berkeley) for PI3Kp110 isoform-specific inhibitors. We thank C. Jorgensen (ICR), R. Williams (UMM), I. Louria-hayon, R. Tian, and E. Petsalaki (SLRI) for critical input, A. James, V. Nguyen, and B. Larsen (SLRI) for technical assistance and M.M. Stacey, C. Chen and J. Jin for comments on the manuscript. Supported by Genome Canada through the Ontario Genomics Institute, the Ontario Research Fund from the Ontario Ministry of Research and Innovation, a Terry Fox Foundation team grant, the Canadian Institutes of Health Research (MOP-13466-6849), and the Canada Foundation for Innovation. M.A.S is supported by a Vanier Canada Graduate Studentship and R.B. is supported by a CIHR postdoctoral fellowship. Support for R.J.D. was from the National Health and Medical Research Council of Australia and Cancer Council New South Wales (NSW), and for D.R.C. from Cancer Institute NSW.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature. sMRM quantification data sets from this study can be found at http://pawsonlab.mshri.on.ca/Shc1_dynamics.
Author contributions Y.Z. conceived and implemented the sMRM approach. C.Z., Y.Z. and L.T. developed and performed sMRM assays. S.A.T. provided technical MS support. Y.Z., M.A.S., N.S.D., D.R.C., and A.Y.D. performed biochemical and functional experiments. R.W.H. generated Shc1-deficient MEFs and Grb2flx/flx MEFs. Y.Z. and A.P. performed the computational analysis. T.P., R.D. and A-C.G. oversaw the project. Y.Z., M.A.S., K.C. and T.P. wrote the paper with input from J.W.D..
The authors declare competing financial interests: S.A.T. is an employee of AB SCIEX. AB SCIEX has provided support for the Ontario Research Fund grant (awarded to T.P.).
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature12308
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4931914
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Colorectal carcinoma progression is not influenced by the pseudokinase PEAK1.
Sci Rep, 14(1):27663, 12 Nov 2024
Cited by: 0 articles | PMID: 39532961 | PMCID: PMC11557890
SHCBP1 promotes cisplatin resistance of ovarian cancer through AKT/mTOR/Autophagy pathway.
Apoptosis, 13 Oct 2024
Cited by: 0 articles | PMID: 39397124
Dissecting phospho-motif-dependent Shc1 interactome using long synthetic protein fragments.
Chem Sci, 12 Aug 2024
Cited by: 0 articles | PMID: 39184293 | PMCID: PMC11342145
Proteomic characterization identifies clinically relevant subgroups of soft tissue sarcoma.
Nat Commun, 15(1):1381, 15 Feb 2024
Cited by: 1 article | PMID: 38360860 | PMCID: PMC10869728
Index analysis: An approach to understand signal transduction with application to the EGFR signalling pathway.
PLoS Comput Biol, 20(2):e1011777, 05 Feb 2024
Cited by: 0 articles | PMID: 38315738 | PMCID: PMC10868873
Go to all (176) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Genes & Proteins
- (1 citation) UniProt - Q5VWQ8
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Signaling adaptor ShcD suppresses extracellular signal-regulated kinase (Erk) phosphorylation distal to the Ret and Trk neurotrophic receptors.
J Biol Chem, 292(14):5748-5759, 17 Feb 2017
Cited by: 7 articles | PMID: 28213521 | PMCID: PMC5392570
Epidermal growth factor receptors containing a single tyrosine in their C-terminal tail bind different effector molecules and are signaling-competent.
J Biol Chem, 292(50):20744-20755, 26 Oct 2017
Cited by: 10 articles | PMID: 29074618 | PMCID: PMC5733609
The adaptor proteins p66Shc and Grb2 regulate the activation of the GTPases ARF1 and ARF6 in invasive breast cancer cells.
J Biol Chem, 289(9):5687-5703, 09 Jan 2014
Cited by: 35 articles | PMID: 24407288 | PMCID: PMC3937643
Tyrosine phosphorylation of Cbl upon epidermal growth factor (EGF) stimulation and its association with EGF receptor and downstream signaling proteins.
J Biol Chem, 271(24):14554-14559, 01 Jun 1996
Cited by: 88 articles | PMID: 8662998
Funding
Funders who supported this work.
CIHR (1)
Grant ID: MOP-13466-6849
Canadian Institutes of Health Research (3)
Grant ID: 13466-4
Grant ID: 13466-2
Grant ID: 13466-3





