Abstract
Importance
Cannabis dependence (CAD) is a serious problem worldwide and is of growing importance in the United States because cannabis is increasingly available legally. Although genetic factors contribute substantially to CAD risk, at present no well-established specific genetic risk factors for CAD have been elucidated.Objective
To report findings for DSM-IV CAD criteria from association analyses performed in large cohorts of African American and European American participants from 3 studies of substance use disorder genetics.Design, setting, and participants
This genome-wide association study for DSM-IV CAD criterion count was performed in 3 independent substance dependence cohorts (the Yale-Penn Study, Study of Addiction: Genetics and Environment [SAGE], and International Consortium on the Genetics of Heroin Dependence [ICGHD]). A referral sample and volunteers recruited in the community and from substance abuse treatment centers included 6000 African American and 8754 European American participants, including some from small families. Participants from the Yale-Penn Study were recruited from 2000 to 2013. Data were collected for the SAGE trial from 1990 to 2007 and for the ICGHD from 2004 to 2009. Data were analyzed from January 2, 2013, to November 9, 2015.Main outcomes and measures
Criterion count for DSM-IV CAD.Results
Among the 14 754 participants, 7879 were male, 6875 were female, and the mean (SD) age was 39.2 (10.2) years. Three independent regions with genome-wide significant single-nucleotide polymorphism associations were identified, considering the largest possible sample. These included rs143244591 (β = 0.54, P = 4.32 × 10-10 for the meta-analysis) in novel antisense transcript RP11-206M11.7;rs146091982 (β = 0.54, P = 1.33 × 10-9 for the meta-analysis) in the solute carrier family 35 member G1 gene (SLC35G1); and rs77378271 (β = 0.29, P = 2.13 × 10-8 for the meta-analysis) in the CUB and Sushi multiple domains 1 gene (CSMD1). Also noted was evidence of genome-level pleiotropy between CAD and major depressive disorder and for an association with single-nucleotide polymorphisms in genes associated with schizophrenia risk. Several of the genes identified have functions related to neuronal calcium homeostasis or central nervous system development.Conclusions and relevance
These results are the first, to our knowledge, to identify specific CAD risk alleles and potential genetic factors contributing to the comorbidity of CAD with major depression and schizophrenia.Free full text

Genome-wide Association Study of Cannabis Dependence Severity, Novel Risk Variants, and Shared Genetic Risks
Abstract
IMPORTANCE
Cannabis dependence (CAD) is a serious problem worldwide and is of growing importance in the United States because cannabis is increasingly available legally. Although genetic factors contribute substantially to CAD risk, at present no well-established specific genetic risk factors for CAD have been elucidated.
OBJECTIVE
To report findings for DSM-IV CAD criteria from association analyses performed in large cohorts of African American and European American participants from 3 studies of substance use disorder genetics.
DESIGN, SETTING, AND PARTICIPANTS
This genome-wide association study for DSM-IV CAD criterion count was performed in 3 independent substance dependence cohorts (the Yale-Penn Study, Study of Addiction: Genetics and Environment [SAGE], and International Consortium on the Genetics of Heroin Dependence [ICGHD]). A referral sample and volunteers recruited in the community and from substance abuse treatment centers included 6000 African American and 8754 European American participants, including some from small families. Participants from the Yale-Penn Study were recruited from 2000 to 2013. Data were collected for the SAGE trial from 1990 to 2007 and for the ICGHD from 2004 to 2009. Data were analyzed from January 2, 2013, to November 9, 2015.
MAIN OUTCOMES AND MEASURES
Criterion count for DSM-IV CAD.
RESULTS
Among the 14 754 participants, 7879 were male, 6875 were female, and the mean (SD) age was 39.2 (10.2) years. Three independent regions with genome-wide significant single-nucleotide polymorphism associations were identified, considering the largest possible sample. These included rs143244591 (β = 0.54, P = 4.32 × 10−10 for the meta-analysis) in novel antisense transcript RP11-206M11.7; rs146091982 (β = 0.54, P = 1.33 × 10−9 for the meta-analysis) in the solute carrier family 35 member G1 gene (SLC35G1); and rs77378271 (β = 0.29, P = 2.13 × 10−8 for the meta-analysis) in the CUB and Sushi multiple domains 1 gene (CSMD1). Also noted was evidence of genome-level pleiotropy between CAD and major depressive disorder and for an association with single-nucleotide polymorphisms in genes associated with schizophrenia risk. Several of the genes identified have functions related to neuronal calcium homeostasis or central nervous system development.
CONCLUSIONS AND RELEVANCE
These results are the first, to our knowledge, to identify specific CAD risk alleles and potential genetic factors contributing to the comorbidity of CAD with major depression and schizophrenia.
After nicotine, cannabis is the most widely abused drug worldwide.1 In the United States, the accelerated decriminalization of cannabis is based on the erroneous perception that it is relatively harmless.2 In fact, cannabis use produces craving,3 dependence,4 and drug-seeking behavior,5 as with the use of other substances. Despite these risks, the prevalence of cannabis use and cannabis use disorders has dramatically increased since 2001,6 and the political momentum to increase availability has continued. Use of cannabis early in life is associated with an increased risk for schizophrenia (SCZ),7 and sets of SCZ-associated risk alleles predict cannabis use.8 Cannabis use is also a risk factor for depressive symptoms,9 and a twin study showed cannabis dependence (CAD) to be associated with an elevated risk for major depressive disorder (MDD).10 Substance use and other psychiatric illnesses may share common genetic risk factors; or reverse causation, self-medication, or confounding by other factors may explain their co-occurrence.
