Abstract
Importance
Plasma genotyping of cell-free DNA has the potential to allow for rapid noninvasive genotyping while avoiding the inherent shortcomings of tissue genotyping and repeat biopsies.Objective
To prospectively validate plasma droplet digital PCR (ddPCR) for the rapid detection of common epidermal growth factor receptor (EGFR) and KRAS mutations, as well as the EGFR T790M acquired resistance mutation.Design, setting, and participants
Patients with advanced nonsquamous non-small-cell lung cancer (NSCLC) who either (1) had a new diagnosis and were planned for initial therapy or (2) had developed acquired resistance to an EGFR kinase inhibitor and were planned for rebiopsy underwent initial blood sampling and immediate plasma ddPCR for EGFR exon 19 del, L858R, T790M, and/or KRAS G12X between July 3, 2014, and June 30, 2015, at a National Cancer Institute-designated comprehensive cancer center. All patients underwent biopsy for tissue genotyping, which was used as the reference standard for comparison; rebiopsy was required for patients with acquired resistance to EGFR kinase inhibitors. Test turnaround time (TAT) was measured in business days from blood sampling until test reporting.Main outcomes and measures
Plasma ddPCR assay sensitivity, specificity, and TAT.Results
Of 180 patients with advanced NSCLC (62% female; median [range] age, 62 [37-93] years), 120 cases were newly diagnosed; 60 had acquired resistance. Tumor genotype included 80 EGFR exon 19/L858R mutants, 35 EGFR T790M, and 25 KRAS G12X mutants. Median (range) TAT for plasma ddPCR was 3 (1-7) days. Tissue genotyping median (range) TAT was 12 (1-54) days for patients with newly diagnosed NSCLC and 27 (1-146) days for patients with acquired resistance. Plasma ddPCR exhibited a positive predictive value of 100% (95% CI, 91%-100%) for EGFR 19 del, 100% (95% CI, 85%-100%) for L858R, and 100% (95% CI, 79%-100%) for KRAS, but lower for T790M at 79% (95% CI, 62%-91%). The sensitivity of plasma ddPCR was 82% (95% CI, 69%-91%) for EGFR 19 del, 74% (95% CI, 55%-88%) for L858R, and 77% (95% CI, 60%-90%) for T790M, but lower for KRAS at 64% (95% CI, 43%-82%). Sensitivity for EGFR or KRAS was higher in patients with multiple metastatic sites and those with hepatic or bone metastases, specifically.Conclusions and relevance
Plasma ddPCR detected EGFR and KRAS mutations rapidly with the high specificity needed to select therapy and avoid repeat biopsies. This assay may also detect EGFR T790M missed by tissue genotyping due to tumor heterogeneity in resistant disease.Free full text

Prospective validation of rapid plasma genotyping as a sensitive and specific tool for guiding lung cancer care
Associated Data
Abstract
Importance
Plasma genotyping of cell-free DNA (cfDNA) has the potential to allow for rapid noninvasive genotyping while avoiding the inherent shortcomings of tissue genotyping and repeat biopsies.
Objective
To prospectively validate plasma droplet digital PCR (ddPCR) for the rapid detection of common EGFR and KRAS mutations as well as the EGFR T790M acquired resistance mutation.
Design
Eligible patients underwent an initial blood draw and immediate plasma ddPCR for EGFR exon 19 del, L858R, T790M and/or KRAS G12X between July 2014 and June 2015. All patients underwent biopsy for tissue genotyping which was used as the reference standard for comparison; rebiopsy was required for patients with acquired resistance to EGFR kinase inhibitors. Test turnaround time (TAT) was measured in business days from blood draw until test reporting.
Setting
National Cancer Institute (NCI) designated comprehensive cancer center.
Participants
Advanced non-squamous NSCLC patients that are either (i) newly diagnosed and planned for initial therapy or (ii) have developed acquired resistance to an EGFR kinase inhibitor and are planned for re-biopsy.
Main Outcome Measure
Plasma ddPCR assay sensitivity, specificity and TAT.
Results
180 patients were enrolled in the study (120 newly diagnosed, 60 with acquired resistance). Tumor genotype included 80 EGFR exon 19/L858R mutants, 35 EGFR T790M, 25 KRAS G12X mutants. Median TAT for plasma ddPCR was 3 days. Tissue genotyping median TAT was 12 days for newly diagnosed patients and 27 days for acquired resistance patients. Plasma ddPCR exhibited a PPV of 100% (95%CI 91-100%) for EGFR 19 del, 100% (95%CI 85-100%) L858R and 100% (95%CI 79-100%) for KRAS, but lower for T790M at 79% (95%CI 62-91%). Sensitivity of plasma ddPCR was 82% (95%CI 69-91%) for EGFR 19 del, 74% (95%CI 55-88%) for L858R and 77% (95%CI 60-90%) for T790M but lower for KRAS at 64% (95%CI 43-82%). Sensitivity for EGFR or KRAS was higher in patients with multiple metastatic sites (p=0.001) and those with hepatic (p=0.001) or bone metastases (p=0.004), specifically.
