Abstract
Free full text

Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease
Abstract
Increasing evidence suggests a role of intestinal dysbiosis in obesity and non-alcoholic fatty liver disease (NAFLD). But it remains unknown in nonobese NAFLD. This prospective, cross-sectional study sought to characterize differences in fecal microbiota between nonobese adult individuals with and without NAFLD and their potential association with metabolic markers of disease progression. A total of 126 nonobese subjects were enrolled: 43 NAFLD and 83 healthy controls (HC). The microbial community was profiled by denaturing gradient gel electrophoresis and examined by 454 pyrosequencing of the 16S ribosomal RNA V3 region. Lower diversity and a phylum-level change in the fecal microbiome were found in NAFLD. Compared with HC, patients had 20% more phylum Bacteroidetes (p =
= 0.005) and 24% less Firmicutes (p
0.005) and 24% less Firmicutes (p =
= 0.002). Within Firmicutes, four families and their 8 genera, which were short-chain fatty acids-producing and 7α-dehydroxylating bacteria, were significantly decreased. Moreover, Gram-negative (G−) bacteria were prevalent in NAFLD (p
0.002). Within Firmicutes, four families and their 8 genera, which were short-chain fatty acids-producing and 7α-dehydroxylating bacteria, were significantly decreased. Moreover, Gram-negative (G−) bacteria were prevalent in NAFLD (p =
= 0.008). Furthermore, a significant correlation with metabolic markers was revealed for disturbed microbiota in NAFLD. This novel study indicated that intestinal dysbiosis was associated with nonobese NAFLD and might increase the risk of NAFLD progression.
0.008). Furthermore, a significant correlation with metabolic markers was revealed for disturbed microbiota in NAFLD. This novel study indicated that intestinal dysbiosis was associated with nonobese NAFLD and might increase the risk of NAFLD progression.
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver condition in Western countries1 and some Asian countries2. NAFLD, ranging from simple non-alcoholic fatty liver to non-alcoholic steatohepatitis (NASH)3, is considered to be the hepatic manifestation of metabolic syndrome. Without an effective available treatment, the prognosis of NASH is poor because of the risk of progressive liver diseases such as cirrhosis and hepatocellular carcinoma. NAFLD is characterized by a broad spectrum of manifestations ranging from simple steatosis, NASH to liver cirrhosis and hepatocellular carcinoma4. Without an effective available treatment, the prognosis of NASH is poor. The human intestinal microbiota (IM) plays an important role in human health and diseases5,6,7 such as obesity8,9, diabetes mellitus10, liver cirrhosis11 and cancer12. Due to its close link with obesity, the pathogenesis of NAFLD has also been proposed to be the result of multiple ‘hits’, including the widely accepted contribution of the altered gut microbiota associated with obesity13,14.
Alterations of microbial communities, particularly a shift associated with obesity, correlates closely with the prevalence and progress of NAFLD15,16,17,18. For example, Zhu et al. has suggested a connection between gut-derived endogenous alcohol and NASH in the pathogenesis of obese children18. Raman et al. have revealed that a significant compositional shift in the IM (such as the decrease of some selected members of Firmicutes) is associated with NAFLD17. However, subsequent studies in adults have shown inconsistent and even contradictory results. For instance, Mouzaki M et al. have found a decrease in Bacteroidetes in obese patients with NASH compared with healthy controls (HCs) but no difference between patients with simple steatosis and HCs16. In these previous studies, all patients had a high body mass index (BMI) (>26), and the BMI and age of the NAFLD patients were significantly higher than those of HCs16,17. BMI could be a major confounder, because BMI has been demonstrated to be a major determinant of compositional changes to microbial communities8. To our knowledge, no studies have directly assessed the relationship between IM composition and nonobese adult patients with NAFLD.
Importantly, NAFLD is not rare in nonobese adults19,20,21,22,23. A recent study in Hong Kong showed that 20% of the nonobese population are NAFLD23. Another study in mainland China showed that the prevalence of NAFLD is 7.27% at baseline, and among these, 8.88% of nonobese subjects developed NAFLD during a 5-year follow-up22. In an adult population in Taiwan, the prevalence of NAFLD has been reported to be 11.5% in nonobese adults19. The study in Korea has reported a prevalence of NAFLD of 23.4% among 768 non-obese, nondiabetic individuals older than 30 years21. A study conducted in India has reported that 75% of the NAFLD patients studied had a BMI <25 Kg/m2, and 54% were neither overweight nor abdominal obesity20. Therefore, it is critically important to understand whether abnormal IM is associated with non-obese patients with NAFLD to better understand and prevent NAFLD.
Kg/m2, and 54% were neither overweight nor abdominal obesity20. Therefore, it is critically important to understand whether abnormal IM is associated with non-obese patients with NAFLD to better understand and prevent NAFLD.
Here, we used metagenomic approaches, including a bacterial community fingerprinting method of denaturing gradient gel electrophoresis (DGGE) and high-throughput 454 pyrosequencing targeting the 16S rRNA gene, to characterize the fecal microbial communities in nonobese adult individuals with NAFLD. Furthermore, to determine which of the thousands of microbial species are crucial for human health and to understand the potential molecular host-microbiome interactions, correlation analysis between the fecal microbiomic profiles and the clinical biochemical markers of NAFLD progression22 was also performed. Our findings provide a more comprehensive understanding of fecal microbiota in nonobese individuals with NAFLD and facilitate future preventive attempts to manipulate the commensal microbiota in nonobese patients with NAFLD.
Results
Clinical data of individuals in the nonobese cohort
Altogether, 126 nonobese subjects with (n =
= 43) or without (n
43) or without (n =
= 83) NAFLD were enrolled in this prospective cross-sectional study. We performed anthropometric and biochemical phenotyping of the individuals (Table 1). Differences in clinical biochemical metabolic markers were compared between individuals with and without NAFLD. These markers are used to provide important information about liver function and to identify potential NAFLD patients. The liver synthetic function of all the individuals was in the normal range, as determined by albumin and International Normalized Ratio levels (data not shown). The age and gender distributions between the individuals with and without NAFLD were similar. All subjects consumed an omnivorous Chinese diet. Although all enrolled subjects were nonobese, the individuals with NAFLD had higher BMI, waist circumference, hip circumference and waist-to-hip ratio values (p
83) NAFLD were enrolled in this prospective cross-sectional study. We performed anthropometric and biochemical phenotyping of the individuals (Table 1). Differences in clinical biochemical metabolic markers were compared between individuals with and without NAFLD. These markers are used to provide important information about liver function and to identify potential NAFLD patients. The liver synthetic function of all the individuals was in the normal range, as determined by albumin and International Normalized Ratio levels (data not shown). The age and gender distributions between the individuals with and without NAFLD were similar. All subjects consumed an omnivorous Chinese diet. Although all enrolled subjects were nonobese, the individuals with NAFLD had higher BMI, waist circumference, hip circumference and waist-to-hip ratio values (p =
= 0.004, 0.0001, 0.006, and 0001, respectively). In consistent with reports by Xu et al. of nonobese NAFLD22, the individuals with NAFLD were characterized by higher levels of alanine aminotransferase (ALT) (p
0.004, 0.0001, 0.006, and 0001, respectively). In consistent with reports by Xu et al. of nonobese NAFLD22, the individuals with NAFLD were characterized by higher levels of alanine aminotransferase (ALT) (p <
< 0.0001), alkaline phosphatase (p
0.0001), alkaline phosphatase (p =
= 0.002), total triglycerides (p
0.002), total triglycerides (p <
< 0.0001), very low-density lipoprotein cholesterol (p
0.0001), very low-density lipoprotein cholesterol (p <
< 0.0001), gamma-glutamyl transpeptidase (γ-GT) (p
0.0001), gamma-glutamyl transpeptidase (γ-GT) (p <
< 0.0001), and serum uric acid (p
0.0001), and serum uric acid (p =
= 0.016). In addition, the patients had markedly lower levels of high-density lipoprotein cholesterol (p
0.016). In addition, the patients had markedly lower levels of high-density lipoprotein cholesterol (p =
= 0.0003) compared with those of HCs. The increased levels of these markers suggest that individuals with NAFLD are featured by metabolic disturbances known to be associated with an increased risk of liver dysfunction and hepatocyte lipid accumulation and decreased β-oxidation energy production24.
0.0003) compared with those of HCs. The increased levels of these markers suggest that individuals with NAFLD are featured by metabolic disturbances known to be associated with an increased risk of liver dysfunction and hepatocyte lipid accumulation and decreased β-oxidation energy production24.
