Abstract
Free full text
Tumor hypoxia causes DNA hypermethylation by reducing TET activity
Summary
Hypermethylation of tumor suppressor gene (TSG) promoters confers growth advantages to cancer cells, but how these changes arise is poorly understood. Here, we report that tumor hypoxia reduces the activity of oxygen-dependent TET enzymes, which catalyze DNA de-methylation through 5-methylcytosine oxidation. This occurs independently of hypoxia-associated alterations in TET expression, proliferation, metabolism, HIF activity or reactive oxygen, but directly depends on oxygen shortage. Hypoxia-induced loss of TET activity increases hypermethylation at gene promoters in vitro. Also in patients, TSG promoters are markedly more methylated in hypoxic tumors, independently of proliferation, stromal cell infiltration and tumor characteristics. Our data suggest cellular selection of hypermethylation events, with almost half of them being ascribable to hypoxia across tumor types. Accordingly, increased hypoxia after vessel pruning in murine breast tumors increases hypermethylation, while restored tumor oxygenation by vessel normalization abrogates this effect. Tumor hypoxia thus acts as a novel regulator underlying DNA methylation.
Mutational processes underlying oncogenesis are well studied. Apart from genetic changes, tumors are also epigenetically distinct from their tissue of origin. Most established are DNA methylation changes, but the mechanisms underlying these are poorly understood1.
In tumors, DNA methylation changes involve global hypomethylation, and local hypermethylation (HM) of CpG-rich gene promoters1. HM frequently affects tumor suppressor genes (TSGs), down-regulating their expression and thus contributing to oncogenesis. How methylation changes arise remains debated. Following an instructive model, genetic changes are a prerequisite for methylation changes2. For instance, BRAF mutations lead to HM in colorectal tumors3. A limitation of this model is that, while pervasive, HM of TSGs can be explained by somatic mutations in only a fraction of tumors. As a striking example, extensive HM was found in ependymomas devoid of somatic mutations4.
In contrast to methylation, DNA de-methylation mechanisms have remained elusive, until recently, when ten-eleven translocation methylcytosine dioxygenases (TET1, TET2 and TET3) were shown to oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC)5. 5hmC and its further oxidized derivatives are subsequently replaced with an unmodified C by base-excision repair to achieve de-methylation6. Reduced 5mC oxidation due to decreased TET activity thus increases DNA methylation. Mutations suppressing TET activity and thus reducing 5hmC are often found in myeloid leukemia and glioblastoma6–9, but less frequently in other tumor types. In contrast, 5hmC loss is pervasive in tumors and even proposed as a cancer hallmark10. Thus, similar to HM, somatic mutations explain the loss of 5hmC in only a fraction of tumors, and it remains unclear which other factors trigger this loss2.
Interestingly, TET enzymes are Fe2+ and α-ketoglutarate-(αKG)-dependent dioxygenases, similar to HIF-prolyl-hydroxylase domain proteins (PHDs)11. The latter are sensitive in their activity to oxygen and act as oxygen sensors: under normoxic conditions PHDs hydroxylate the HIF transcription factors, targeting them for proteasomal degradation, whereas under hypoxia they fail to hydroxylate, leading to HIF stabilization and hypoxia response activation12. Expanding tumors continuously become disconnected from their vascular supply, resulting in vicious cycles of hypoxia followed by HIF activation and tumor vessel formation13. Consequently, hypoxia pervades in solid tumors, with oxygen levels ranging from 5% to anoxia, and about a third of tumor areas containing <0.5% oxygen14. Although DNA HM and hypoxia are well-recognized cancer hallmarks, the impact of hypoxia on TET hydroxylase activity and subsequent DNA (de)methylation has not been assessed. We here hypothesize that a hypoxic micro-environment decreases TET hydroxylase activity in tumors, leading to an accumulation of 5mC and acquisition of HM.
Impact of hypoxia on DNA hydroxymethylation activity
To assess whether hypoxia affects TET activity, we exposed 10 human and 5 murine cell lines with detectable 5hmC levels for 24 hours to 21% or 0.5% O2, a level commonly observed in tumors14. Hypoxia induction was verified and DNA was extracted and profiled for nucleotide composition using LC/MS. 11 cell lines, including eight cancer cell lines, displayed 5hmC loss (Figure 1a). However, this did not translate into global 5mC increases (Extended data figure 1), presumably because 5mC is more abundant and at many sites not targeted by TETs15. The effect of hypoxia was concentration- and time-dependent: a dose-response revealed gradual reductions from 1-2% O2 onwards and a time course respectively, a 20% and 40% reduction after 15 and >24 hours (Figure 1b-c). Loss of 5hmC was not secondary to increased 5hmC oxidation to 5fC16, as hypoxia also decreased 5fC levels in ES cells (Extended data figure 1).