Despite knowledge of the neurobiology of the endocannabinoid system and its response to tetrahydrocannabinol, little is known about specific genetic factors influencing susceptibility to CAD or cannabis abuse. A twin study showed that several aspects of cannabis use are heritable, including an early opportunity to use (h2 = 72%), early onset of use (h2 = 80%), lifetime use of cannabis 11 or more times (h2 = 76%), and cannabis abuse or dependence (h2 = 21%–72%), where h2 is hertiability.11–13 Possible evidence of linkage of CAD on chromosome 1614 and linkage and association encompassing the neuregulin 1 gene (NRG1 [OMIM 142445]; known as a possible SCZ risk gene15) on chromosome 816 have been found. Despite several genome-wide association studies (GWAS) on cannabis-related traits, no genome-wide significant (GWS) associations were observed for initiation of use17 or for CAD.18 Herein we report on findings for DSM-IV CAD criteria from association analyses performed in large cohorts of African American and European American participants from 3 studies of substance use disorder genetics who underwent genotyping with genome-wide microarrays. The primary cohort has been used in previous studies to identify genes associated with opioid (OD),19 cocaine (CD),20 alcohol (AD),21 and nicotine (ND) dependence22 and posttraumatic stress disorder.23
Methods
Participants and Diagnostic Procedures
The samples included 6000 African American and 8754 European American participants (race was assigned based on genetic data; eMethods in the Supplement) from the following 3 studies: (1) the Yale-Penn Study cohort of small nuclear families and unrelated individuals (2020 individuals in 850 families and 6951 unrelated individuals), collected to study the genetics of substance dependence19–21; (2) the GWAS data set from the Study of Addiction: Genetics and Environment (SAGE),24–27 collected to study the genetics of AD, ND, and CD (183 individuals in 89 families and 3707 unrelated individuals); and (3) the GWAS International Consortium on the Genetics of Heroin Dependence (ICGHD),28,29 a collaboration formed to identify genes associated with heroin dependence risk (66 individuals in 33 families and 1827 unrelated individuals). The SAGE and ICGHD data sets are publicly available via application. The present study received institutional review board approval from all participating institutions, and written informed consent was obtained from all study participants.
Participants from the Yale-Penn Study were recruited from 2000 to 2013. These participants were administered the Semi-Structured Assessment for Drug Dependence and Alcoholism30 to derive DSM-IV diagnoses of lifetime CAD and other major psychiatric traits. Data were collected for the SAGE trial from 1990 to 2007, and participants underwent phenotyping with the Semi-Structured Assessment for the Genetics of Alcoholism.31 Data were collected for the ICGHD from 2004 to 2009, and participants completed a comprehensive psychiatric diagnostic interview based on the Semi-Structured Assessment of the Genetics of Alcoholism–Australia.31 The method of phenotyping was similar across the 3 samples. Additional information about recruitment, genotyping, imputation, and quality control for the study cohorts is provided in eMethods in the Supplement.
Statistical Analysis
Data were analyzed from January 2, 2013, to November 9, 2015. Association analyses were performed using a count of DSM-IV CAD criteria (0–7) as the outcome variable and the imputed minor allele dosage (adjusted for sex, age, and the first 3 ancestry principal components) as a predictor variable. This ordinal trait model has greater power to detect genetic associations than a univariate model based on disease status because of greater information content and improved specificity of the dependence measure. Association tests were performed using linear association models embedded in generalized estimating equations to correct for correlations among related individuals.32 Analyses were performed separately within each data set and population group, and the results were combined by meta-analysis using the inverse variance method implemented in the program METAL.33 Genomic inflation factors (λ) were calculated within each subpopulation, and P values were corrected accordingly. We performed a second correction for the λ factor calculated after the meta-analysis.
For the primary analysis, individuals were included regardless of cannabis exposure. As secondary analyses, individuals who reported never having used cannabis were excluded, and the primary model was repeated adjusting for the criterion counts for AD, CD, and OD. Participants from 2 genotyping batches in the Yale-Penn cohort (Yale-Penn 1 and Yale-Penn 2) were combined with the SAGE sample to form a discovery data set. A sample consisting of the ICGHD data and additional samples from the Yale-Penn cohort who did not undergo genotyping at the time of the discovery analyses (Yale-Penn 3) were used to replicate the top associations.
Cross-Disorder Analysis
We attempted to uncover shared genetic variation between CAD and 5 psychiatric disorders, including SCZ, MDD, bipolar affective disorder, attention-deficit/hyperactivity disorder, and autism spectrum disorder using the GWAS analysis reported herein and publicly available GWAS results from the Psychiatric Genomics Consortium (http://www.med.unc.edu/pgc/).34 To explore cross-disorder genetic relationships, we used stratified quintile-quintile (QQ) plots to evaluate the relative enrichment of single-nucleotide polymorphisms (SNPs) associated with both disorders. The QQ plots, which contrast the observed distribution of P values with the expected distribution under the null hypothesis (uniform in GWAS), were used to assess P value inflation in the GWAS results. Grouping associated SNPs for one disorder and comparing (across groups) the QQ plots of another disorder, however, could also reveal the enrichment of GWAS signals between disorders, which made them suitable for cross-disorder enrichment screening.
We also applied a statistical framework for pleiotropy analysis, Genetic Analysis Incorporating Pleiotropy and Annotation (GPA).35 The GPA was built as a mixture model with parameters estimated using an efficient expectation-maximization algorithm, where associated SNPs were modeled with a β [α, 1] distribution and unassociated SNPs with a uniform [0, 1] distribution. A likelihood ratio test assessed the significance of pleiotropy between disorders. The GPA also detected the SNPs that were pleiotropic by calculating the posterior probability of association with both disorders.
Results
Participant demographic characteristics and the correlation between the criterion counts for CAD and other substance use disorder traits are shown in Table 1. The DSM-IV CAD criterion counts were significantly (P < .05) correlated with the criteria counts for AD, CD, OD, and ND. The correlations varied by sample and population and ranged from r2 = 0.15 for OD to r2 = 0.61 for CD criteria. The CAD criterion counts were significantly heritable in European American (19%–25%; P = .006) but not African American (10%–11%; P = .08) participants. eFigure 1 in the Supplement shows a histogram of the CAD criterion count in African American and European American participants in each cohort; 3 or more criteria indicate a diagnosis of CAD. The criterion count distribution is very similar in African American and European American participants. In the Yale-Penn sample, where comorbid psychiatric diagnoses were available, CAD was significantly associated with MDD in African American participants (odds ratio, 1.07; P = .006) but not SCZ, bipolar affective disorder, attention-deficit/hyperactivity disorder, or autism spectrum disorder. Cannabis dependence was not associated with any of these disorders in European American participants.