Conclusion
Plasma ddPCR detects EGFR and KRAS mutations rapidly with the high specificity needed to select therapy and avoid repeat biopsies. This assay may also detect EGFR T790M missed by tissue genotyping due to tumor heterogeneity in resistant disease. This is the first prospective study to demonstrate the utility of ddPCR-based plasma genotyping in advanced NSCLC.
INTRODUCTION
Plasma genotyping utilizes tumor-derived cell-free DNA (cfDNA) to allow for rapid noninvasive genotyping of tumors. This technology is currently poised to transition into a treatment decision-making tool in multiple cancer types. It is particularly relevant to the treatment of advanced non-small cell lung cancer (NSCLC) where therapy hinges upon rapid and accurate detection of targetable EGFR, ALK and ROS1 alterations.1-6 Plasma genotyping is capable of circumventing many limitations of standard tissue genotyping including slow turnaround time (TAT), limited tissue for testing and the potential for failed biopsies. It may be particularly useful in directing the rapid use of new targeted therapies for acquired resistance in advanced EGFR mutant NSCLC where the need for repeat biopsy and heterogeneity of resistance mechanisms has amplified the inherent limitations of traditional genotyping.7,8
The need to carefully validate the test characteristics of each of the myriad individual plasma genotyping assays before use in clinical decision-making is paramount. We have previously reported the development of a quantitative droplet digital polymerase chain reaction (ddPCR)-based assay for the detection of EGFR kinase mutations and KRAS codon 12 mutations in plasma.9 The detection of these mutations has the potential to guide treatment by either facilitating targeted therapy with an EGFR tyrosine kinase inhibitor (TKI) or ruling out the presence of other potentially targetable alteration in the case of KRAS.5 Alternative platforms including Cobas, peptide nucleic acid-mediated PCR, multiplexed next-generation sequencing (NGS), high performance liquid chromatography (HPLC) and Scorpion-ARMS have also been examined in retrospective analyses of patient samples.10-22 The test characteristics of these assays have been variable and may be attributable to differences in testing platforms as well as the retrospective nature of these studies, their smaller size and the timing of blood collection with respect to disease progression and therapy initiation. The absence of reliable prospective data on the use of specific plasma genotyping assays in advanced NSCLC has left key aspects of its utility largely undefined and slowed its uptake as a tool for clinical care in both newly diagnosed and EGFR acquired resistance patients.
We have conducted the first prospective study of the use of ddPCR-based plasma genotyping for the detection of EGFR and KRAS mutations (NCT02279004). This study was performed in the two settings where we anticipate clinical adoption of this assay: (i) newly diagnosed advanced NSCLC patients and (ii) those with acquired resistance to EGFR kinase inhibitors. The primary aim of this study was to prospectively evaluate the feasibility and accuracy of this assay for the detection of EGFR/KRAS mutations in newly diagnosed patients and EGFR T790M in acquired resistance patients in a clinical setting. Additional endpoints included test TAT and the effect of sample treatment conditions on test accuracy.
METHODS
Trial Design
Advanced NSCLC patients were prospectively enrolled onto an institutional review board (IRB)-approved plasma genotyping study protocol (Dana-Farber/Harvard Cancer Center protocol #14-147, NCT02279004). Patients were eligible for the study if they had biopsy-proven advanced or recurrent non-squamous NSCLC and were either treatment naive (cohort 1) or had acquired resistance to an EGFR TKI (cohort 2). All patients must have been planned to begin new systemic therapy and have either tissue available for standard genotyping or a planned repeat biopsy. All patients had radiographic evidence of disease, were 18 years of age or older and signed written informed consent before any study-related procedure. Participant-defined race was recorded given known associations between race and the frequency of EGFR mutant tumors.
All patients underwent an initial paired blood collection after study enrollment. These two tubes of blood subsequently underwent plasma isolation, cfDNA extraction and ddPCR-based genotyping. One tube of blood was processed and analyzed immediately as per standard operating procedures and the second subjected to pre-planned variations in specimen handling designed to simulate real-world testing conditions including (i) standard EDTA tube shipped overnight on ice and (ii) Streck tube shipped at room temperature. Comparison between paired samples was made on the basis of sample quality, total DNA as determined by PicoGreen assay and quantitative ddPCR result. Differences between paired tubes were analyzed by paired T-test. If more than two weeks elapsed before initiating planned systemic therapy, the blood draw was repeated. TAT for plasma genotyping was measured in business days from the date of blood draw until reporting of results to the study investigator.
Patient samples from cohort 1 underwent ddPCR-based plasma genotyping for EGFR exon 19 del, L858R and KRAS codon 12 mutations. Cohort 2 samples underwent testing for EGFR exon 19 del, L858R and T790M. Plasma genotyping results were compared to tissue genotyping results from initial biopsy (cohort 1) or re-biopsy at acquired resistance (cohort 2) as the reference standard.