Table 1
| Variables | With NAFLD (n = = 43) 43) | Without NAFLD (n = = 83) 83) | p-value |
|---|---|---|---|
| Sex (male/female) | 43 (36/7) | 83 (70/13) | — |
| Age (years) | 47.0 (34.5–61.0) | 40.5 (33.0–52.0) | 0.31 |
| Body mass index (kg/m2) | 23.19 (22.19–24.22) | 21.77 (20.7–23.38) | 0.004 |
| Waist circumference (cm) | 89 (84–91.75) | 79.5 (72.5–85) | <0.0001 |
| Hip circumference (cm) | 98 (94.75–100) | 95 (91–97) | 0.0058 |
| Waist-to-hip ratio | 0.90 (0.89–0.92) | 0.84 (0.80–0.89) | <0.0001 |
| Systolic blood pressure (mm Hg) | 127.0 (111.5–136.0) | 119 (110–129) | 0.19 |
| Diastolic blood pressure (mm Hg) | 75 (70.5–82.5) | 72 (67–80) | 0.36 |
| ALT (U/l) | 29 (20.5–39.5) | 14.5 (12–20.75) | 0.0001 |
| AST (U/l) | 22 (18–25.5) | 20 (18.0–21.75) | 0.06 |
| Alkaline phosphatase (g/l) | 78 (63–91) | 65 (54–78.75) | 0.002 |
| Total bilirubin (μmol/l) | 13 (11–18) | 12 (10–17) | 0.182 |
| Direct bilirubin (μmol/l) | 4 (3–5) | 4 (3–5) | 0.95 |
| Indirect bilirubin (μmol/l) | 10 (8.5–13) | 9 (6.25–12) | 0.12 |
| Total triglycerides (mmol/l) | 1.83 (1.43–2.6) | 1.04 (0.77–1.45) | 0.0001 |
| Total cholesterol (mmol/l) | 4.9 (4.25–5.36) | 4.68 (4.1–5.11) | 0.12 |
| High-density lipoprotein cholesterol (mmol/l) | 1.12 (0.89–1.30) | 1.4 (1.19–1.59) | 0.0003 |
| Low-density lipoprotein cholesterol (mmol/l) | 2.54 (2.14–2.95) | 2.50 (2.04–2.79) | 0.40 |
| Very low-density lipoprotein cholesterol (mmol/l) | 1.01 (0.84–1.42) | 0.7 (0.47–0.94) | <0.0001 |
| Fasting glucose (mmol/l) | 4.96 (4.74–5.47) | 4.87 (4.40–5.20) | 0.132 |
| γ-Glutamyltransferase (U/l) | 37 (23.5–62.5) | 20 (14–28) | <0.0001 |
| Serum uric acid (μmol/l) | 383 (310–423) | 340.5 (392.3–387.3) | 0.016 |
| Hemogobin (g/l) | 158 (153–165) | 154 (144–164) | 0.07 |
| Platelet count (×109/l) | 214 (183–250) | 199 (168.5–234) | 0.08 |
Note: Data are expressed as the mean (SEM.) or median (IQR, interquartile range).
Differed pattern of fecal microbiota between individuals with and without NAFLD
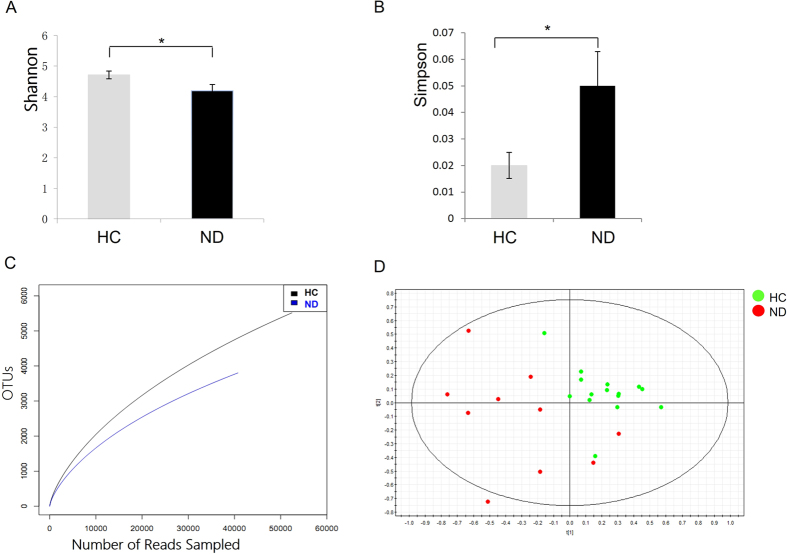
Compositional differences in the microbiota of this nonobese cohort were first assessed by DGGE patterns (Supplementary Figure S1A) combined with several clustering methods. As shown in Supplementary Figure S1B, according to multidimensional scaling analysis, an unconstrained clustering method, microbiota composition differed significantly between the two groups. Moreover, PLS-DA (Fig. 1A–C and supplementary Table S1), an supervised analysis method, also allowed for the detection of differences between individuals with and without NAFLD independent of their ALT level, indicating distinctive fecal microbial communities between HC and NAFLD groups. Furthermore, PLS-DA showed that NAFLD individuals with or without normal liver enzyme levels were also associated with clearly different microbiota (Fig. 1D). The validation plot (Supplementary Figure S2) demonstrated that the PLS-DA models were valid: the Q2 regression line had a negative intercept, and all of the permuted R2 values to the left of the intercept were lower than the original point to the right. These findings suggested the role of gut microbiota in NAFLD and the potential of using the gut microbiota as a marker to differentiate among individuals with or without NAFLD.

(A) NAFLD patients vs. HCs, (B) ND vs. HCs, (C) NS vs. HCs, and (D) ND vs. NS. HCs (n =
= 55), healthy controls; ND (n
55), healthy controls; ND (n =
= 20), individuals with NAFLD; NS (n
20), individuals with NAFLD; NS (n =
= 14), NAFLD patients with elevated serum liver enzymes.
14), NAFLD patients with elevated serum liver enzymes.
Quantification of fecal microbial changes associated with the presence of NAFLD by 454 pyrosequencing
To assess quantitative changes in the fecal microbiota associated with the presence of NAFLD, parallel pyrosequencing was performed in a selected subgroup of the nonobese cohort. The demographic and clinical information of the selected subjects and the characteristics of pyrosequencing data were listed in Supplementary Tables S2 and S3, respectively. The distribution of BMI, waist circumference, gender, smoking, alcohol consumption, and dietary habits were not statistically different between individuals with and without NAFLD in the subgroup. In agreement with the findings of DGGE profiling, decreased ecological diversities and altered fecal microbiome compositions of NAFLD patients (ND, n =
= 8; NS, n
8; NS, n =
= 2) were exhibited compared with those of HCs (n
2) were exhibited compared with those of HCs (n =
= 15) (Figs 2 and and33 and Supplementary Table S3). As shown in Fig. 2A, the Shannon diversity index (H) was significantly reduced in the NAFLD group (4.19
15) (Figs 2 and and33 and Supplementary Table S3). As shown in Fig. 2A, the Shannon diversity index (H) was significantly reduced in the NAFLD group (4.19 vs. 4.71, p
vs. 4.71, p =
= 0.03), whereas the Simpson’s diversity index (0.05
0.03), whereas the Simpson’s diversity index (0.05 vs. 0.02, p
vs. 0.02, p =
= 0.02) was increased (Fig. 2B), suggesting decreased diversity of the IM in patients with NAFLD. Reduced richness in bacterial diversity was also observed in individuals with NAFLD by the rarefaction curve trend (Fig. 2C). Additionally, the Good’s coverage of each sample for genera was greater than 80%, demonstrating that these sequences may represent major constituents in the bacterial community. Based on the pyrosequencing data, the fecal microbial communities of individuals with or without NAFLD showed clear separation in the PLS-DA score plot (R2X
0.02) was increased (Fig. 2B), suggesting decreased diversity of the IM in patients with NAFLD. Reduced richness in bacterial diversity was also observed in individuals with NAFLD by the rarefaction curve trend (Fig. 2C). Additionally, the Good’s coverage of each sample for genera was greater than 80%, demonstrating that these sequences may represent major constituents in the bacterial community. Based on the pyrosequencing data, the fecal microbial communities of individuals with or without NAFLD showed clear separation in the PLS-DA score plot (R2X =
= 0.466, R2Y
0.466, R2Y =
= 0.76, Q2(cum)
0.76, Q2(cum) =
= 0.239) (Fig. 2D), indicating that NAFLD was associated with the significant changes in diversity and composition of the fecal microbial community compared with those in HCs.