Table 1
Demographic Characteristics of the Study Cohorts
| Samplea | Age, Mean (SD), y | Female Sex, No./Total No. | DSM-IV CAD Criterion Count, Mean (SD) | CAD Diagnosis, No./Total No. of Participants | Correlation With DSM-IV CAD Criterion Count, r2 | |||
|---|---|---|---|---|---|---|---|---|
| AD | CD | ND | OD | |||||
| Yale-Penn African American | 41.3 (9.7) | 2209/4750 | 1.7 (2.2) | 1296/4750 | 0.40 | 0.30 | 0.36 | 0.15 |
| Yale-Penn European American | 38.6 (11.5) | 1712/4221 | 2.0 (2.3) | 1388/4221 | 0.27 | 0.32 | 0.31 | 0.23 |
| SAGE African American | 39.9 (7.3) | 638/1250 | 1.4 (2.2) | 276/1250 | 0.43 | 0.43 | 0.39 | 0.17 |
| SAGE European American | 38.4 (9.7) | 1478/2640 | 1.0 (1.9) | 434/2640 | 0.51 | 0.61 | 0.40 | 0.41 |
| ICGHD | 36.2 (9.1) | 838/1893 | 3.2 (2.5) | 1062/1893 | 0.24 | 0.24 | 0.25 | 0.33 |
Abbreviations: AD, alcohol dependence; CAD, cannabis dependence; CD, cocaine dependence; ICGHD, International Consortium on the Genetics of Heroin Dependence; ND, nicotine dependence; OD, opioid dependence; SAGE, Study of Addiction: Genetics and Environment.
GWAS Results
Manhattan and QQ plots for the meta-analysis discovery GWAS results for African American and European American Yale-Penn 1 and 2 and SAGE cohorts are displayed in eFigures 2 and 3 in the Supplement. We found little evidence of P value inflation. Table 2 shows associations in the discovery sample with P < 1.0 × 10−5 in African American or European American participants or the combined meta-analysis, trimmed for linkage disequilibrium. eTable 1 in the Supplement shows the same results, together with additional information about each SNP, including the results within each discovery sample subgroup, after excluding individuals with no cannabis exposure, and after adjusting for comorbid substance use disorders. We identified GWS associations with reliably imputed SNPs in 3 distinct regions (Table 2), 2 specific to African American participants and 1 in the combined sample. First, rs186825689 (P = 1.86 × 10−8 for the African American meta-analysis) is located 12.4 kb upstream from the gene encoding S100 calcium binding protein (S100B) with contributions from both informative African American samples. Second, rs143244591 (P = 2.18 × 10−8 for the African American meta-analysis) maps to a novel antisense transcript RP11-206M11.7 (Havana gene: OTTHUMG00000159583) located in the gene of the same name on chromosome 3 with at least nominally significant evidence in each of the 3 African American samples. Third, rs77378271(P = 2.76 × 10−8 for the European American meta-analysis) is an intronic SNP in the CUB and Sushi multiple domains 1 gene (CSMD1 [OMIM 608397]) with evidence of association in 3 of the 6 samples. We also identified consistent, non-GWS evidence of association in the combined sample of European American and African American participants with a large block of SNPs in and around the phosphatidylinositol 4-kinase type 2β gene (PI4K2B [OMIM 612101]), with consistent effect direction in every European American and African American population tested (minimum P = 1.74 × 10−7 for the meta-analysis). This signal was GWS when individuals without cannabis exposure were excluded (minimum P = 2.98 × 10−8 for the meta-analysis).
Table 2
SNPs Associated With DSM-IV CAD at P < 1.0 × 10−5 in the Discovery Meta-analysis Trimmed for SNPs in Linkage Disequilibrium
| Chromosome | Base Pair Positiona | Effect Allele | Reference Allele | SNP | Gene | Effect Allele Frequency | P Value for Meta-analysis
| |||
|---|---|---|---|---|---|---|---|---|---|---|
| African American Cohort | European American Cohort | African American Cohort | European American Cohort | All Participants | ||||||
| 1 | 88729683 | C | T | rs74823926 | NA | 0.96 | 0.97 | 5.26 × 10−7 | 6.36 × 10−1 | 1.40 × 10−5 |
|
| ||||||||||
| 2 | 39166173 | T | G | rs114383460 | ARHGEF33 | 0.96 | NA | 1.09 × 10−6 | NA | NA |
|
| ||||||||||
| 2 | 78028838 | T | A | rs12621150 | NA | 0.85 | 0.90 | 7.67 × 10−1 | 1.05 × 10−6 | 2.91 × 10−4 |
|
| ||||||||||
| 2 | 100451676 | T | C | rs7586604 | AFF3 | 0.39 | 0.16 | 4.15 × 10−5 | 6.19 × 10−3 | 1.06 × 10−6 |
|
| ||||||||||
| 2 | 103764414 | G | A | rs144605126 | NA | 0.96 | NA | 8.67 × 10−7 | NA | NA |
|
| ||||||||||
| 2 | 118490901 | G | A | rs150064803 | NA | 0.95 | NA | 3.41 × 10−7 | NA | NA |
|
| ||||||||||
| 2 | 167214714 | G | A | rs143020225 | SCN9A | 0.95 | NA | 7.19 × 10−7 | NA | NA |
|
| ||||||||||
| 3 | 149013935 | G | A | rs143244591b | RP11-206M11.7 | 0.96 | NA | 2.18 × 10−8b | NA | NA |
|
| ||||||||||
| 4 | 25201318 | A | T | rs73252553 | PI4K2B | 0.96 | 0.90 | 1.18 × 10−3 | 2.28 × 10−5 | 1.66 × 10−7 |
|
| ||||||||||