Patients from both cohorts that had a mutation detected by ddPCR-based genotyping subsequently underwent two follow-up blood draws at 1-2 weeks and 4-6 weeks after beginning systemic therapy. These samples underwent serial quantitative genotyping by ddPCR for the detected mutation.
Plasma Genotyping
Samples from venous blood draws were collected in EDTA tubes and underwent centrifugation within 1 hour of sample collection and plasma preparation as previously described.9 Immediate extraction of cfDNA was then performed using the QIAmp circulating nucleic acid kit (QIagen) according to the manufacturer’s protocol. DNA was eluted in 100 uL of AVE buffer and stored at −80C until genotyping was performed. Genotyping of cfDNA was performed by ddPCR (BioRad) and primer/probes were custom-ordered from Life Technologies. The development of this assay has been previously described.9 Briefly, cfDNA is emulsified into approximately 20 000 droplets, mixed with appropriate primer/probe mixes and then undergoes PCR to endpoint. Droplets are then read in a flow cytometer and fluorescence signal quantified in order to determine the number of copies of mutant allele per mL (eMethods). To simulate standard clinical practice, the assay was performed twice weekly (Monday and Thursday). Laboratory personnel performing plasma ddPCR were blinded to tissue genotyping results.
Tissue Genotyping
Clinical tumor genotyping was performed for all patients on initial biopsy material (cohort 1) or re-biopsy material following development of acquired resistance (cohort 2). Turnaround time for tissue genotyping was measured from the date of the initial genotyping order until the reporting of the final genotyping result. In cases where a repeat biopsy was required to successfully complete tissue genotyping, the time required to perform the repeat biopsy was included in the TAT measurement.
Statistical Analysis
From a total of 120 NSCLC patients studied in cohort 1, we estimated that 24 and 30 would have EGFR and KRAS mutations, respectively, based upon prior data at our institution. Concordance between tumor and plasma genotyping results had at least 80% power to detect a kappa statistic of 0.85 (compared to a null of 0.6) while controlling for a one-sided type 1 error rate of 0.15.
For the 60 patients with acquired resistance that were planned for cohort 2, we estimated that half would harbor T790M detected in their resistance biopsy. An expanded target of 80 patients was originally planned but was revised to 60 patients due to feasibility concerns. Concordance between tumor and plasma genotyping results for T790M had 88% power to detect a kappa coefficient of 0.85 (compared to a null of 0.6) while controlling for a one-sided type 1 error rate of 0.05. Categorical variables were compared using Fisher’s exact test, and continuous variables were compared using the Wilcoxon rank sum test or Kruskal-Wallis test. No adjustments have been made for multiple comparisons.
RESULTS
Patients
A total of 180 patients with advanced NSCLC were enrolled in the study with either newly diagnosed disease (n=120) or acquired resistance to an EGFR TKI (n=60). The majority of patients had adenocarcinoma histology (94%) and only a minority had either NSCLC NOS (3%) or adenosquamous histology (3%) (Table 1). Patients were predominantly female (62%) and primarily Caucasian (83%) and Asian (10%). Patients who did not complete their initial blood draw or any tissue genotyping were excluded from analysis (eFigure 1). An additional 28 patients did not have sufficient tissue available for KRAS G12X testing after completion of initial EGFR testing and were excluded from the KRAS G12X analysis.