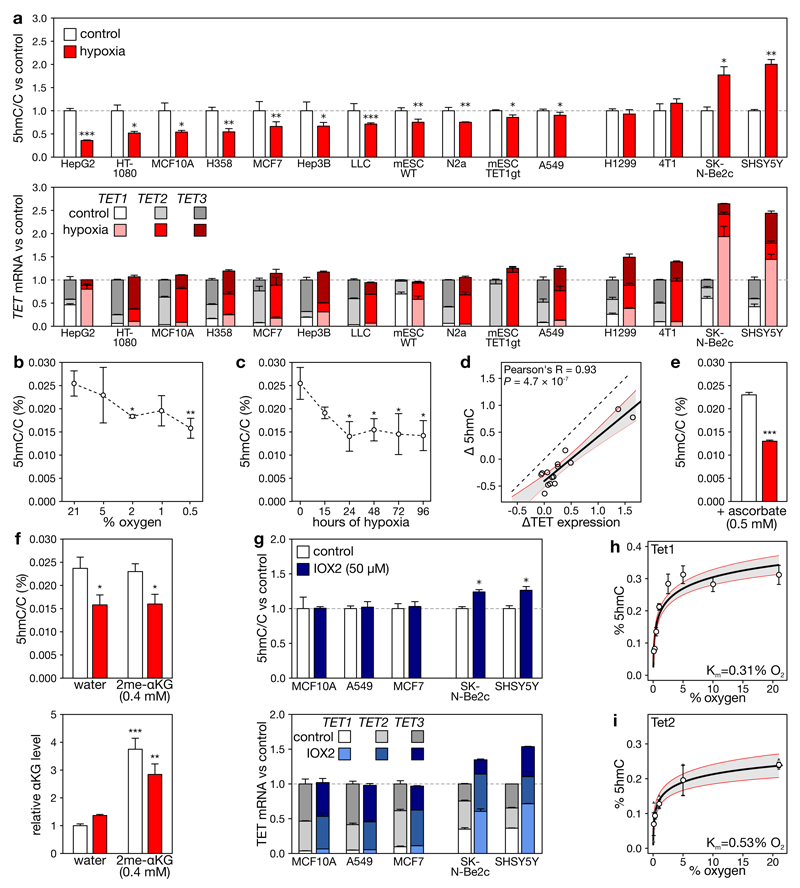
a, Levels of 5hmC (upper), and overall TET expression (lower) in cell lines grown for 24 h under 21% or 0.5% O2. RNA expression is expressed relative to the combined estimated level of all 3 TET paralogues under 21% O2., b-c, 5hmC/C levels in MCF7 cells exposed to different O2 levels for 24 h (b), or 0.5% oxygen for indicated times (c). d, Correlation of changes in overall TET expression and 5hmC upon hypoxia. Each circle represents a cell line, the full line the correlation. e-f, Levels of 5hmC (e, f) and α-ketoglutarate (f) in MCF7 cells grown with ascorbate (e), water or dimethyl-α-ketoglutarate (f) under 21% or 0.5% O2 (white or red). α-ketoglutarate changes are relative to matching water controls. g, As (a), but for cells exposed to IOX2. h-i, Michaelis-Menten curve of Tet1 (h) and Tet2 (i, n=3) for O2. Error bars denote s.e.m., grey areas: 95% c.i., n = 5 replicates for panels (a-h), *P<0.05, **P<0.01, ***P<0.001 by t-test (b, c, e) or ANOVA with post-hoc Tukey HSD (f).
In some cell lines, 5hmC failed to decrease under hypoxia. Particularly, 5hmC was unaffected in H1299 and 4T1, and even increased in SHSY5Y and SK-N-Be2c neuroblastoma cells, as reported previously17 (Figure 1a). When profiling TET expression, neuroblastoma cells displayed potent hypoxia-induction of TET1 and TET2, H1299 and 4T1 exhibited intermediate increases, and all other cell lines no or only modest increases of some TET paralogues (Figure 1a). Tet expression changes were confirmed at the protein level in murine cell lines, and HIF1β-ChIP-seq further confirmed that HIF binds near the promoters of TETs that are upregulated, but not near those that are unaltered (Extended data figure 2a-b), in keeping with the cell-type specificity of the hypoxia response12. Importantly, no cell line showed decreased TET expression, indicating that 5hmC loss is not due to reduced TET expression.