| 4 | 119716950 | A | C | rs28595532 | SEC24D | 0.92 | 0.94 | 2.02 × 10−7 | 1.08E-01 | 1.13 × 10−6 |
|
| ||||||||||
| 5 | 11892384 | T | C | rs114311699 | CTNND2 | 0.96 | NA | 3.78 × 10−7 | NA | NA |
|
| ||||||||||
| 5 | 177746600 | G | C | rs10066744 | COL23A1 | 0.96 | NA | 4.82 × 10−7 | NA | NA |
|
| ||||||||||
| 6 | 51221457 | A | G | rs17665889 | NA | 0.94 | 0.90 | 1.51 × 10−1 | 9.41 × 10−8 | 2.58 × 10−4 |
|
| ||||||||||
| 7 | 84952631 | A | G | rs12534830 | NA | 0.88 | 0.66 | 7.76 × 10−3 | 1.54 × 10−5 | 4.52 × 10−7 |
|
| ||||||||||
| 8 | 3073489 | A | G | rs77378271b | CSMD1 | 0.96 | 0.94 | 2.13 × 10−1 | 2.76 × 10−8b | 4.60 × 10−8b |
|
| ||||||||||
| 9 | 29364327 | G | T | rs10969106 | NA | NA | 0.97 | NA | 7.39 × 10−8 | NA |
|
| ||||||||||
| 10 | 31981385 | T | C | rs115553536 | NA | 0.94 | NA | 6.46 × 10−7 | NA | NA |
|
| ||||||||||
| 10 | 43592809 | G | C | rs74400468 | RET | 0.96 | 0.97 | 5.53 × 10−2 | 1.59 × 10−6 | 6.46 × 10−7 |
|
| ||||||||||
| 10 | 70490106 | A | T | rs12218439 | CCAR1 | 0.96 | 0.96 | 1.01 × 10−6 | 2.01 × 10−1 | 1.13 × 10−4 |
|
| ||||||||||
| 10 | 95659958 | A | G | rs146091982 | SLC35G1 | 0.95 | NA | 1.95 × 10−7 | NA | NA |
|
| ||||||||||
| 11 | 20561010 | C | G | rs73443003 | NA | 0.75 | NA | 1.31 × 10−6 | NA | NA |
|
| ||||||||||
| 11 | 81433204 | A | AAAG | rs200453611 | NA | 0.91 | 0.84 | 9.43 × 10−7 | 6.24 × 10−1 | 6.81 × 10−4 |
|
| ||||||||||
| 11 | 108899423 | GTA | G | rs200391037 | NA | 0.96 | 0.88 | 1.02 × 10−1 | 3.72 × 10−6 | 1.32 × 10−6 |
|
| ||||||||||
| 12 | 56274155 | T | C | rs193047854 | NA | 0.97 | NA | 7.06 × 10−7 | NA | NA |
|
| ||||||||||
| 20 | 21706604 | A | AT | rs199783889 | NA | 0.94 | NA | 3.32 × 10−7 | NA | NA |
|
| ||||||||||
| 21 | 18019319 | T | C | rs78068107 | NA | 0.96 | NA | 1.02 × 10−6 | NA | NA |
|
| ||||||||||
| 21 | 48006053 | A | C | rs186825689b | NA | 0.96 | NA | 1.86 × 10−8b | NA | NA |
Replication Results
The SNPs in Table 2 were tested for CAD association in the 2 replication samples (ICGHD and Yale-Penn 3). Table 3 shows the replication cohort-specific results for these SNPs, with the meta-analysis results from the discovery phase and the discovery + replication phase. The smallest P value in the ICGHD cohort among the 13 SNPs that could be reliably imputed and analyzed (this cohort was European Australian) was at rs74823926 (P = .064) in an intergenic region on chromosome 1. Several associations, however, were replicated in the Yale-Penn 3 sample (Table 3). The P values for 2 of the 3 GWS SNPs improved after meta-analysis with the replication cohorts (rs143244591 in RP11-206M11.7, from 1.38 × 10−8 to 4.32 × 10−10; rs77378271 in CSMD1, from 2.84 × 10−8 to 2.13 × 10−8), as did the P value for another SNP, rs146091982 in the solute carrier family 35 member G1 (SLC35G1 [Ensembl ENSG00000176273]) (from 1.31 × 10−7 to 1.33 × 10−9). The signal in PI4K2B also improved (P = 5.57 × 10−8 for the full meta-analysis). However, rs186825689 near S100B was no longer GWS (P = 8.27 × 10−8) in the full meta-analysis. The Figure shows Manhattan plots for the regions encompassing RP11-206M11.7 (Figure, A), SLC35G1 (Figure, B), CSMD1 (Figure, C), and PI4K2B (Figure, D) in the discovery sample and after meta-analysis with the replication samples.
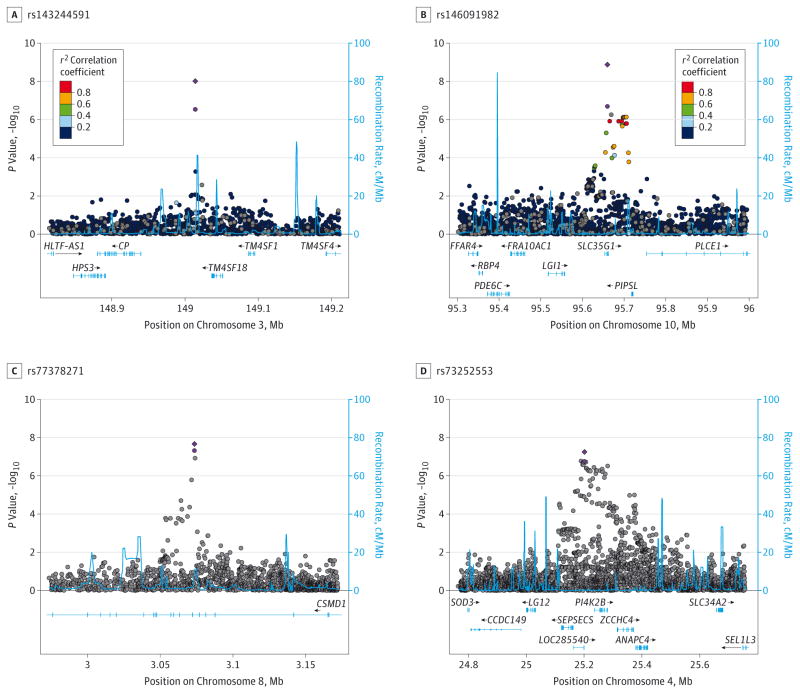
Association results from single-nucleotide polymorphisms (SNPs) in 4 regions. A, The 148.8- to 149.2-MB region encompassing RP11-206M11.7 on chromosome 3 in the Yale-Penn and Study of Addiction: Genetics and Environment (SAGE) African American participants. B, The 95.3- to 96-MB region encompassing SLC35G1 on chromosome 10 in the Yale-Penn and SAGE African American participants. C, The 2.8- to 4.8-MB region on chromosome 8 encompassing CSMD1 in the Yale-Penn, SAGE, and International Consortium on the Genetics of Heroin Dependence (ICGHD) African American and European American participants. D, The 25.07- to 25.43-MB region encompassing PI4K2B on chromosome 4 in the Yale-Penn, SAGE, and ICGHD African American and European American participants. In A and B, the SNPs are color coded according to the correlation coefficient (r2) in the 1000 Genomes African samples with the most significant SNP. In C and D, results from the African American and European American participants were combined, and no linkage disequilibrium information was displayed. The light purple circle represents the −log10 P value for the most significant regional SNP in the meta-analysis of the discovery samples; the purple diamond, the result for that SNP after meta-analysis with the replication sample(s). The light blue line and right y-axis show the observed recombination rate.