Table 1
Patient Characteristics
| Cohort 1 | Cohort 2 | ||||||
|---|---|---|---|---|---|---|---|
| Total | Newly Diagnosed | Acquired Resistance | |||||
| Patients | 180* | 120 | 60 | ||||
| Sex | Male | 68 | 38% | 50 | 42% | 18 | 30% |
| Female | 112 | 62% | 70 | 58% | 42 | 70% | |
|
| |||||||
| Median Age | 62 | 64 | 58 | ||||
|
| |||||||
| Race | White | 152 | 84% | 109 | 91% | 43 | 72% |
| Asian | 20 | 11% | 7 | 6% | 13 | 22% | |
| Black | 5 | 3% | 2 | 2% | 3 | 5% | |
| Hispanic | 3 | 2% | 2 | 2% | 1 | 1% | |
|
| |||||||
|
Stage at
diagnosis | Recurrent | 5 | 3% | 5 | 4% | 0 | - |
| IIIB | 3 | 2% | 3 | 3% | 0 | - | |
| IV | 172 | 95% | 112 | 93% | 60 | 100% | |
|
| |||||||
|
Number of
Metastatic Sites | 1 | 72 | 40% | 55 | 46% | 17 | 28% |
| 2 | 62 | 34% | 41 | 34% | 21 | 35% | |
| 3 | 23 | 13% | 15 | 12% | 8 | 13% | |
| ≥4 | 23 | 13% | 9 | 8% | 14 | 23% | |
|
| |||||||
|
Site of
biopsy utilized for genotyping | Lung | 47 | 39% | 29 | 53% | ||
| Pleural biopsy, fluid | 12 | 10% | 3 | 5% | |||
| Liver | 7 | 6% | 6 | 10% | |||
| Lymph node | 28 | 23% | 8 | 14% | |||
| Other | 26 | 22% | 10 | 18% | |||
|
| |||||||
|
Tissue
Genotype | EGFR exon 19 del | 50 | 28% | 14 | 12% | 37 | 62% |
| EGFR L858R | 32 | 18% | 13 | 11% | 18 | 30% | |
| Rare EGFR | 5 | 3% | 0 | 0% | 5 | 8% | |
| EGFR T790M | - | - | - | - | 35 | 58%† | |
| KRAS G12X | 26 | 14% | 26 | 22% | - | - | |
| EGFR/KRAS WT | 64 | 36% | 64 | 53% | - | - | |
| Failed | 3 | 1% | 3 | 3% | 5 | 8%‡ | |
|
| |||||||
|
Tissue
genotyping method | Sanger sequencing | 6 | 5% | 0 | 0% | ||
| PCR | 29 | 25% | 43 | 78% | |||
| Targeted NGS | 12 | 10% | 12 | 22% | |||
| PCR and NGS | 70 | 60% | 0 | 0% | |||
|
| |||||||
|
Additional
Biopsy Required | 34 | 19% | 22 | 19% | 12 | 21% | |
|
| |||||||
| Histology | AdenoCa | 169 | 94% | 112 | 93% | 57 | 95% |
| Adenosquamous | 6 | 3% | 3 | 3% | 3 | 5% | |
| NSCLC NOS | 5 | 3% | 5 | 4% | 0 | - | |
The confirmed tissue genotypes of the 115 eligible newly diagnosed patients included 14 EGFR exon 19 del, 13 EGFR L858R, 26 KRAS G12X and 62 EGFR/KRAS wild-type (Table 1The 54 eligible patients with acquired resistance possessed a range of EGFR sensitizing mutations (37 EGFR exon 19 del, 18 EGFR L858R, 5 rare) and 35 (58%) of these patients were EGFR T790M positive according to tissue genotyping performed on re-biopsy specimens.
Turnaround Time & Repeat Biopsy
Plasma genotyping was completed successfully in all patients. The median turnaround time from blood collection to report delivery was 3 business days in newly diagnosed patients and 2 business days in acquired resistance patients (range 1-7 days). In comparison, the median turnaround time for tissue genotyping in newly diagnosed patients was significantly longer at 12 business days (range 1-54 days, p<0.001). The median turnaround time for tissue genotyping was similarly longer in patients with acquired resistance to EGFR kinase inhibitors at 27 business days (range 1-146 days). A repeat biopsy was required in 22 (19%) newly diagnosed patients in order to obtain sufficient tissue to complete genotyping. Similarly, 12 (21%) acquired resistance patients required multiple repeat biopsies in order to obtain sufficient tissue for EGFR T790M genotyping. Turnaround time measurements included the time required to obtain an additional biopsy if necessary due to failure of one or more biopsy attempts.
Assay Characteristics
The accuracy of the EGFR exon 19 del, L858R and KRAS G12 X assays were studied first in newly diagnosed patients (n=115) (eFigure 1), Plasma ddPCR exhibited high specificity for the detection of EGFR exon 19 del (100%, 101/101), L858R (100%, 102/102) and KRAS G12X (100%, 62/62). Positive predictive value was similarly high for all assays at 100% (Table 2). Assay sensitivity was more modest for EGFR exon 19 del (86%, 12/14), L858R (69%, 9/13) and KRAS G12X (64%, 16/25) (Table 2). Concordance was 0.91 (p=0.01) for EGFR exon 19 del, 0.8 (p=0.08) for L858R and 0.72 (p=0.13) for KRAS G12X. Assay sensitivity among newly diagnosed and acquired resistance patients was similar for the detection of EGFR exon 19 del (82%, 41/50) and L858R (74%, 23/31) (Table 2). A single false positive result was initially reported for EGFR exon 19 del testing (132 copies/mL) which occurred in a young, never smoker with a scant tumor biopsy that was negative for any EGFR mutations. Repeat biopsy was then performed and subsequent tumor genotyping confirmed an EGFR exon 19 del mutation.