Since hypoxia differentially affects TET expression, we correlated hypoxia-associated changes in overall TET expression (the combined abundances of TET1, TET2 and TET3) with changes in 5hmC levels. Hypoxia reduced 5hmC on average by 44% (P=0.0097) in each cell line (Figure 1d), independently of TET expression changes. Nevertheless, changes in TET expression also determined 5hmC levels. This was confirmed by siRNA knockdown of TET2, which constitutes ~60% of all TET expression in MCF7 cells: this reduced 5hmC levels also by ~60% (Extended data figure 2c). Likewise, Tet1-KO ES cells displayed lower 5hmC levels than wild-type ES cells, in which Tet1 is the predominantly expressed Tet paralogue, both under 21% or 0.5% O2 (Figure 1a, Extended data figure 2d).
Hence, 5hmC levels after hypoxia appear to be determined by altered oxygen availability and by changes in TET abundance. This explains why cell lines without hypoxia-induced upregulation of TETs display 5hmC loss, whereas cell lines strongly upregulating TETs compensate this, resulting in equal or increased 5hmC levels.
Changes secondary to hypoxia do not affect DNA hydroxymethylation
Apart from gene expression, TET activity is affected by a variety of cellular processes, including changes in reactive oxygen species (ROS), Krebs cycle metabolites and proliferation7,11,17,18. Since such changes might also occur secondary to hypoxia, we investigated whether they underlie 5hmC reductions in hypoxia.
Firstly, ROS could affect TETs in the nucleus through inactivation of Fe2+ in their catalytic domain. Although ROS was overall increased upon hypoxia, no increase in nuclear ROS was detected by a nucleus-specific ROS probe or 8-oxo-guanine quantification (Extended data figure 3a-f). Ascorbate supplementation to counteract ROS increases19, moreover failed to rescue 5hmC loss (Figure 1e).
Secondly, changes in metabolites such as succinate and fumarate affect TET function by competing with its cofactor αKG7. The concentration of these metabolites was however not increased in hypoxic MCF10A or ES cells, and only 3-4-fold in MCF7 cells (Extended data figure 3g-i). The onco-metabolite 2-hydroxyglutarate was also increased in hypoxic MCF7 and MCF10A cells, but levels were only ~5-10% of αKG (Extended data figure 3h,j), and therefore unlikely to affect TET activity, as affinity of these competing metabolites for hydroxylases is lower or similar to αKG7,20. Indeed, culturing MCF7 cells in glutamine-free medium to decrease these metabolite concentrations did not alter 5hmC levels (Extended data figure 3k). Exogenously adding cell-permeable αKG under hypoxia to counteract putative competing metabolites likewise did not rescue the 5hmC loss (Figure 1f). This excludes that metabolite competition underlies hypoxia-associated 5hmC loss.
Thirdly, increases in cell proliferation have been linked to 5hmC loss21. However, cell growth was unaffected or decreased upon exposure to hypoxia in all cell lines tested, indicating that increased proliferation does not underlie 5hmC reduction (Extended data figure 3l).
Fourthly, to exclude cellular changes secondary to HIF activation, we pharmacologically activated the hypoxia response program by exposing 5 cell lines grown in atmospheric conditions to IOX2, a small molecule inhibitor displaying high specificity for PHDs22 (Extended data figure 3m). Cell lines not characterized by hypoxia-induced TET expression changes (i.e., MCF10A, A549 and MCF7) showed no change in 5hmC under IOX2, while SK-N-Be2c and SHSY5Y, characterized by TET upregulation, did show an increase in 5hmC (Figure 1g). Thus, upon IOX2 exposure, 5hmC changes mirrored changes in TET transcription. We also prepared nuclear protein extracts from MCF7 cells grown under hypoxic and atmospheric conditions, and then compared their 5mC oxidative capacities at the same oxygen tension in vitro; these were however identical (Extended data figure 3n). Loss of 5hmC was therefore not secondary to activation of the hypoxia response program.
In a final experiment, we assessed the effect of varying oxygen concentrations on the activity of recombinant purified Tet1 or Tet2, by measuring conversion of 5mC to 5hmC on double-stranded genomic DNA. We observed a dose-dependent loss of 5hmC production with decreasing oxygen concentration. Importantly, under the hypoxic conditions applied in this study (0.5% O2), Tet1 and Tet2 activity were reduced by 45±7% and 52±8% (P=0.01; Figure 1h-i).