Table 3
Association Results in the Discovery and Replication Samples for the SNPs Shown in Table 2
| SNP | P Value for Replication Cohort
| P Value for Meta-analysis
| Directiond | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Yale-Penn 3 Cohorta
| ICGHD Cohort | Discovery Cohortb
| Discovery + Replication Cohortc
| All Participants | |||||
| African American | European American | African American | European American | African American | European American | ||||
| rs74823926 | 9.40 × 10−3 | 7.54 × 10−1 | 6.36 × 10−2 | 5.26 × 10−7 | 6.36 × 10−1 | 3.34 × 10−6 | 1.59 × 10−1 | 8.62 × 10−6 | +x++++−++ |
|
| |||||||||
| rs114383460 | 7.22 × 10−1 | NA | NA | 1.09 × 10−6 | NA | 1.93 × 10−6 | NA | 1.93 × 10−6 | +x + xxx−xx |
|
| |||||||||
| rs12621150 | 9.68 × 10−1 | 1.15 × 10−1 | 3.61 × 10−1 | 7.67 × 10−1 | 1.05 × 10−6 | 7.75 × 10−1 | 7.92 × 10−6 | 5.13 × 10−4 | ++++−+−−+ |
|
| |||||||||
| rs7586604 | 2.80 × 10−1 | 2.78 × 10−1 | 6.04 × 10−1 | 4.15 × 10−5 | 6.19 × 10−3 | 1.76 × 10−5 | 1.69 × 10−2 | 1.81 × 10−6 | −−−−−−−+− |
|
| |||||||||
| rs144605126 | 1.18 × 10−1 | NA | NA | 8.67 × 10−7 | NA | 3.84 × 10−6 | NA | 3.84 × 10−6 | +x + x + x−xx |
|
| |||||||||
| rs150064803 | 1.74 × 10−1 | NA | NA | 3.41 × 10−7 | NA | 1.41 × 10−6 | NA | 1.41 × 10−6 | +x + x + x−xx |
|
| |||||||||
| rs143020225 | 1.53 × 10−1 | NA | NA | 7.19 × 10−7 | NA | 1.95 × 10−7 | NA | 1.95 × 10−7 | +x + xxx + xx |
|
| |||||||||
| rs143244591 | 3.24 × 10−3 | NA | NA | 2.18 × 10−8e | NA | 4.32 × 10−10e | NA | 4.32 × 10−10e | +x + x + x+xx |
|
| |||||||||
| rs73252553 | 6.40 × 10−1 | 8.32 × 10−2 | 4.10 × 10−1 | 1.18 × 10−3 | 2.28 × 10−5 | 2.25 × 10−3 | 5.45 × 10−6 | 5.57 × 10−8 | ++++++−++ |
|
| |||||||||
| rs28595532 | 8.06 × 10−1 | 6.07 × 10−2 | 2.24 × 10−1 | 2.02 × 10−7 | 1.08 × 10−1 | 1.89 × 10−7 | 1.52 × 10−1 | 3.60 × 10−6 | ++++++++− |
|
| |||||||||
| rs114311699 | 7.30 × 10−1 | NA | NA | 3.78 × 10−7 | NA | 2.75 × 10−7 | NA | 2.75 × 10−7 | +x + x + x+xx |
|
| |||||||||
| rs10066744 | 3.89 × 10−1 | NA | NA | 4.82 × 10−7 | NA | 2.27 × 10−7 | NA | 2.27 × 10−7 | +x + xxx + xx |
|
| |||||||||
| rs17665889 | 2.17 × 10−1 | 1.17 × 10−1 | 7.65 × 10−1 | 1.51 × 10−1 | 9.41 × 10−8 | 2.69 × 10−1 | 5.39 × 10−6 | 8.41 × 10−4 | −+++−++−+ |
|
| |||||||||
| rs12534830 | 5.85 × 10−1 | 8.67 × 10−1 | 3.99 × 10−1 | 7.76 × 10−3 | 1.54 × 10−5 | 1.05 × 10−2 | 3.40 × 10−5 | 1.12 × 10−6 | ++++++−−+ |
|
| |||||||||
| rs77378271 | 9.25 × 10−2 | 4.19 × 10−2f | 7.95 × 10−1 | 2.13 × 10−1 | 2.76 × 10−8e | 1.07 × 10−1 | 5.16 × 10−8 | 2.13 × 10−8e | +−++x+++− |
|
| |||||||||
| rs10969106 | NA | 5.50 × 10−1 | 8.47 × 10−1 | NA | 7.39 × 10−8 | NA | 1.74 × 10−7 | 1.74 × 10−7 | x + x + x+xx+ |
|
| |||||||||
| rs115553536 | 1.80 × 10−1 | NA | NA | 6.46 × 10−7 | NA | 2.14 × 10−6 | NA | 2.14 × 10−6 | +x + x + x−xx |
|
| |||||||||
| rs74400468 | 3.49 × 10−2 | 6.61 × 10−1 | 5.66 × 10−1 | 5.53 × 10−2 | 1.59 × 10−6 | 1.22 × 10−2 | 2.46 × 10−5 | 9.82 × 10−7 | ++−+++++− |
|
| |||||||||
| rs12218439 | 2.29 × 10−1 | 1.01 × 10−1 | 3.21 × 10−1 | 1.01 × 10−6 | 2.01 × 10−1 | 4.91 × 10−6 | 2.13 × 10−1 | 4.20 × 10−4 | x++−++−−+ |
|
| |||||||||
| rs146091982 | 8.84 × 10−4 | NA | NA | 1.95 × 10−7 | NA | 1.33 × 10−9e | NA | 1.33 × 10−9e | +x + x + x+xx |
|
| |||||||||
| rs73443003 | 2.53 × 10−2 | NA | NA | 1.31 × 10−6 | NA | 1.20 × 10−7 | NA | 1.20 × 10−7 | −x−x−x−xx |
|
| |||||||||
| rs200453611 | 7.29 × 10−1 | 5.20 × 10−1 | 9.40 × 10−1 | 9.43 × 10−7 | 6.24 × 10−1 | 1.56 × 10−6 | 5.34 × 10−1 | 1.12 × 10−3 | ++++++−++ |
|
| |||||||||
| rs200391037 | 9.28 × 10−1 | 3.09 × 10−2f | 4.19 × 10−1 | 1.02 × 10−1 | 3.72 × 10−6 | 1.04 × 10−1 | 4.32 × 10−5 | 1.10 × 10−5 | ++++++−−+ |
|
| |||||||||
| rs193047854 | 6.74 × 10−1 | NA | NA | 7.06 × 10−7 | NA | 5.51 × 10−7 | NA | 5.51 × 10−7 | +x + xxx + xx |
|
| |||||||||
| rs199783889 | 4.38 × 10−1 | NA | NA | 3.32 × 10−7 | NA | 1.12 × 10−6 | NA | 1.12 × 10−6 | +x + xxx−xx |
|
| |||||||||
| rs78068107 | 2.90 × 10−2 | NA | NA | 1.02 × 10−6 | NA | 1.31 × 10−5 | NA | 1.31 × 10−5 | +x + xxx−xx |
|
| |||||||||
| rs186825689 | 4.51 × 10−1 | NA | NA | 1.86 × 10−8e | NA | 8.27 × 10−8 | NA | 8.27 × 10−8 | +x + xxx−xx |
Abbreviations: ICGHD, International Consortium on the Genetics of Heroin Dependence; NA, not applicable; SAGE, Study of Addiction: Genetics and Environment; SNP, single-nucleotide polymorphism.