0.239) (Fig. 2D), indicating that NAFLD was associated with the significant changes in diversity and composition of the fecal microbial community compared with those in HCs.
 =
= 10) compared with individuals without NAFLD (n
10) compared with individuals without NAFLD (n =
= 15).
15).(A) The Shannon diversity index, (B) the Simpson diversity index, and (C) the rarefaction curve trend between patients with NAFLDs and HCs; (D) PLS-DA score plots based on the relative abundances of microbial genera of the first two components (t [1] =
= 20.9%, t [2]
20.9%, t [2] =
= 16.1%) showed the trend that the two groups were well separated.
16.1%) showed the trend that the two groups were well separated.

(A) Ratios of the phyla Firmicutes and Bacteroidetes, (B) phyla Firmicutes and Bacteroidetes, (C) classes clostridia and Bacteroidia, (D) families Lachnospiraceae and Ruminococcaceae; (E) Lactobacillaceae and Peptostreptococcaceae; (F) G+ and G− bacteria; (G) The ratios of G− to G+ bacteria.
Further analysis of microbiome data showed that NAFLD was associated with changes in the fecal microbiota at the phylum, class, family and genus levels (Fig. 3, Table 2 and Supplementary Figure S3). As shown in Fig. 3A, a significant decrease in the ratio of Firmicutes to Bacteroidetes (F/B) (0.47 vs. 1.4, p
vs. 1.4, p =
= 0.003) was found between the groups. In addition, individuals with NAFLD had higher fecal levels of the phylum Bacteroidetes (66.0% vs. 46.4%, p
0.003) was found between the groups. In addition, individuals with NAFLD had higher fecal levels of the phylum Bacteroidetes (66.0% vs. 46.4%, p =
= 0.005) and lower levels of Firmicutes (27.5% vs. 51.9%, p
0.005) and lower levels of Firmicutes (27.5% vs. 51.9%, p =
= 0.002) compared with HCs (Fig. 3B). Furthermore, the predominant class Bacteroidia in the phylum Bacteroidetes was enriched in patients with NAFLD (65.93% vs. 46.18%, p
0.002) compared with HCs (Fig. 3B). Furthermore, the predominant class Bacteroidia in the phylum Bacteroidetes was enriched in patients with NAFLD (65.93% vs. 46.18%, p =
= 0.004), whereas the class Clostridia within the phylum Firmicutes was depleted in individuals with NAFLD (27.02% vs. 50.91%, p
0.004), whereas the class Clostridia within the phylum Firmicutes was depleted in individuals with NAFLD (27.02% vs. 50.91%, p =
= 0.001) (Fig. 3C). Furthermore, the marked depletion of Firmicutes in NAFLD group was mostly explained by decreases in four families: Lachnospiraceae (18.67% vs. 34.26%, p
0.001) (Fig. 3C). Furthermore, the marked depletion of Firmicutes in NAFLD group was mostly explained by decreases in four families: Lachnospiraceae (18.67% vs. 34.26%, p =
= 0.002), Ruminococcaceae (6.69% vs. 13.67%, p
0.002), Ruminococcaceae (6.69% vs. 13.67%, p =
= 0.018), Lactobacillaceae (0.04% vs. 0.73%, p
0.018), Lactobacillaceae (0.04% vs. 0.73%, p =
= 0.0008), and Peptostreptococcaceae (0.09% vs. 0.44%, p
0.0008), and Peptostreptococcaceae (0.09% vs. 0.44%, p =
= 0.002) (Fig. 3D,E).
0.002) (Fig. 3D,E).
Table 2
| NO. | Genus | Family | Phylum | VIP | With ND | Without ND | pvalue |
|---|---|---|---|---|---|---|---|
| 1 | Bacteroides | Bacteroidaceae | Bacteroidetes | 5.9 | 29% ± ± 4% 4% | 20% ± ± 1.8% 1.8% | 0.103 |
| 2 | Prevotella | Prevotellaceae | Bacteroidetes | 3.8 | 8% ± ± 4% 4% | 3% ± ± 1% 1% | 0.789 |
| 3 | Pseudobutyrivibrio | Lachnospiraceae | Firmicutes | 1.9 | 0.45% ± ± 0.2% 0.2% | 2.16% ± ± 0.6% 0.6% | 0.02 |
| 4 | Anaerotruncs | Ruminococcaceae | Firmicutes | 1.9 | 0.19% ± ± 0.2% 0.2% | 0.96% ± ± 0.4% 0.4% | 0.004 |
| 5 | Lactobacillus | Lactobacillaceae | Firmicutes | 1.7 | 0.04% ± ± 0.03% 0.03% | 0.74% ± ± 0.4% 0.4% | 0.003 |
| 6 | Roseburia | Lachnospiraceae | Firmicutes | 1.6 | 1.48% ± ± 0.6% 0.6% | 2.59% ± ± 0.4% 0.4% | 0.01 |
| 7 | Coprococcus | Lachnospiraceae | Firmicutes | 1.6 | 0.34% ± ± 0.1% 0.1% | 2.4% ± ± 0.6% 0.6% | 0.009 |
| 8 | Streptococcus | Streptococcaceae | Firmicutes | 1.5 | 0.28% ± ± 0.01% 0.01% | 0.2% ± ± 0.1% 0.1% | 0.24 |
| 9 | Ruminococcus | Ruminococcaceae | Firmicutes | 1.4 | 1.03% ± ± 0.7% 0.7% | 1.59% ± ± 0.3% 0.3% | 0.02 |
| 10 | Moryella | Lachnospiraceae | Firmicutes | 1.4 | 0.05% ± ± 0.02% 0.02% | 0.35% ± ± 0.1% 0.1% | 0.02 |
| 11 | Anaerosporabacter | Lachnospiraceae | Firmicutes | 1.4 | 1.08% ± ± 0.1% 0.1% | 2.02% ± ± 0.1% 0.1% | 0.02 |
| 12 | Oscillibacter | Ruminococcaceae | Firmicutes | 1.2 | 0.8% ± ± 0.4% 0.4% | 1.4% ± ± 0.3% 0.3% | 0.05 |
| 13 | Bifidobacterium | Bifidobacteriaceae | Actinobacteria | 1.2 | 0.36% ± ± 0.2% 0.2% | 0.22% ± ± 0.1% 0.1% | 0.82 |
| 14 | Escherichia | Enterobacteriaceae | Proteobacteria | 1 | 0.32% ± ± 0.3% 0.3% | 0.1% ± ± 0.05% 0.05% | 0.78 |
Note: Data are expressed as the mean (SEM.); VIP, the variable importance in projection.