Together, these data demonstrate that decreased oxygen availability directly diminishes the oxidative activity of TETs, independently of changes in HIF activity, competing metabolites, proliferation, nuclear ROS or TET expression.
Genomic loci displaying differential DNA hydroxymethylation
To analyze where in the genome hypoxia reduces 5hmC, DNA from hypoxic and control MCF7 cells was immunoprecipitated using antibodies targeting 5mC or 5hmC, and subjected to high-throughput sequencing (DIP-seq). We detected 290,382 sites enriched for 5hmC. Upon hypoxia, 10,001 of these peaks exhibited a decrease in 5hmC (5% FDR), versus only 18 exhibiting an increase, thereby confirming the global 5hmC loss (Figure 2a; Supplementary table 1). Genomic annotation of these peaks using chromHMM23 revealed they were predominantly found at gene promoters, but also at enhancers and actively transcribed regions, in line with known TET binding (Figure 2b)15. For example, 5hmC was decreased near transcription start sites of NSD1, FOXA1 and CDKN2A (Extended data figure 4). Analysis of 5mC-DIP signals at these 10,001 regions highlighted that, in 724 out of 875 altered regions at P<0.05, the 5mC content was increased, although only 1 of these sites survived 5% FDR correction (Figure 2c; Supplementary table 2). Increases in 5mC were thus more subtle than decreases observed for 5hmC.
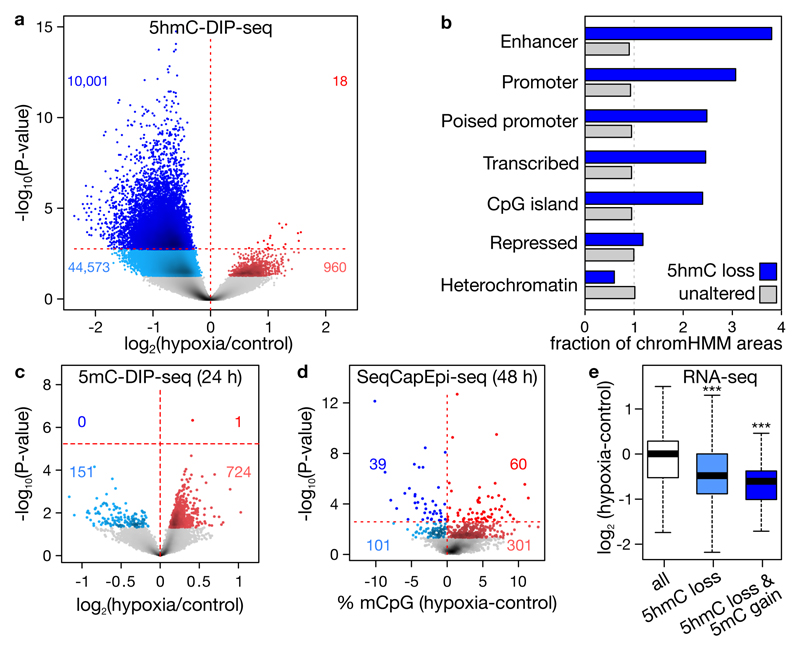
a, Changes in 5hmC at 290,382 peaks detected using 5hmC-DIP-seq. Peaks gaining and loosing 5hmC (red and blue) are highlighted at P<0.05 and 5% FDR adjustment (lighter and darker). b, Observed/expected fraction of 5hmC peaks overlapping with chromHMM chromatin states and exhibiting hypoxia-associated 5hmC loss (n=10,001, blue) or not (n=280,381, grey). c-d, Changes in 5mC after 24 (c) or 48 (d) hours of 0.5% O2, assessed by 5mC-DIP-seq at 10,001 hypohydroxymethylated peaks upon hypoxia (c) or by BS-seq at 1,894 regions capture-selected using SeqCapEpi (d). e, Expression changes of genes in hypohydroxymethylated, and both hypohydroxymethylated and hypermethylated peaks. Plots depict 3 (a, e), 4 (c) or 5 (d) replicates, P-values by negative binomial generalized linear models (a, c), Fisher’s exact (d) or t-test (e, ***P<0.001).