Cross-Disorder Analysis Results
The QQ plots of 5 Psychiatric Genomics Consortium traits (SCZ, bipolar affective disorder, autism spectrum disorder, attention-deficit/hyperactivity disorder, and MDD) were stratified based on our CAD GWAS results at significance levels of P < .05, P < .01, P < 1 × 10−3, and P < 1 × 10−4. We observed enrichment of the MDD GWAS signal in the CAD GWAS (eFigure 4 in the Supplement) in European American participants, but no clear enrichment for the other 4 psychiatric disorders in either population group (eFigure 5 in the Supplement).
We used GPA to test the significance of pleiotropy between CAD and the same 5 psychiatric disorders (eMethods in the Supplement). For each disease pair, we estimated the percentage of SNPs shared by 2 diseases and tested the significance of pleiotropy (eTable 2 in the Supplement). The European American population yielded significant evidence of CAD-MDD pleiotropy (P = 2.39 × 10−5); genome wide, 1.7% of all imputed SNPs were estimated to be associated with both CAD and MDD. Of these, rs10954732 in P450 oxidoreductase(POR[OMIM124015]) had the largest posterior probability (although not significant) of association with both traits (P = 2.59 × 10−6 for CAD; P = .02 for MDD; posterior probability, 0.70).
Discussion
We report herein the first GWS results for CAD to our knowledge. The sample includes a large proportion (18%–36%, depending on race and cohort) of individuals with CAD from 2 ancestral populations in 3 independent cohorts. We identified 3 regions with GWS SNPs imputed to the 1000 Genomes reference panel that implicate several biological processes and provide insight into the biology of CAD, including evidence of an inflammatory component in the disorder, which may also mediate risk for SCZ36 and MDD.37,38 The smallest P value observed (P = 4.32 × 10−10) was at rs143244591 in RP11-206M11.7. Little is known about this antisense transcript or which, if any, genes it regulates. Minor alleles were protective. The next most significant locus was SLC35G1 (rs146091982, P = 1.33 × 10−9), a potential member of the drug/metabolite transporter superfamily (EamA, previously DUF6). Ubiquitously expressed, SLC35G1 binds stromal interaction molecule 1, a calcium sensor that communicates the calcium load within the endoplasmic reticulum to store-operated channels in the plasma membrane39 when calcium stores in the endoplasmic reticulum are depleted.40 The SLC35G1–stromal interaction molecule 1 complex likely regulates the activity of the transporters that coordinate cytosolic calcium through modulation of pump activities.40 The third GWS locus, CSMD1 (rs77378271; P = 2.13 × 10−8), is highly expressed in the growth cones of developing central nervous system neurons, where it likely acts as a regulator of complement activation and inflammation.41 Different SNPs in CSMD1 have been associated with SCZ at the GWS level.42 Thus, CSMD1 is the second gene to be implicated in both disorders (after NRG116) and may explain at least part of their shared genetic susceptibility.
Two other established SCZ risk genes, RIMS1 (OMIM 606629) (minimum SNP, P = 1.59 × 10−5) and MEF2C (minimum SNP, P = 5.22 × 10−5), showed suggestive association with CAD. MEF2C is highly expressed in developing mammalian neurons and is thought to mediate calcium-dependent survival of neurons that have made the appropriate synaptic connections.43 From a biological perspective, RIMS1 is immediately relevant; RIMS1 acts as a scaffold protein that regulates synaptic vesicle exocytosis, affecting cannabinoid receptor 1 (CR1)–mediated long-term suppression of γ-aminobutyric acid release, ultimately mediating presynaptic forms of long-term plasticity.44 Minor alleles at rs142305709 in RIMS1 were associated with fewer CAD criteria in African American participants. We observed at least a nominally significant signal in both Yale-Penn African American analysis subsets and a non-significant trend in SAGE African American participants.
Limitations of the GWAS findings should be noted. One of the significant SNPs identified (rs143244591 on chromosome 3) has little supportive evidence for association from other SNPs in the region, possibly owing to low linkage disequilibrium. However, despite stringent imputation quality thresholds for including SNPs in the analysis (r2≥0.8) and evidence of an association in the replication sample, this signal may represent an imputation artifact. Second, although none of the GWS SNPs identified in the full GWAS analysis are rare, they could be described as infrequent, with minor allele frequencies in a range sometimes associated with false-positive results (4%–6%). Also, of the GWS regions, only CSMD1 showed evidence of associations in European American and African American participants. The region containing PI4K2B, which became GWS after excluding unexposed individuals (see below), was also at least nominally associated with CAD in both populations. The 2 African American–specific SNPs were rare or monomorphic in European American participants. The lack of association in European American participants could be owing to different linkage disequilibrium patterns or the absence of causal variants. The Yale-Penn samples who underwent genotyping on the HumanOmni1-Quad and Human Core Exome chips showed more consistent results than the corresponding SAGE population, which is not surprising insofar as SAGE participants were recruited from different areas and ascertained using different criteria (AD, CD, and OD in Yale-Penn and primarily AD and ND in SAGE). The difference in ascertainment criteria (use of licit vs illicit drugs) across studies likely explains the fact that the proportion of cannabis-exposed individuals varied significantly across cohorts (2293 in SAGE population [76.9%] and 7626 in the Yale-Penn population [85.0%]). The limitations of phenotypic distribution and population differences are more relevant to the Australian ICGHD replication cohort and may explain the lack of replication in this cohort. Despite this, we obtained statistically significant evidence for formal replication for the SNP in SLC35G1 and stronger evidence for association at many of the top SNPs after including the replication samples. Finally, these cohorts have higher rates of polysubstance dependence than the general population and may not be generalizable to individuals who only use cannabis.