Moreover, at the genus level, 14 key microbial genera were identified as key microbes with PLS-DA. These genera were responsible for the separation between non-obese individuals with and without NAFLD (Table 2), and well-known gut bacteria such as Bacteroides, Pseudobutyrivibrio and Lactobacillus. Interestingly, differences in the fecal microbiota of individuals with NAFLD were observed only in genera belonging to Firmicutes (Table 2 and Supplementary Figure S3). For example, within the Lachnospiraceae family, Coprococcus (mean 0.34% vs. 2.4%, p =
= 0.009), Pseudobutyrivibrio (0.45% vs. 2.16%, p
0.009), Pseudobutyrivibrio (0.45% vs. 2.16%, p =
= 0.02), Moryella (0.05% vs. 0.35%, p
0.02), Moryella (0.05% vs. 0.35%, p =
= 0.02), Roseburia (1.48% vs. 2.59%, p
0.02), Roseburia (1.48% vs. 2.59%, p =
= 0.01) and Anaerosporobacter (1.08% vs. 2.02%, p
0.01) and Anaerosporobacter (1.08% vs. 2.02%, p =
= 0.02) were decreased in individuals with NAFLD. Within the family Ruminococcaceae, a similar decrease was observed for Anaerotruncus (0.19% vs. 0.96%, p
0.02) were decreased in individuals with NAFLD. Within the family Ruminococcaceae, a similar decrease was observed for Anaerotruncus (0.19% vs. 0.96%, p =
= 0.004) and Ruminococcus (1.03% vs. 1.59%, p
0.004) and Ruminococcus (1.03% vs. 1.59%, p =
= 0.02). Notably, Lactobacillus, the main genus of the Lactobacillaceae family, was markedly reduced in both abundance (0.04% vs. 0.74%, p
0.02). Notably, Lactobacillus, the main genus of the Lactobacillaceae family, was markedly reduced in both abundance (0.04% vs. 0.74%, p =
= 0.003) and prevalence (30% vs. 82%, p
0.003) and prevalence (30% vs. 82%, p =
= 0.008) in NAFLD patients compared with HCs (Supplementary Figure S4). Furthermore, to explore the potential role of the IM in the pathogenesis of NAFLD, the relative abundances of G- bacteria and Gram-positive (G+) bacteria based on the pyrosequencing data were assessed. Importantly, in individuals with NAFLD compared with HCs, G- bacteria were markedly enriched (85.21% vs. 71.8%, p
0.008) in NAFLD patients compared with HCs (Supplementary Figure S4). Furthermore, to explore the potential role of the IM in the pathogenesis of NAFLD, the relative abundances of G- bacteria and Gram-positive (G+) bacteria based on the pyrosequencing data were assessed. Importantly, in individuals with NAFLD compared with HCs, G- bacteria were markedly enriched (85.21% vs. 71.8%, p =
= 0.009), and G+ bacteria were decreased (14.79% vs. 28.2%, p
0.009), and G+ bacteria were decreased (14.79% vs. 28.2%, p =
= 0.01) (Fig. 3F). Additionally, the ratios of G−/G+ bacteria in individuals with NAFLD were enriched compared with those in HCs (5.72
0.01) (Fig. 3F). Additionally, the ratios of G−/G+ bacteria in individuals with NAFLD were enriched compared with those in HCs (5.72 vs. 3.52, p
vs. 3.52, p =
= 0.009) (Fig. 3G).
0.009) (Fig. 3G).
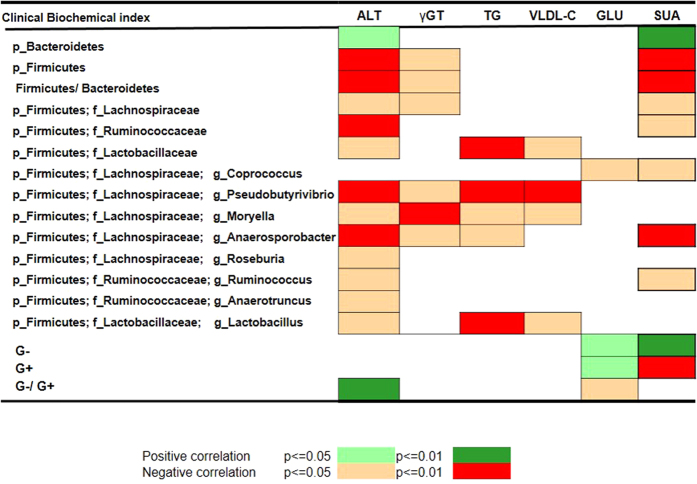
Potential correlation between intestinal dysbiosis and liver biochemical markers
We also attempted to assess the correlation between differences in microbial composition and clinical liver biochemical markers of NAFLD in the non-obese cohort. Interestingly, a clear significant correlation with the set of liver biochemistry indices in plasma was found for NAFLD-induced disturbed microbiota at the phylum and family and genus level by Spearmen’s rank correlation analysis (Fig. 4 and Supplementary Figures S5 and S6). These liver biochemistry markers are well-known clinical biochemical markers for NAFLD diagnosis1,2,22. At the global level, significant correlations with markers of liver status, such as ALT and uric acid, were found for the two dominant phyla Firmicutes, and Bacteroidetes. In addition, a significant correlation with γ-GT, markers of liver injury, was also found for Firmicutes. Additionally, at the family level, further significant correlations were found for plasma ALT with 3 families (Lachnospiraceae, Ruminococcaceae, Lactobacillaceae), and for plasma γ-GT with Lachnospiraceae. Moreover, both plasma total triglycerides and very low-density lipoprotein cholesterol were significantly correlated with Lactobacillaceae (p <
< 0.0001).
0.0001).
Furthermore, the correlations between the set of liver biochemistry markers and disturbed microbial genera were also revealed (Fig. 4 and Supplementary Figure S7). For instance, one metabolic marker, fasting glucose, revealed only a single microbial association with Coprococcus. Other markers had multiple correlations. For example, the increase of plasma ALT was statistically linked with seven depleted genera (Pseudobutyrivibrio, Moryella, Anaerosporobacter, Roseburia, Ruminococcus, Anaerotruncus, and Lactobacillus). These findings indicated a close interrelationship between the intestinal microbial composition and liver biochemical state, highlighting that alterations of the IM are implicated in the presence and development of NAFLD.
Discussion
To our knowledge, this is the first study assessing the gut microbiota in nonobese individuals with NAFLD compared with those without NAFLD and specifically correlating the microbial composition of individuals with clinical indices. Here, we identified the significant distinguishing fecal microbiota at all taxonomic levels in non-obese individuals with NAFLD compared with those without NAFLD; the former were characterized by a more pronounced Firmicutes-poor microbiota and marked lower overall microbial richness. Further analyses revealed that the altered microbiota associated with the presence of NAFLD significantly correlated with liver biochemical markers in the nonobese cohort.
Classifying NAFLD individuals based on BMI is crucial for deciphering the potential role of gut microbiota in NAFLD, because BMI represents a major determinant of compositional changes in microbial communities25. Moreover, non-obese patients with NAFLD also represent a considerable portion of NAFLD patients. Notably, in spite of the lower BMI and lower prevalence of obesity in the Asia-Pacific region, people in this region still tend to develop NAFLD. Because both NAFLD and obesity are believed to be influenced by the gut microbiota, comparisons between non-obese individuals with and without NAFLD allowed us to further address the effects of the IM on the liver function.
In human IM, Firmicutes and Bacteroidetes are the dominant bacteria, accounting for approximately 99% of the whole microbiota6,8. In our cohort, a significant lower abundance of Firmicutes and a higher abundance of Bacteroidetes were exhibited in individuals with NAFLD compared with those without NAFLD. This finding was partly in agreement with the findings of Zhu et al. and Schwiertz A et al., who have demonstrated lower Firmicutes and higher Bacteroidetes abundances in obese patients compared with lean healthy controls18,26. But Zhu et al. did not find the similar difference between obese and NASH microbiome18. The novelty of our study is that we provide the first report of a direct association between lower Firmicutes and NAFLD in non-obese individuals. Interestingly, our findings were significantly different with those of a recent report of Mouzaki et al., who have shown lower Bacteroidetes levels in NASH patients with higher BMI than in patients with simple steatosis and controls with lower BMI16. These results indicated that obese NASH and nonobese NAFLD have different fecal microbiome. And the discrepancy between the findings might partly reflect the imbalance of uncontrolled effects of factors between the groups, such BMI and different detection techniques. The decreased proportion of Bacteroidetes has also been demonstrated in patients with high BMI in previously published literature in the field of obesity8,9. Thus, the high BMI of subjects may have affected the results of the study by Zhu et al., although the linear regression, adjusting for BMI, was performed to theoretically limit the potential confounding effect of BMI. In addition, the different detection techniques also affected these results between the two study cohorts, which was evidenced by previous studies by our group11 and Larsen et al.27. In our study cohort, all recruited individuals were non-obese adults, which could avoid the confounding factor of BMI. It should be noted that in our non-obese cohort, our observations suggested that a slight increase in BMI, even within the normal range, in people with BMI <25 kg/m2 was significantly associated with an increased risk of NAFLD. However, it is unlikely that there are significant BMI-dependent variations in the IM composition within the nonobese spectrum16,18. And in our quantification study of fecal microbial change, we only selected the nonobese individuals with the median value of BMI to ensure the similar distribution of BMI and waist circumference between two groups. In addition, unbiased metagenomics approaches were applied in this study to characterize the whole IM, including some unknown but potentially functional bacteria28.
kg/m2 was significantly associated with an increased risk of NAFLD. However, it is unlikely that there are significant BMI-dependent variations in the IM composition within the nonobese spectrum16,18. And in our quantification study of fecal microbial change, we only selected the nonobese individuals with the median value of BMI to ensure the similar distribution of BMI and waist circumference between two groups. In addition, unbiased metagenomics approaches were applied in this study to characterize the whole IM, including some unknown but potentially functional bacteria28.