Table 2
Plasma ddPCR assay sensitivity, specificity and positive predictive value.
| Sensitivity analysis | Specificity analysis | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Assay | Sensitivity | True positive (tissue + plasma +) | False negative (tissue + plasma −) | Specificity | True negative (tissue − plasma −) | False positive (tissue− plasma +) |
Positive
predictive value | |
|
| ||||||||
|
EGFR
exon 19 del | Newly diagnosed | 86% (57-98%) | 12 | 2 | 100% (96-100%) | 101 | 0 | 100% (74-100%) |
| Acquired resistance | 81% (64-92%) | 29 | 7 | 100% (85-100%) | 23 | 0 | 100% (88-100%) | |
| Overall | 82% (69-91%) | 41 | 9 | 100% (97-100%) | 124 | 0 | 100% (91-100%) | |
|
| ||||||||
|
EGFR
L858R | Newly diagnosed | 69% (39-91%) | 9 | 4 | 100% (96-100%) | 102 | 0 | 100% (66-100%) |
| Acquired resistance | 78% (52-94%) | 14 | 4 | 100% (91-100%) | 41 | 0 | 100% (77-100%) | |
| Overall | 74% (55-88%) | 23 | 8 | 100% (97-100%) | 143 | 0 | 100% (85-100%) | |
|
| ||||||||
|
EGFR
T790M | Acquired resistance | 77% (60-90%) | 27 | 8 | 63% (38-84%) | 12 | 7 | 79% (62-91%) |
|
| ||||||||
| KRAS G12X | Newly diagnosed | 64% (43-82%) | 16 | 9 | 100% (94-100%) | 62 | 0 | 100% (79-100%) |
|
| ||||||||
Note: 95% exact binomial confidence intervals noted below each value.
Accuracy of the EGFR T790M assay was studied in patients with acquired resistance to EGFR TKI. The detection of this resistance mutation by plasma ddPCR exhibited a lower specificity (63%, 12/19) and positive predictive value (79%, 27/34) than was seen for EGFR sensitizing mutations when compared to tumor genotyping of the resistance biopsy, thus, concordance was also lower for the detection of EGFR T790M (kappa statistic 0.4, p=0.097). Sensitivity of this assay was similar to that observed for EGFR sensitizing mutations (77%, 27/35) (Table 2). The test characteristics for the detection of EGFR sensitizing mutations were similar in acquired resistance patients compared to newly diagnosed patients (Table 2).
Predictors of Test Sensitivity and Dynamic Range
Patient and disease characteristics potentially associated with increased test sensitivity were examined using a composite test sensitivity variable combining both EGFR and KRAS assay sensitivity results. Of the variables listed in Table 1, a significant association was demonstrated between test sensitivity and the presence of hepatic metastases (p=0.001), bone metastases (p=0.007), increasing number of metastatic sites (p=0.001) (Figure 1).
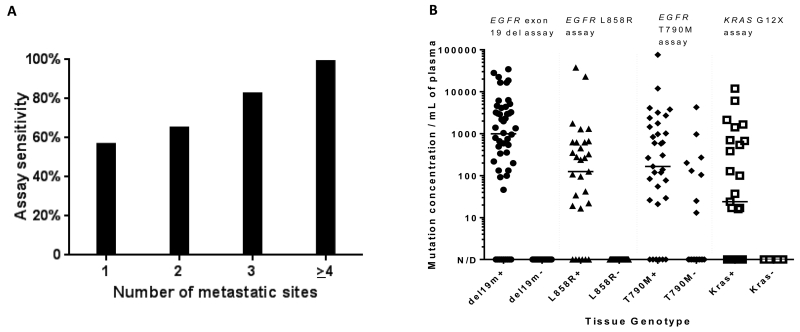
(A) The sensitivity of plasma ddPCR for the detection of EGFR and KRAS mutations increases directly with the number of metastatic sites present in a given patient (p<0.001). (B) Dynamic range of plasma genotyping using a validated ddPCR-based assay. Wide dynamic range and the absence of false positives are noted for the detection of KRAS G12X and EGFR sensitizing mutations. A small number of false positives are seen with the EGFR T790M assay – potentially secondary to tumor heterogeneity with respect to acquired resistance mechanisms (n=174).
The relationship between detected mutant EGFR or KRAS cfDNA copy number and clinical characteristics was next examined as a marker of tumor cfDNA shed. Given the wide dynamic range noted with this assay (Figure 1), significant associations between clinical characteristics and log10-transformed mutant cfDNA copy number in patients with detected mutant cfDNA were sought. Only increasing number of metastatic sites was associated with a higher mutant cfDNA copy number (p=0.03).
Paired Analysis
Multiple real-world sample treatment conditions were tested using paired samples drawn from the same patient at the same point in time. The use of an EDTA tube that was shipped on ice overnight before processing revealed identical qualitative assay results and not significantly different total DNA (p=0.38) and mutant allele copy number (p=0.26) compared to immediate processing (n=25 patients). Similarly, use of a Streck DNA-preservation tube shipped at room temperature overnight yielded identical qualitative assay results and there was no significant difference in total DNA (p=0.25) or mutant allele copy number (p=0.32) compared to standard processing (n=20 patients) (eFigure 2).