Effect of Exposure Status and Comorbidity
Because the inclusion of genetically at-risk individuals who never initiated cannabis use might have influenced our results, we repeated the primary analyses in the discovery cohort after removing unexposed individuals. Two of the 3 regions identified remained GWS (eTable 1 in the Supplement). The P value for rs143244591 on chromosome 3 improved slightly (P = 1.13 × 10−8, meta-analysis exposed) and was associated at P ≤ .02 in each of the African American subgroups. The signal at rs77378271 in CSMD1 was almost identical (P = 2.95 × 10−8, meta-analysis exposed) and showed association at P < 5.07 × 10−3 in 2 of the 3 European American subgroup and at P = 4.46 × 10−4 in 1 of the African American subgroups. In addition, the block of SNPs in and around PI4K2B became GWS with a consistent effect direction (minor alleles being protective) in every European American and African American population tested and became GWS (minimum P = 2.98 × 10−8, meta-analysis exposed, at rs147170184). The evidence for pleiotropy between CAD and MDD was attenuated substantially (P = .60) after excluding unexposed participants. That the removal of unexposed individuals from the analysis had a relatively minor effect on the primary findings and actually improved the strength of some suggests that any loss in power owing to the smaller sample was offset by an increase in phenotypic precision. In the pleiotropy analysis, which relies on genome-level association results and is not limited to the most significantly associated SNPs, the power loss apparently outweighed any increase in precision. The significance of each of the top SNPs was modestly attenuated after adjusting for the DSM-IV criterion counts for AD, CD, and OD (eTable 1 in the Supplement).
Ion Homeostasis and Addiction
The previously published GWAS of OD19 and CD20 in a subset of this sample each identified risk genes and pathways involved in the regulation of neuronal calcium and potassium, and the pathway involving synaptic long-term potentiation was also identified for OD. Also, a cross-disorder analysis identified calcium signaling in neurons as a pathway mediating 5 psychiatric diseases, including SCZ and MDD.34 The GWS association in SLC35G1 and GWS (in the discovery sample only) associations in and around S100B suggest ion homeostasis may play a role in CAD risk.
Shared Risk for CAD and Other Psychiatric Disorders
Many previous studies7,8,45,46 have focused on the relationship between CAD and SCZ, whereas the correlation between CAD and MDD has received much less attention. Although depressive disorders are highly comorbid with CAD in clinical settings,47 to our knowledge no previous genomics study has explored CAD-MDD pleiotropy. We found some evidence for genetic correlation between the risks for CAD and MDD. The existence of shared genetic factors for CAD-MDD is supported by the overlap in SNPs nominally associated with both traits, although we found no significant evidence of pleiotropy at any single SNP. We also found limited support for the possibility that such a relationship exists for CAD and SCZ based on relatively strong signals for both traits with variants in CSMD1 (although not the same variants). Nongenetic explanations such as patients with SCZ or MDD mediating the symptoms of these disorders with cannabis use might also explain the comorbidity. These analyses are exploratory, and follow-up studies to validate and extend these findings are necessary.
Conclusions
This study provided the first GWS evidence to our knowledge for SNPs associated with CAD via GWAS in 3 distinct genomic locations. These findings will lead our understanding of genetic vulnerability to CAD in new directions that can inform our understanding of the biology of CAD. We obtained entirely novel evidence of genetic overlap between CAD and MDD and conclude that CSMD1 may be a candidate gene that affects the risk for CAD and SCZ, a topic of considerable research interest.7,48–50 These results also suggest that common pathways (nervous system development, inflammation, and ion homeostasis) mediate the risk for multiple psychiatric disorders and dependence on multiple substances, including cannabis.
Acknowledgments
Funding/Support: This study was supported by grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, and R01 AA017535 from the National Institutes of Health (NIH), the Veterans Affairs Connecticut Healthcare Center, and the Philadelphia Veterans Affairs Mental Illness Research, Education and Clinical Center. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided by grant U01 HG004422 from the NIH Genes, Environment and Health Initiative. SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies under the Genes, Environment and Health Initiative. The portion of the genotyping that was performed at the Johns Hopkins University Center for Inherited Disease Research was supported by grant U01HG004438 from the NIH Genes, Environment and Health Initiative, and by contract HHSN268200782096C (High Throughput Genotyping for Studying the Genetic Contributions to Human Disease) from the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, NIH. Support for collection of datasets and samples was provided by grant U10 AA008401 (Collaborative Study on the Genetics of Alcoholism), grant P01 CA089392 (Collaborative Genetic Study of Nicotine Dependence), and grant R01 DA013423 (Family Study of Cocaine Dependence) from the NIH. The International Consortium on the Genetics of Heroin Dependence (principal investigator, Elliot Nelson, MD) was funded by HHSN268200782096C (NIH contract: High Throughput Genotyping for Studying the Genetic Contributions to Human Disease) and HHSN268201100011I (NIH contract: High Throughput Genotyping for Studying the Genetic Contributions to Human Disease).
Footnotes
Additional Information: The publicly available datasets used for analysis were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/gap) through accession numbers phs000092.v1.p and phs000277.v1.p1.
Author Contributions: Dr Sherva had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Kranzler, Koesterer, Farrer, Gelernter.Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Sherva, Wang.
Critical revision of the manuscript for important intellectual content: Sherva, Kranzler, Zhao, Koesterer, Herman, Farrer, Gelernter.
Statistical analysis: Sherva, Wang, Zhao, Koesterer, Farrer.
Obtained funding: Kranzler, Gelernter.
Administrative, technical, or material support: Kranzler, Farrer, Gelernter.
Study supervision: Kranzler, Zhao, Farrer, Gelernter.
Conflict of Interest Disclosures: Dr Kranzler reports being a consultant or an advisory board member for Alkermes, Indivior, Lundbeck, and Otsuka (unrelated to the present study) and being a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which is supported by AbbVie, Ethypharm, Lilly, Lundbeck, and Pfizer. No other disclosures were reported.