Bacterial phylotypes decrease in nonobese patients with NAFLD was mostly associated with Firmicutes. The families Lachnospiraceae and Ruminococcaceae are two predominant members of Firmicutes. These bacteria are short-chain fatty acids (SCFAs)-producing bacteria that are well known for their health-promoting functions, including the production of nutrients for the host and an energy supply for the colonic epithelium, the modulation of colonic pH29, and the maintenance of host immune homeostasis30. A decrease in SCFAs might result in deteriorated intestinal integrity and increased intestinal permeability, thus making decreased SCFAs a very important pathogenic factor in NAFLD31. In addition, these bacteria are also capable of performing 7-α dehydroxylation and deconjugation in bile acid metabolism, which have been demonstrated to play a critical role in the pathogenesis of chronic liver diseases such as inflammation, cirrhosis and hepatocellular carcinoma12,32. Indeed, in Jone’s metagenomics study, the function bile salt hydrolases had been identified in all major bacterial phyla and archaeal species in the gut33, supporting that IM might modulate the bile acid metabolism in NAFLD patients. In addition, it has been evidenced that IM could modulate the fecal bile acids profiles in patients with cirrhosis32 and even promote hepatocellular carcinoma development through modulating the enterohepatic circulation of BAs12. Furthermore, we showed that fecal microbial ecological diversity was significantly reduced in patients with NAFLD. Thus far, the exact reason for decreased bacterial diversity in NAFLD remains unclear. It has been demonstrated during antibiotic treatment34 and in diseases such as liver cirrhosis19 and obesity35. Such alterations in the IM have recently been reported for a variety of liver diseases19,32,36. Thus, our finding of general dysbiosis in NAFLD raises the possibility that the compositional change could result in imbalanced microbial ecology, which might itself play a role in increasing the susceptibility to liver diseases.
In this study, we also found a higher abundance of lipopolysaccharide-producing G- bacteria in non-obese individuals with NAFLD. In agreement with our study, through pyrosequencing analysis, De Minicis et al. have also revealed an increase in G- bacteria in mice with bile duct ligation fed a high-fat diet37. And a significant difference in Escherichia was exhibited between obese children with and without NASH in the study by Zhu et al.18. Additionally, evidence from the literature supports a causal role for endotoxin produced by G- bacteria in NAFLD/NASH14,38,39. Here, a significant positive correlation between G bacteria and fasting glucose levels was also found, highly suggesting the potential contribution of predictive markers for assessing the presence and development of NAFLD in non-obese individuals. It would be interesting to assess differences in the circulating levels of endotoxin between individuals with and without NAFLD in a nonobese cohort.
Notably, we first demonstrated that the health-promoting Lactobacillaceae family and its most abundant genus Lactobacillus showed marked reductions in both abundance and prevalence in patients with NAFLD compared with HCs. Probiotic bacteria such as Lactobacillus and Bifidobacterium promote beneficial effects, likely through anti-inflammatory actions and stabilization of the intestinal barrier, thereby attenuating liver pathologies40. As concluded by many clinical studies, these probiotic bacteria reduce features of NAFLD in humans41,42,43 and liver injury in mouse models44. For example, some strains of Lactobacillus lead to substantial reductions in the levels of ALT in 10 patients with NASH43. Consistently with the above findings, a negative correlation between Lactobacillus and a wide range of biochemical indices of NAFLD progression, including ALT, and TG, was also found, highly facilitating future efforts to better manipulate the gut microbiota for the treatment of NAFLD.
It is widely accepted that the gut microbiota is crucial for NAFLD31, which is considered as the hepatic manifestation of metabolic syndrome. However, to date, little is understood about the molecular host-microbiome interactions that influence host metabolic pathways. Recent studies both in adult patients with NAFLD45 and mice13 supported that gut dysbiosis exerts a critical influence in the progression of NAFLD. In this study a notable finding was the clear correlation between the microbiome composition and well-known clinical indices of NAFLD progression, such as higher blood ALT, γ-GT, TG, VLDL-C, GLU and SUA, suggesting that the microbiota might be closely involved in the pathogenesis of the progression of NAFLD. For example, the blood level of ALT and γ-GT, the inflammatory indices, were significantly correlated with the distinguishing bacteria between individuals with and without NAFLD. In consistent with our findings, Boursier et al. also demonstrated that gut dysbiosis associated with NAFLD severity is accompanied by a shift in the metabolic functions of gut microbiota45. In addition, the correlation between IM and SUA was also firstly revealed in this study. The level of SUA is elevated significantly in patients with NAFLD, which is an independent risk factor of NAFLD46. Uric acid is the major end product of purine metabolism, and has been proposed as a natural scavenger of peroxynitrite and peroxynitrite-derived radicals47,48. Despite the fact that it is not clear how IM regulates these opposing roles of uric acid on redox balance, our study added new clues that IM might promote steatosis to steatohepatitis in NAFLD by its metabolic pathways and products.
A considerable number of studies support the very important role of the microbiome in the pathogenesis of metabolic disturbance in NAFLD18,30,35,49,50. Recently, Mouzaki et al. have reported a potential negative association between Bacteroidetes and insulin resistance when controlling for BMI18. Compared with previous reports, our study presents stronger evidence supporting these findings, because we did assess differences in the gut microbiota between nonobese individuals with and without NAFLD; additionally, our study suggests a potential correlation between alterations in fecal microbiota and metabolic disturbance in NAFLD. Future studies are required to establish the mechanistic relevance of our novel findings to the pathogenesis of NAFLD. For instance, it will be necessary to reveal the presence of gut-derived metabolites (i.e., lipopolysaccharide or SCFAs) in the peripheral or portal blood as a medium for the systemic effects of these compounds in human beings or gut microbiome biomarkers associated with the progression of NAFLD and/or NASH.
There were several limitations in this study. First, NAFLD was diagnosed by ultrasonographic methods, which could not determine the severity of NAFLD51. Nevertheless, ultrasonography is reported with reasonable accuracy and widely used for population-based studies22,51. Here, all included subjects had no history of any disease and were recruited at the time of their annual physical examination, it was impractical and unethical to perform liver biopsy in this study. And we performed all the ultrasonographic diagnosis by a single experienced radiographer and the diagnostic performance of the ultrasonographer was evaluated by one senior ultrasonographer meanwhile. Second, this study design did not allow for the examination of insulin resistance, although limited research has found insulin resistance might be a risk factor of NAFLD among nonobese individuals23. Future studies are necessary to investigate the correlation between insulin resistance and IM in nonobese NAFLD. Despite these limitations, our conceptually novel study provided important insights into the IM in nonobese adult patients with NAFLD and may have significant clinical importance for NAFLD manipulation.
In summary, our findings firstly showed that the non-obese patients with NAFLD were characterized by a decrease in gut microbial diversity, the depletion of Firmicutes including health-promoting bacteria such as SCFAs-producing Lachnospiraceae, 7α-dehydroxylating Ruminococcaceae and beneficial Lactobacillaceae, and the increase of potentially opportunistic pathogenic lipopolysaccharide-producing bacteria. In addition, we established a link between the altered microbiota and well-known liver biochemical indices of NAFLD progression. This novel study may pave the way for new prevention and therapeutic principles. To establish clinical feasible diagnostic and therapeutic methods, further studies utilizing the integrative approach of metabonomics and metagenomics with computational technology6 are required to elucidate the key microbes and its metabolic pathway associated with NAFLD.