Exploratory patterns of mutant cfDNA changes in response to systemic therapy
Patients with a detectable mutation by plasma ddPCR underwent serial blood draws on treatment. A total of 50 patients completed at least one follow-up blood draw on treatment. Serial quantitative plasma ddPCR among these patients revealed clear changes in the level of detectable mutant allele frequency during treatment (Figure 2). Changes in detectable mutation by plasma ddPCR fell into several recurrent, descriptive patterns including complete resolution of detectable mutant cfDNA either at initial repeat blood draw (Figure 2A) or subsequently (Figure 2B), residual detectable mutant cfDNA following initial decrease (Figure 2C), initial decrease followed by increase (Figure 2D) or initial increase that was either transient (Figure 2E) or maintained (Figure 2F). Patients with complete resolution of mutant cfDNA at either 2 or 6 weeks exhibited a treatment discontinuation rate of 0% (0/23) at initial and 4% (1/23) second re- imaging assessment. Patients without complete resolution exhibited a treatment discontinuation rate of 33% (9/27) at initial re-imaging and 56% (15/27) at second re-imaging assessment. Treatment discontinuation decisions were made by treating clinicians who were blinded to serial plasma genotyping results. Objective data on overall survival and progression-free survival are presently immature. These patterns of change in plasma response are exploratory at present, but provide a potential framework for future analysis of the correlation between changes in detectable mutant cfDNA and response to therapy.

Distinct patterns of ddPCR plasma response emerge in patients undergoing serial plasma genotyping at 2 weeks and 6 weeks post-treatment. Mutant cfDNA was observed to either become undetectable at 2 weeks (A), decrease and then become undetectable at 6 weeks (B), decrease progressively but remain detectable at 6 weeks (C), increase initially and then decrease at 6 weeks (D), decrease at 2 weeks and then rebound at 6 weeks (E), or progressively increase (F). Patients with complete resolution of mutant cfDNA (A-B) exhibited a treatment discontinuation rate of 0% (0/23) and 4% (1/23) at initial and second restaging CT scans. Alternatively, patients without complete resolution (C-F) had a treatment discontinuation rate of 33% (9/27) at initial re-imaging and 56% (15/27) at second re-imaging assessment. Patient genotypes included EGFR sensitizing alone (--), EGFR sensitizing in the presence of T790M (--) and KRAS G12X (--).
DISCUSSION
In this prospective study, we demonstrate the highly specific and rapid nature of plasma genotyping. No false positives were seen for driver mutations in EGFR or KRAS, and turnaround time from specimen draw to result was a matter of days. This assay exhibited 100% positive predictive value for the detection of these mutations. Sensitivity was more modest and was directly correlated with both number of metastatic sites and the presence of liver or bone metastases. This newly demonstrated relationship is likely related to increased cfDNA shed in the setting of more extensive disease where tumor cfDNA shed is the chief driver of assay sensitivity and determines its upper limit. The characteristics of plasma ddPCR prospectively demonstrated in this study are similar or improved compared to previous retrospective reports of other cfDNA genotyping assays.10-13,15,16,23,24 These retrospective studies are smaller, frequently examine a mix of tumor types/stages and lack the careful prospective design needed to demonstrate the readiness of this technology to transition to a tool for selecting therapy. Studies that utilize retrospective samples from clinical trials that enrolled only EGFR mutant patients are further limited by an inability to both blind laboratory investigators to tissue genotype and to generalize their assay test characteristics to a genetically heterogeneous real-world patient population.11 These differences and the multiple platforms examined previously have led to variable test characteristics and uncertainty regarding the clinical application of these technologies. This study is the first to prospectively demonstrate the ability of a ddPCR-based plasma genotyping assay to rapidly and accurately detect EGFR and KRAS mutations in a real-world clinical setting with the rigor necessary to support that this assay is capable of directing clinical care.
Even with a diagnostic sensitivity of less than 100%, such a rapid assay with 100% positive predictive value carries the potential for immense clinical utility. The 2-3 day TAT contrasts starkly with the 27 day TAT for tumor genotyping seen in patients needing a new tumor biopsy. This long turnaround time is due largely to the practical reality that many newly diagnosed patients require a repeat biopsy to obtain tissue for genotyping as do all acquired resistance patients. Consider the case of one subject that participated in this study, an octgenarian patient with metastatic NSCLC who had developed acquired resistance to erlotinib with painful bone metastases (Figure 3). Due to the patient’s age and comorbidities, significant concerns existed about the risks of a biopsy and further systemic therapy. Plasma was drawn on study and within 24 hours ddPCR demonstrated 806 copies/mL of EGFR T790M. A confirmatory lung biopsy was performed which confirmed EGFR T790M. A third-generation EGFR kinase inhibitor, osimertinib, was subsequently initiated and the patient had a partial response to therapy which was maintained for over 1 year. The potential of this technology to avoid repeat biopsy in both newly diagnosed patients with insufficient tissue as well as patients with acquired resistance is considerable.