Role of the Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Additional Contributions: Work in recruitment and assessment was performed at Yale University School of Medicine and the APT Foundation by James Poling, PhD; at McLean Hospital by Roger Weiss, MD; at the Medical University of South Carolina by Kathleen Brady, MD, PhD, and Raymond Anton, MD; and at the University of Pennsylvania by David Oslin, MD. Genotyping services for a part of our genome-wide association study were provided by the Center for Inherited Disease Research and Yale University (Center for Genome Analysis), which is fully funded by Federal contract N01-HG-65403 from the NIH to The Johns Hopkins University. Ann Marie Lacobelle, BS, Catherine Aldi, BS, and Christa Robinson, BS, provided technical assistance. The Semi-structured Assessment for Drug Dependence and Alcoholism interviewers, led by Yari Nuñez and Michelle Slivinsky, devoted substantial time and effort to phenotype the study sample. John Farrell, PhD, Section of Biomedical Genetics, Boston University School of Medicine, provided database management assistance. None of these individuals were compensated for this contribution to this study. Assistance with phenotype harmonization, genotype cleaning, and general study coordination was provided by the Gene Environment Association Studies Coordinating Center (supported by grant U01 HG004446 from the NIH). Assistance with data cleaning was provided by the National Center for Biotechnology Information.
References
Full text links
Read article at publisher's site: https://doi.org/10.1001/jamapsychiatry.2016.0036
Read article for free, from open access legal sources, via Unpaywall:
https://jamanetwork.com/journals/jamapsychiatry/articlepdf/2504223/yoi160002.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Cannabis use disorder: from neurobiology to treatment.
J Clin Invest, 134(20):e172887, 15 Oct 2024
Cited by: 0 articles | PMID: 39403927 | PMCID: PMC11473150
Review Free full text in Europe PMC
A genome-wide investigation into the underlying genetic architecture of personality traits and overlap with psychopathology.
Nat Hum Behav, 12 Aug 2024
Cited by: 0 articles | PMID: 39134740
The genetic landscape of substance use disorders.
Mol Psychiatry, 29(11):3694-3705, 29 May 2024
Cited by: 0 articles | PMID: 38811691 | PMCID: PMC11541208
Review Free full text in Europe PMC
Application of polygenic scores to a deeply phenotyped sample enriched for substance use disorders reveals extensive pleiotropy with psychiatric and somatic traits.
Neuropsychopharmacology, 49(13):1958-1967, 23 Jul 2024
Cited by: 0 articles | PMID: 39043921 | PMCID: PMC11480112
Genetic and non-genetic predictors of risk for opioid dependence.
Psychol Med, 54(8):1779-1786, 06 Feb 2024
Cited by: 1 article | PMID: 38317430
Go to all (100) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
dbGaP - The database of Genotypes and Phenotypes
- (1 citation) dbGaP - phs000092
Diseases (4)
- (1 citation) OMIM - 606629
- (1 citation) OMIM - 612101
- (1 citation) OMIM - 142445
- (1 citation) OMIM - 608397
Ensembl Genome Browser
- (1 citation) Ensembl - ENSG00000176273
SNPs (Showing 29 of 29)
- (5 citations) dbSNP - rs143244591
- (4 citations) dbSNP - rs77378271
- (3 citations) dbSNP - rs74823926
- (3 citations) dbSNP - rs146091982
- (2 citations) dbSNP - rs200453611
- (2 citations) dbSNP - rs78068107
- (2 citations) dbSNP - rs73252553
- (2 citations) dbSNP - rs10969106
- (2 citations) dbSNP - rs114311699
- (2 citations) dbSNP - rs114383460
- (2 citations) dbSNP - rs186825689
- (2 citations) dbSNP - rs28595532
- (2 citations) dbSNP - rs143020225
- (2 citations) dbSNP - rs10066744
- (2 citations) dbSNP - rs150064803
- (2 citations) dbSNP - rs73443003
- (2 citations) dbSNP - rs12621150
- (2 citations) dbSNP - rs144605126
- (2 citations) dbSNP - rs193047854
- (2 citations) dbSNP - rs12218439
- (2 citations) dbSNP - rs200391037
- (2 citations) dbSNP - rs199783889
- (2 citations) dbSNP - rs74400468
- (2 citations) dbSNP - rs17665889
- (2 citations) dbSNP - rs115553536
- (2 citations) dbSNP - rs7586604
- (2 citations) dbSNP - rs12534830
- (1 citation) dbSNP - rs147170184
- (1 citation) dbSNP - rs142305709
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genetic Risk Variants Associated With Comorbid Alcohol Dependence and Major Depression.
JAMA Psychiatry, 74(12):1234-1241, 01 Dec 2017
Cited by: 47 articles | PMID: 29071344 | PMCID: PMC6331050
Association of OPRM1 Functional Coding Variant With Opioid Use Disorder: A Genome-Wide Association Study.
JAMA Psychiatry, 77(10):1072-1080, 01 Oct 2020
Cited by: 82 articles | PMID: 32492095 | PMCID: PMC7270886
Linkage analysis followed by association show NRG1 associated with cannabis dependence in African Americans.
Biol Psychiatry, 72(8):637-644, 19 Apr 2012
Cited by: 30 articles | PMID: 22520967 | PMCID: PMC3699339
S100A10 identified in a genome-wide gene × cannabis dependence interaction analysis of risky sexual behaviours.
J Psychiatry Neurosci, 42(4):252-261, 01 Jun 2017
Cited by: 6 articles | PMID: 28418321 | PMCID: PMC5487272
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1 TR001863
NCI NIH HHS (2)
Grant ID: P30 CA016359
Grant ID: P01 CA089392
NHGRI NIH HHS (4)
Grant ID: U01 HG004446
Grant ID: U01 HG004422
Grant ID: N01HG65403
Grant ID: U01 HG004438
NHLBI NIH HHS (3)
Grant ID: HHSN268200782096C
Grant ID: HHSN268201100011C
Grant ID: HHSN268201100011I
NIAAA NIH HHS (3)
Grant ID: U10 AA008401
Grant ID: R01 AA017535
Grant ID: R01 AA011330
NIDA NIH HHS (5)
Grant ID: RC2 DA028909
Grant ID: R01 DA012690
Grant ID: R01 DA012849
Grant ID: R01 DA013423
Grant ID: R01 DA018432