Methods
Patients and study design
The study protocol was approved by the Ethical Review Board of the first affiliated hospital of the medical school of Zhejiang University and conformed to the ethical guidelines of the 1975 Declaration of Helsinki. Volunteer subjects were randomly recruited during their annual health survey. Exclusion criteria included the following: BMI ≥25 kg/m2; excess alcohol consumption (>20
kg/m2; excess alcohol consumption (>20 g per day for men and >10
g per day for men and >10 g per day for women); a history of cancer, respiratory problems, renal diseases, or endocrine disorder; a history of viral hepatitis, autoimmune hepatitis, genetic hemochromatosis, primary biliary cirrhosis, primary sclerosing cholangitis, α1-antitrypsin deficiency, Wilson’s disease, drug-induced hepatotoxicity, or other known causes of chronic liver disease; an intake history of any medicine, antibiotics or probiotics within the preceding 3 months; and a history of prescription medications, dietary restrictions or lifestyle modifications through diet and exercise at enrollment. Written informed consent and individual information were obtained from all of the volunteer subjects. Data on cigarette smoking, alcohol consumption, physical activity, dietary habits, and a family history of hypertension, diabetes or NAFLD were obtained through a questionnaire. A positive smoking history was defined as currently smoking or with a history of chronic/regular smoking52. The positive physical activity habit was defined as having at least 120
g per day for women); a history of cancer, respiratory problems, renal diseases, or endocrine disorder; a history of viral hepatitis, autoimmune hepatitis, genetic hemochromatosis, primary biliary cirrhosis, primary sclerosing cholangitis, α1-antitrypsin deficiency, Wilson’s disease, drug-induced hepatotoxicity, or other known causes of chronic liver disease; an intake history of any medicine, antibiotics or probiotics within the preceding 3 months; and a history of prescription medications, dietary restrictions or lifestyle modifications through diet and exercise at enrollment. Written informed consent and individual information were obtained from all of the volunteer subjects. Data on cigarette smoking, alcohol consumption, physical activity, dietary habits, and a family history of hypertension, diabetes or NAFLD were obtained through a questionnaire. A positive smoking history was defined as currently smoking or with a history of chronic/regular smoking52. The positive physical activity habit was defined as having at least 120 min exercise per week53. All subjects were required to avoid strenuous physical exercise and high-energy food with the 24
min exercise per week53. All subjects were required to avoid strenuous physical exercise and high-energy food with the 24 hours preceding the examination and sampling.
hours preceding the examination and sampling.
Clinical physical examinations were performed in all enrolled subjects by trained medical staff using a standardized procedure. Briefly, a detailed medical history and health habit inventory were taken by a physician. Then, physical examination, anthropometric measurements, abdominal (including hepatic) ultrasonic examination, and clinical biochemical measurements were performed. The diagnosis of NAFLD was made based on evidence of fatty liver upon ultrasonography and the exclusion of other known etiologies of chronic liver disease in accordance with the established clinical and ultrasound criteria54. And the individuals with diagnosis of metabolic diseases such as diabetes, hypertension and dyslipidemia were excluded. Finally, a total of 126 nonobese subjects were recruited. The microbial community was profiled by DGGE in all subjects (43 NAFLD, 83 HCs) and further examined by 454 pyrosequencing in the sub-cohort (10 NAFLD, 15 HCs).
Hepatic ultrasonic examination was conducted using a Sequoia 512 Acuson sonography machine with a 1.5-MHz probe (Siemens Healthcare, Erlangen, German) and performed by two experienced ultrasonographer, who were blinded to the study design and clinical data. The ultrasound criteria for the diagnosis of fatty liver were based on the suggestions by the Chinese Liver Disease Association55. Briefly, fatty liver was defined as a diffuse enhancement of the near-field echo in the hepatic region and gradual attenuation of the far-field echo if combined with any of the following: (1)unclear display of intrahepatic lacuna structure; (2)mild to moderate hepatomegaly with a round and blunt border; or (3)color Doppler ultrasonography showing a reduction in the blood flow signal in the liver or a blood flow signal that is difficult to display even when the distribution of blood flow is normal22.
Fresh fecal samples were placed in sterile tubes under anaerobic conditions (GENbox anaer, BioMérieux), transferred to the laboratory immediately in an ice box, and stored at −70 °C after preparation within 15
°C after preparation within 15 min6. Blood samples collected early in the morning on the same day were collected and sent to the diagnostic testing laboratory for detection of the twenty-two clinical biochemical parameters. Laboratory tests included ALT, aspartate aminotransferase, albumin, platelet count, γ-GT, fasting glucose, and the lipid profile (cholesterol and triglycerides). NAFLD patients with elevated serum liver enzymes (ALT and γ-GT) were highly suspected of patients with NASH56.
min6. Blood samples collected early in the morning on the same day were collected and sent to the diagnostic testing laboratory for detection of the twenty-two clinical biochemical parameters. Laboratory tests included ALT, aspartate aminotransferase, albumin, platelet count, γ-GT, fasting glucose, and the lipid profile (cholesterol and triglycerides). NAFLD patients with elevated serum liver enzymes (ALT and γ-GT) were highly suspected of patients with NASH56.
Fecal microbial DNA Extraction
Fecal genomic DNA was isolated from stool samples by using a QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA) according to the kit protocol with modifications, as previously described by us6. Briefly, the fecal samples were lysed by incubating the samples in lysis buffer, and this was followed by bead beating with zirconium beads (0.1 mm) with a FastPrep instrument (MP Biomedicals, Carlsbad, CA) for 60
mm) with a FastPrep instrument (MP Biomedicals, Carlsbad, CA) for 60 seconds at the speed of 4
seconds at the speed of 4 m/s, purified with spin columns, and eluted with 200
m/s, purified with spin columns, and eluted with 200 μl of buffer AE. The quality of extracted genomic DNA was checked by using the 0.8% agarose gel, and the DNA concentrations were determined with a Nano-Drop 1000 spectrophotometer (NanoDrop Technologies). The extracted DNA was stored at −20
μl of buffer AE. The quality of extracted genomic DNA was checked by using the 0.8% agarose gel, and the DNA concentrations were determined with a Nano-Drop 1000 spectrophotometer (NanoDrop Technologies). The extracted DNA was stored at −20 °C.
°C.
PCR-DGGE profiling
The dominant gut bacterial community structure from the fecal microbiota was illustrated by PCR-DGGE analysis, as previously described by us6. Isolated fecal DNA was used as a template for amplification of the 16S rRNA-V3 region using a universal primer and the hot-start touchdown protocol described by Muyzer et al.57. The reaction mixture contained 1 unit of TaKaRa (Takara, Dalian, China) rTaq polymerase, 2.5
unit of TaKaRa (Takara, Dalian, China) rTaq polymerase, 2.5 μl of 10
μl of 10 ×
× PCR buffer, 2
PCR buffer, 2 μl of dNTP mixture (2.5
μl of dNTP mixture (2.5 mM), 0.3
mM), 0.3 μl of each primer (20
μl of each primer (20 pmol/μl), and 20
pmol/μl), and 20 ng of extracted fecal DNA in a total volume of 25
ng of extracted fecal DNA in a total volume of 25 μl. Reactions were run in a Veriti 96-well thermal cycler (Applied Biosystems, Foster City, CA). To reduce PCR bias, two separate PCR reactions of each sample were pooled for DGGE profiling. The quality of PCR product was checked by a 1% agarose gel and a NanoDrop 1000 spectrophotometer (NanoDrop Technologies). PCR-DGGE fingerprinting was performed using a D-code system (Bio-Rad, USA) with a house-made pipeline, as previously reported24. The DGGE images and their profiles were analyzed by using Bionumerics software (Applied Maths, Belgium), including multidimensional scaling, which is a grouping technique that allows for a convenient visual interpretation. The intensity and position of bands in each lane in the DGGE image were digitalized into a spectrum of 832 variables. In addition, the acquired spectra were linearly aligned, in which the bands in the marker lane were selected as the standard to rectify mobile shifts of the bands in different DGGE profiles. Then, multivariate data analysis, including principal component analysis and partial least-squares-latent structure discriminate analysis (PLS-DA), was performed by using Simca-P 12.0 (Umetrics AB, Sweden) on the data set to observe the structure of the fecal DGGE profiles, such as clustering and outliers6,58.
μl. Reactions were run in a Veriti 96-well thermal cycler (Applied Biosystems, Foster City, CA). To reduce PCR bias, two separate PCR reactions of each sample were pooled for DGGE profiling. The quality of PCR product was checked by a 1% agarose gel and a NanoDrop 1000 spectrophotometer (NanoDrop Technologies). PCR-DGGE fingerprinting was performed using a D-code system (Bio-Rad, USA) with a house-made pipeline, as previously reported24. The DGGE images and their profiles were analyzed by using Bionumerics software (Applied Maths, Belgium), including multidimensional scaling, which is a grouping technique that allows for a convenient visual interpretation. The intensity and position of bands in each lane in the DGGE image were digitalized into a spectrum of 832 variables. In addition, the acquired spectra were linearly aligned, in which the bands in the marker lane were selected as the standard to rectify mobile shifts of the bands in different DGGE profiles. Then, multivariate data analysis, including principal component analysis and partial least-squares-latent structure discriminate analysis (PLS-DA), was performed by using Simca-P 12.0 (Umetrics AB, Sweden) on the data set to observe the structure of the fecal DGGE profiles, such as clustering and outliers6,58.