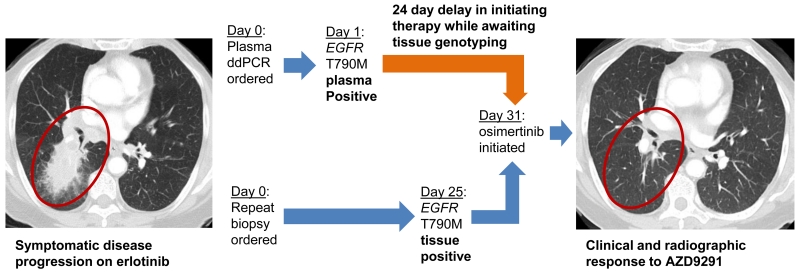
Case Report: A patient in their eighties with metastatic EGFR mutant NSCLC developed acquired resistance to erlotinib. Symptomatic progression of pulmonary and bone metastases were noted (primary lung lesion labelled). Empiric single-agent chemotherapy or best supportive care alone were considered given the patient’s age and comorbidities. However, plasma ddPCR was performed and the result returned the next day revealing 806 copies/mL of EGFR T790M. The patient underwent rebiopsy which confirmed EGFR T790M and the patient was able to start therapy with osimertinib – a novel third generation EGFR kinase inhibitor with excellent clinical and radiographic response. Importantly, the plasma ddPCR T790M result was returned 24 days before the results of the repeat tissue biopsy were available.
A key limitation of plasma ddPCR is that while this method is extremely adept at rapidly detecting specific targetable mutations, it cannot easily detect copy number alterations and rearrangements. The ddPCR panel assessed in this study thus cannot currently detect targetable alterations in either ALK or ROS1. This limitation may potentially be addressed by utilizing targeted NGS of cfDNA for broad, multiplexed detection of complex genomic including ALK and ROS1 rearrangements although this method is potentially slower than ddPCR-based methods and has been less thoroughly evaluated.25 The potential exists to utilize these technologies in tandem in advanced NSCLC to facilitate rapid initiation of therapy. Tissue genotyping and repeat biopsy would be specifically used to direct therapy in cases where plasma genotyping was uninformative due to limitations of assay sensitivity. This approach would be particularly useful in cases of EGFR acquired resistance where a repeat biopsy for T790M testing could be avoided entirely in many patients. Beyond detecting targetable alterations in order to drive therapy, and the identification of non-targetable oncogenic drivers such as KRAS mutations that preclude the presence of other targetable alterations may guide a clinician to rapidly initiate alternative therapies such as chemotherapy or immunotherapy.5 The finding that assay sensitivity is highest in patients with more extensive metastatic disease suggests that those patients most in need of rapid treatment initiation would also be least likely to have false negative results.
One surprising result of our study was evidence of recurrent false positive results for EGFR T790M in patients with acquired resistance, despite no false positives for other mutations studied. The sensitivity of the EGFR T790M assay was comparable to that of the EGFR sensitizing mutation assays and similarly related to both disease burden and the presence of liver or bone metastases which are likely predictive of increased tumor cfDNA shed. We hypothesize that the lower assay specificity is due to the genomic heterogeneity whereby the T790M status of the biopsied site is not representative of all metastatic sites in a patient, a phenomenon supported by mounting evidence in the acquired resistance setting.26,27 This is consistent with the finding that a minority of apparently EGFR T790M tissue negative patients respond to therapy with third generation EGFR kinase inhibitors.7,8,28 These observations raise questions regarding the fallibility of tissue based genotyping as the reference standard for T790M status. The use of plasma genotyping to detect EGFR T790M thus has great potential to identify patients that would benefit from newly approved third generation EGFR kinase inhibitors but would be unable to access them based on falsely negative tissue genotyping results. Indeed, plasma genotyping may allow more reliable assessment of both T790M status as well as the biology of resistance across all sites of a heterogeneous cancer as opposed to a tissue biopsy and is likely to be an essential tool for future trials targeting drug resistance. The potential to avoid repeat biopsy entirely in patients where plasma ddPCR detects T790M further strengthens the utility of this technology, although repeat biopsy would still be required in patients with uninformative plasma ddPCR due to limitations with respect to assay sensitivity.