Pyrosequencing and data analyses
Pyrosequencing technology is a powerful tool for the comprehensive analysis of IM28 in patients with liver diseases such as liver cirrhosis11 and NASH18. As previously described, after PCR products amplified and quantified, equimolar concentrations were pooled and sequenced on a 454 Life Sciences Genome Sequencer FLX system (Roche) according to the manufacturer’s recommendations11. All reads were deposited in GeneBank (SRA056280), quality trimmed, analyzed and grouped into operational taxonomic units at a sequence similarity level of 97%. These operational taxonomic units were used for diversity (Shannon, Simpson), richness (Chao 1), and rarefaction curve analysis. And bacterial coverage for each library was calculated by Good’s coverage11.
The table of normalized genera counts was fed into Simca-P 12.0 for principal component analysis and PLS-DA11,59. Prior to multivariate data statistics analysis, the normalized data were pareto-scaled. And the supervised models were validated with a permutation test that was repeated 200 times to prevent model overfitting58. In addition, the variable importance in projection (VIP) index was used to select the discriminating bacteria and reflected the influence of each microbial genus in the different group. Variables with VIP >
> 1 are important contributors to generation of the model. The Mann-Whitney U test was used to evaluate group differences, and Spearman’s correlation coefficients were used to assess bivariate relationships between variables by using SPSS Version 16.0 (SPSS Inc., Chicago, IL) and GraphPad Prism 5 (GraphPad Software, Inc.). And p values had been corrected for multiple testing by using Bonferroni correction.
1 are important contributors to generation of the model. The Mann-Whitney U test was used to evaluate group differences, and Spearman’s correlation coefficients were used to assess bivariate relationships between variables by using SPSS Version 16.0 (SPSS Inc., Chicago, IL) and GraphPad Prism 5 (GraphPad Software, Inc.). And p values had been corrected for multiple testing by using Bonferroni correction.
Additional Information
How to cite this article: Wang, B. et al. Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease. Sci. Rep. 6, 32002; 10.1038/srep32002 (2016).
Acknowledgments
We thank Prof. Jiliang He of Zhejiang University and Prof. Honglei Weng of Heidelberg University for the helpful and thoughtful comments for the manuscript. This study was supported by the Zhejiang Provincial and National Natural Science Foundation of China (R16H260001, 30901190, 81172702), National Program on Key Basic Research Project (2013CB531401), and High-tech R & D Program of China (No. 2012AA020204).
Footnotes
Author Contributions B.H.W. and L.J.L. designed the project; B.H.W. managed the project. B.H.W., X.Y.J., M.C., J.P.G., Q.L.B., L.L.T., Y.C. and L.J.L. performed the experiment, analyzed the data, and drafted the manuscript.
References
- Williams C. D. et al.. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology 140, 124–131, 10.1053/j.gastro.2010.09.038 (2011). [Abstract] [CrossRef] [Google Scholar]
- Fan J. G. & Farrell G. C. Epidemiology of non-alcoholic fatty liver disease in China. Journal of hepatology 50, 204–210, 10.1016/j.jhep.2008.10.010 (2009). [Abstract] [CrossRef] [Google Scholar]
- Matteoni C. A. et al.. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology 116, 1413–1419 (1999). [Abstract] [Google Scholar]
- Moschen A. R., Kaser S. & Tilg H. Non-alcoholic steatohepatitis: a microbiota-driven disease. Trends in endocrinology and metabolism: TEM 24, 537–545, 10.1016/j.tem.2013.05.009 (2013). [Abstract] [CrossRef] [Google Scholar]
- Abu-Shanab A. & Quigley E. M. The role of the gut microbiota in nonalcoholic fatty liver disease. Nature reviews. Gastroenterology & hepatology 7, 691–701, 10.1038/nrgastro.2010.172 (2010). [Abstract] [CrossRef] [Google Scholar]
- Li M. et al.. Symbiotic gut microbes modulate human metabolic phenotypes. Proceedings of the National Academy of Sciences of the United States of America 105, 2117–2122, 10.1073/pnas.0712038105 (2008). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wang B. & Li L. Who determines the outcomes of HBV exposure? Trends in microbiology 23, 328–329, 10.1016/j.tim.2015.04.001 (2015). [Abstract] [CrossRef] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S. & Gordon J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023, 10.1038/4441022a (2006). [Abstract] [CrossRef] [Google Scholar]
- Turnbaugh P. J. et al.. A core gut microbiome in obese and lean twins. Nature 457, 480–484, 10.1038/nature07540 (2009). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Qin J. et al.. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60, 10.1038/nature11450 (2012). [Abstract] [CrossRef] [Google Scholar]
- Chen Y. et al.. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 54, 562–572, 10.1002/hep.24423 (2011). [Abstract] [CrossRef] [Google Scholar]
- Yoshimoto S. et al.. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101, 10.1038/nature12347 (2013). [Abstract] [CrossRef] [Google Scholar]
- Henao-Mejia J. et al.. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185, 10.1038/nature10809 (2012). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Seki E. et al.. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nature medicine 13, 1324–1332, 10.1038/nm1663 (2007). [Abstract] [CrossRef] [Google Scholar]
- Le Roy T. et al.. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 62, 1787–1794, 10.1136/gutjnl-2012-303816 (2013). [Abstract] [CrossRef] [Google Scholar]
- Mouzaki M. et al.. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 58, 120–127, 10.1002/hep.26319 (2013). [Abstract] [CrossRef] [Google Scholar]
- Raman M. et al.. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association 11, 868–875 e861–863, 10.1016/j.cgh.2013.02.015 (2013). [Abstract] [CrossRef] [Google Scholar]
- Zhu L. et al.. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57, 601–609, 10.1002/hep.26093 (2013). [Abstract] [CrossRef] [Google Scholar]
- Chen C. H. et al.. Prevalence and risk factors of nonalcoholic fatty liver disease in an adult population of taiwan: metabolic significance of nonalcoholic fatty liver disease in nonobese adults. Journal of clinical gastroenterology 40, 745–752 (2006). [Abstract] [Google Scholar]
- Das K. et al.. Nonobese population in a developing country has a high prevalence of nonalcoholic fatty liver and significant liver disease. Hepatology 51, 1593–1602, 10.1002/hep.23567 (2010). [Abstract] [CrossRef] [Google Scholar]
- Kim H. J. et al.. Metabolic significance of nonalcoholic fatty liver disease in nonobese, nondiabetic adults. Archives of internal medicine 164, 2169–2175, 10.1001/archinte.164.19.2169 (2004). [Abstract] [CrossRef] [Google Scholar]
- Xu C. et al.. Prevalence and risk factors for the development of nonalcoholic fatty liver disease in a nonobese Chinese population: the Zhejiang Zhenhai Study. The American journal of gastroenterology 108, 1299–1304, 10.1038/ajg.2013.104 (2013). [Abstract] [CrossRef] [Google Scholar]
- Wei J. L. et al.. Prevalence and Severity of Nonalcoholic Fatty Liver Disease in Non-Obese Patients: A Population Study Using Proton-Magnetic Resonance Spectroscopy. The American journal of gastroenterology 110, 1306-1314 ; quiz 1315, 10.1038/ajg.2015.235 (2015). [Abstract] [CrossRef] [Google Scholar]
- Rodriguez-Gallego E. et al.. Mapping of the circulating metabolome reveals alpha-ketoglutarate as a predictor of morbid obesity-associated non-alcoholic fatty liver disease. International journal of obesity 39, 279–287, 10.1038/ijo.2014.53 (2015). [Abstract] [CrossRef] [Google Scholar]
- Le Chatelier E. et al.. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546, 10.1038/nature12506 (2013). [Abstract] [CrossRef] [Google Scholar]
- Schwiertz A. et al.. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 18, 190–195, 10.1038/oby.2009.167 (2010). [Abstract] [CrossRef] [Google Scholar]
- Larsen N. et al.. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS one 5, e9085, 10.1371/journal.pone.0009085 (2010). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Margulies M. et al.. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380, 10.1038/nature03959 (2005). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Wong J. M., de Souza R., Kendall C. W., Emam A. & Jenkins D. J. Colonic health: fermentation and short chain fatty acids. Journal of clinical gastroenterology 40, 235–243 (2006). [Abstract] [Google Scholar]
- Bohmig G. A. et al.. n-butyrate downregulates the stimulatory function of peripheral blood-derived antigen-presenting cells: a potential mechanism for modulating T-cell responses by short-chain fatty acids. Immunology 92, 234–243 (1997). [Abstract] [Google Scholar]
- Schnabl B. & Brenner D. A. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 146, 1513–1524, 10.1053/j.gastro.2014.01.020 (2014). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Kakiyama G. et al.. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. Journal of hepatology 58, 949–955, 10.1016/j.jhep.2013.01.003 (2013). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Jones B. V., Begley M., Hill C., Gahan C. G. & Marchesi J. R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proceedings of the National Academy of Sciences of the United States of America 105, 13580–13585, 10.1073/pnas.0804437105 (2008). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Dethlefsen L., Huse S., Sogin M. L. & Relman D. A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS biology 6, e280, 10.1371/journal.pbio.0060280 (2008). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Turnbaugh P. J. et al.. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031, 10.1038/nature05414 (2006). [Abstract] [CrossRef] [Google Scholar]
- Bajaj J. S. et al.. Linkage of gut microbiome with cognition in hepatic encephalopathy. American journal of physiology. Gastrointestinal and liver physiology 302, G168–175, 10.1152/ajpgi.00190.2011 (2012). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- De Minicis S. et al.. Dysbiosis contributes to fibrogenesis in the course of chronic liver injury in mice. Hepatology 59, 1738–1749, 10.1002/hep.26695 (2014). [Abstract] [CrossRef] [Google Scholar]
- Miele L. et al.. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 49, 1877–1887, 10.1002/hep.22848 (2009). [Abstract] [CrossRef] [Google Scholar]
- Ruiz A. G. et al.. Lipopolysaccharide-binding protein plasma levels and liver TNF-alpha gene expression in obese patients: evidence for the potential role of endotoxin in the pathogenesis of non-alcoholic steatohepatitis. Obesity surgery 17, 1374–1380, 10.1007/s11695-007-9243-7 (2007). [Abstract] [CrossRef] [Google Scholar]
- Ritze Y. et al.. Lactobacillus rhamnosus GG protects against non-alcoholic fatty liver disease in mice. PloS one 9, e80169, 10.1371/journal.pone.0080169 (2014). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Aller R. et al.. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: a double blind randomized clinical trial. European review for medical and pharmacological sciences 15, 1090–1095 (2011). [Abstract] [Google Scholar]
- Loguercio C. et al.. Beneficial effects of a probiotic VSL#3 on parameters of liver dysfunction in chronic liver diseases. Journal of clinical gastroenterology 39, 540–543 (2005). [Abstract] [Google Scholar]
- Wong V. W. et al.. Treatment of nonalcoholic steatohepatitis with probiotics. A proof-of-concept study. Annals of hepatology 12, 256–262 (2013). [Abstract] [Google Scholar]
- Li Z. et al.. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology 37, 343–350, 10.1053/jhep.2003.50048 (2003). [Abstract] [CrossRef] [Google Scholar]
- Boursier J. et al.. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 63, 764–775, 10.1002/hep.28356 (2016). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Li Y., Xu C., Yu C., Xu L. & Miao M. Association of serum uric acid level with non-alcoholic fatty liver disease: a cross-sectional study. Journal of hepatology 50, 1029–1034, 10.1016/j.jhep.2008.11.021 (2009). [Abstract] [CrossRef] [Google Scholar]
- Nieto F. J., Iribarren C., Gross M. D., Comstock G. W. & Cutler R. G. Uric acid and serum antioxidant capacity: a reaction to atherosclerosis? Atherosclerosis 148, 131–139 (2000). [Abstract] [Google Scholar]
- Waring W. S., Webb D. J. & Maxwell S. R. Systemic uric acid administration increases serum antioxidant capacity in healthy volunteers. Journal of cardiovascular pharmacology 38, 365–371 (2001). [Abstract] [Google Scholar]
- Vijay-Kumar M. et al.. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231, 10.1126/science.1179721 (2010). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Vrieze A. et al.. The environment within: how gut microbiota may influence metabolism and body composition. Diabetologia 53, 606–613, 10.1007/s00125-010-1662-7 (2010). [Europe PMC free article] [Abstract] [CrossRef] [Google Scholar]
- Angulo P. Nonalcoholic fatty liver disease. The New England journal of medicine 346, 1221–1231, 10.1056/NEJMra011775 (2002). [Abstract] [CrossRef] [Google Scholar]
- Liu S. et al.. Polyomavirus Replication and Smoking Are Independent Risk Factors for Bladder Cancer After Renal Transplantation. Transplantation, 10.1097/TP.0000000000001260 (2016). [Abstract] [CrossRef] [Google Scholar]
- Johnson N. A. & George J. Fitness versus fatness: moving beyond weight loss in nonalcoholic fatty liver disease. Hepatology 52, 370–381, 10.1002/hep.23711 (2010). [Abstract] [CrossRef] [Google Scholar]
- Farrell G. C., Chitturi S., Lau G. K. & Sollano J. D. & Asia-Pacific Working Party on, N. Guidelines for the assessment and management of non-alcoholic fatty liver disease in the Asia-Pacific region: executive summary. Journal of gastroenterology and hepatology 22, 775–777, 10.1111/j.1440-1746.2007.05002.x (2007). [Abstract] [CrossRef] [Google Scholar]
- Zeng M. D. et al.. Guidelines for the diagnosis and treatment of nonalcoholic fatty liver diseases. Journal of digestive diseases 9, 108–112, 10.1111/j.1751-2980.2008.00331.x (2008). [Abstract] [CrossRef] [Google Scholar]
- Kunde S. S., Lazenby A. J., Clements R. H. & Abrams G. A. Spectrum of NAFLD and diagnostic implications of the proposed new normal range for serum ALT in obese women. Hepatology 42, 650–656, 10.1002/hep.20818 (2005). [Abstract] [CrossRef] [Google Scholar]
- Muyzer G., de Waal E. C. & Uitterlinden A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and environmental microbiology 59, 695–700 (1993). [Europe PMC free article] [Abstract] [Google Scholar]
- Wang B. et al.. Metabonomic profiles discriminate hepatocellular carcinoma from liver cirrhosis by ultraperformance liquid chromatography-mass spectrometry. Journal of proteome research 11, 1217–1227, 10.1021/pr2009252 (2012). [Abstract] [CrossRef] [Google Scholar]
- Zhang C. et al.. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. The ISME journal 4, 232–241, 10.1038/ismej.2009.112 (2010). [Abstract] [CrossRef] [Google Scholar]
Articles from Scientific Reports are provided here courtesy of Nature Publishing Group
Citations & impact
Impact metrics
Article citations
Zhi-Kang-Yin formula attenuates high-fat diet-induced metabolic disorders through modulating gut microbiota-bile acids axis in mice.
Chin Med, 19(1):145, 18 Oct 2024
Cited by: 0 articles | PMID: 39425211 | PMCID: PMC11490013
Protective effects of Nogo-B deficiency in NAFLD mice and its multiomics analysis of gut microbiology and metabolism.
Genes Nutr, 19(1):17, 24 Aug 2024
Cited by: 0 articles | PMID: 39182019 | PMCID: PMC11344411
Innovative treatments for obesity and NAFLD: A bibliometric study on antioxidants, herbs, phytochemicals, and natural compounds.
Heliyon, 10(16):e35498, 08 Aug 2024
Cited by: 0 articles | PMID: 39220898 | PMCID: PMC11365328
Review Free full text in Europe PMC
Gut microbes, diet, and genetics as drivers of metabolic liver disease: a narrative review outlining implications for precision medicine.
J Nutr Biochem, 133:109704, 17 Jul 2024
Cited by: 1 article | PMID: 39029595
Review
Vinpocetine and Lactobacillus improve fatty liver in rats: role of adiponectin and gut microbiome.
AMB Express, 14(1):89, 02 Aug 2024
Cited by: 0 articles | PMID: 39095672 | PMCID: PMC11297008
Go to all (179) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease.
Hepatobiliary Pancreat Dis Int, 16(4):375-381, 01 Aug 2017
Cited by: 248 articles | PMID: 28823367
An altered fecal microbiota profile in patients with non-alcoholic fatty liver disease (NAFLD) associated with obesity.
Rev Esp Enferm Dig, 111(4):275-282, 01 Apr 2019
Cited by: 31 articles | PMID: 30810328
Microbiome Signatures Associated With Steatohepatitis and Moderate to Severe Fibrosis in Children With Nonalcoholic Fatty Liver Disease.
Gastroenterology, 157(4):1109-1122, 27 Jun 2019
Cited by: 127 articles | PMID: 31255652 | PMCID: PMC6756995
Gut microbiota and non-alcoholic fatty liver disease.
Hepatobiliary Pancreat Dis Int, 14(6):572-581, 01 Dec 2015
Cited by: 36 articles | PMID: 26663004
Review