This study also examined the potential of the quantitative nature of ddPCR-based plasma genotyping to allow for the early prediction of treatment response. Distinct patterns of change in mutant allele copy number were observed as early as 2 weeks after treatment and were similar to those reported in other tumor types.19,20 We hypothesize that these distinct patterns of change in this study will correlate with specific patterns of radiographic response and emergence of acquired resistance and plan to report these data once mature. The observed differences in treatment discontinuation rates observed in this study comparing patients with complete resolution of detectable mutant cfDNA compared to those with incomplete resolution support this hypothesis. The use of this technology to monitor disease status in real-time has potential utility for both routine clinical care as well as use as an integrated biomarker in early-phase clinical trials.10
In conclusion, ddPCR-based plasma genotyping is a technology that is ready to be employed for clinical decision-making in advanced NSCLC patients. This assay is capable of rapidly detecting EGFR and KRAS mutations with minimal false positives and with the robustness needed for real-world testing. It has great utility for the detection of actionable genomic alterations in patients who are unable to undergo repeat biopsies and may even detect mutations missed by standard tissue genotyping due to tissue heterogeneity. As third generation EGFR T790M inhibitors come into clinical use, the need for re-biopsy and potential role of plasma genotyping will expand dramatically. Further, the potential combination of rapid ddPCR-based plasma genotyping assays with plasma NGS assays for more comprehensive noninvasive genotyping may represent a new paradigm for clinical genotyping.
Acknowledgments
Supported in part by the United States Department of Defense, the National Cancer Institute of the National Institutes of Health (grants R01CA135257, R01CA114465 and P50CA090578), the Phi Beta Psi Sorority, the Stading-Younger Cancer Foundation, the International Association for the Study of Lung Cancer, the Canadian Institutes of Health Research, the Canadian Association of Medical Oncologists, the Gallup Research Fund and the Kaplan Research Fund. These funding organizations were not directly involved in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. AGS, SD, CPP and GRO had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. AGS, SD, CP and GRO were responsible for the data analysis contained in this study.
GRO is a consultant/advisory board member for Ariad, AstraZeneca, Boehringer Ingelheim, Clovis Oncology, and Sysmex; and has received honoraria from AstraZeneca, Boehringer Ingelheim and Chugai. PAJ is a consultant for Boehringer Ingelheim, AstraZeneca, Genentech, Pfizer, Merrimack Pharmaceuticals, Clovis Oncology, Roche, Sanofi and Chugai; and has stock ownership in Gatekeeper Pharmaceuticals. AGS has received travel funding from AstraZeneca and Genentech-Roche. CPP has received travel funding and honoraria from BioRAD Laboratories and Clovis Oncology. GRO, CPP, and PAJ are inventors on a pending patent related to findings described in this manuscript.
Footnotes
SD, RA, AM, NF and SM report no conflicts of interest.
References
Full text links
Read article at publisher's site: https://doi.org/10.1001/jamaoncol.2016.0173
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4982795?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1001/jamaoncol.2016.0173
Article citations
Detection of EGFR mutations in patients with suspected lung cancer using paired tissue-plasma testing: a prospective comparative study with plasma ddPCR assay.
Sci Rep, 14(1):25701, 28 Oct 2024
Cited by: 0 articles | PMID: 39465302 | PMCID: PMC11514293
Bibliometric analysis of bone metastases from lung cancer research from 2004 to 2023.
Front Oncol, 14:1439209, 06 Aug 2024
Cited by: 0 articles | PMID: 39165682 | PMCID: PMC11333251
Review Free full text in Europe PMC
Clinical application of ctDNA in early diagnosis, treatment and prognosis of patients with non-small cell lung cancer.
Future Oncol, 20(29):2213-2224, 29 Jul 2024
Cited by: 1 article | PMID: 39073412 | PMCID: PMC11514542
Review Free full text in Europe PMC
Patient-centric thresholding of Cobas® EGFR mutation Test v2 for surveillance of EGFR-mutated metastatic non-small cell lung cancer.
Sci Rep, 14(1):18191, 06 Aug 2024
Cited by: 1 article | PMID: 39107402 | PMCID: PMC11303541
Optimising primary molecular profiling in non-small cell lung cancer.
PLoS One, 19(7):e0290939, 31 Jul 2024
Cited by: 0 articles | PMID: 39083479 | PMCID: PMC11290658
Go to all (322) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Picoliter-Droplet Digital Polymerase Chain Reaction-Based Analysis of Cell-Free Plasma DNA to Assess EGFR Mutations in Lung Adenocarcinoma That Confer Resistance to Tyrosine-Kinase Inhibitors.
Oncologist, 21(2):156-164, 14 Jan 2016
Cited by: 36 articles | PMID: 26768482 | PMCID: PMC4746084
Detection of the T790M mutation of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study).
Oncotarget, 7(36):58492-58499, 01 Sep 2016
Cited by: 42 articles | PMID: 27542267 | PMCID: PMC5295446
A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance.
Cancer Med, 6(1):154-162, 20 Dec 2016
Cited by: 59 articles | PMID: 28000387 | PMCID: PMC5269560
EGFR mutations in lung cancer: from tissue testing to liquid biopsy.
Future Oncol, 11(11):1611-1623, 01 Jan 2015
Cited by: 57 articles | PMID: 26043215
Review
Funding
Funders who supported this work.
NCI NIH HHS (4)
Grant ID: R01 CA135257
Grant ID: P50 CA090578
Grant ID: R01 CA114465
Grant ID: U01 CA210171