Several days may be required for 5hmC changes to cause 5mC changes19. We therefore cultured cells for 48 (instead of 24) hours under hypoxia, and used targeted bisulfite-sequencing (BS-seq) to obtain base-resolution quantitation of 5mC at ~85Mb of promoters and enhancers. Using this approach, we could assess increases in 5mC for 1,894 of the 10,001 regions displaying 5hmC loss. As observed upon 5mC-DIP-seq, out of 402 altered sites (P<0.05), 301 displayed increased methylation. Likewise, 60 out of 99 altered sites at 5% FDR were increased (P=2.8×10-3; Figure 2d; Supplementary table 3). ChromHMM annotation revealed that these 60 sites were predominantly in gene promoters and enhancers. To assess the impact of HM on gene expression, we performed RNA-seq on hypoxic MCF7 cells. Genes depleted in 5hmC and at the same time increased in 5mC, were characterized by decreased expression upon hypoxia (Figure 2e; P=2.5×10-42 and 7.4×10-4, respectively for 3,660 genes with 5hmC loss and 55 genes with both 5hmC loss and 5mC gain; Supplementary table 4). Reduced TET activity thus leads to an accumulation of 5mC, decreasing expression of associated genes.
Selection of HM events in hypoxic tumors
We next analyzed whether 5hmC loss and concomitant 5mC gain also occur in vivo. We focused on gene promoters as they are more frequently affected upon hypoxia, and directly linked to gene expression. Moreover, as cancer cells go through multiple rounds of sustained hypoxia14, we hypothesized that changes in 5mC might be enriched for, as they provide a substrate for cellular selection of cancer cells, similar to somatic mutations. First, we assessed 5hmC levels in three patient-derived tumor xenografts, wherein we marked hypoxic areas with pimonidazole (Extended data figure 5a). Immunofluorescence analysis revealed decreased 5hmC in hypoxic areas, linking tumor hypoxia to 5hmC loss in vivo.
To model whether hypoxia-associated HM contributes to the oncogenic process, we analyzed tumors profiled in the pan-cancer study of The Cancer Genome Atlas (TCGA)24. We selected 8 solid tumor types (3,141 tumors) for which both DNA methylation (450K array) and gene expression (RNA-seq) data were available for >100 samples, and classified each as hypoxic, normoxic or intermediate using an established gene signature (Extended data figure 5b)25. Next, we analyzed tumor-associated DNA HM in each tumor type by performing unsupervised clustering of 1,000 CpGs that displayed the strongest HM in tumor versus normal tissue (Extended data figure 5c). In the 3 first clusters, displaying low, intermediate and high average HM, we analysed the enrichment of hypoxic tumors. For all 8 tumor types, hypoxic tumors predominated in the hypermethylated cluster and normoxic tumors in the hypomethylated cluster (Figure 3a; P=2×10-4), suggesting that hypoxia leads to increased methylation in tumors.

a, Observed/expected number of hypoxic versus normoxic tumors in 3 methylation clusters for 1,000 CpGs hypermethylated in tumor versus normal tissue. b, Percentage of HM events in promoters of frequently HM genes. n = 3,141 tumors, *P<0.05, **P<0.01, ***P<0.001 by Cochran-Armitage (a), generalized linear model per tumor type corrected for co-variates (Supplementary table 8) (b).
Whereas the above analysis identifies uniform increases in methylation based on average changes, it poorly captures exceptional increases in HM known to occur in a subset of tumors1,26. We therefore also modeled tumor HM by annotating increases in CpG methylation at gene promoters using a stringent threshold (Bonferroni-corrected P<0.05) as HM events. In each tumor type the promoters of 187±38 out of 29,649 genes frequently displayed HM events (Supplementary table 5). Importantly, hypoxic tumors had on average 4.8-fold more HM events in these genes than normoxic tumors (Figure 3b; P=4.1×10-13). These events were functional, reducing gene expression in tumors carrying these HM events (Extended data figure 5d). They primarily affected promoters with a high or intermediate CpG content, in line with TET target preference (Extended data figure 5e)15. Furthermore, they were not restricted to a small subset: 77±6.5%, 49±9.3% or 39±9.1% of hypoxic tumors was affected by ≥1, ≥10 or ≥20 HM events. When considering HM frequencies in normoxic tumors as baseline, up to 48% of HM events were hypoxia-related.
As HM can also be genetically-encoded, mutations in some genes correlated positively with HM (e.g. IDH1, TET1, TET3 and BRAF; Supplementary table 6). Importantly, hypoxia predicted HM independently of the mutation status (P=6.1×10-12). Mutations inhibiting TET activity were moreover infrequent (~1.8% of tumors), indicating that HM is not genetically-encoded in most tumors. TET-mutant tumors were also not more hypoxic, suggesting that hypoxia induces HM, and not vice versa (Extended data figure 5f). Hypoxia-associated HM events occurred independently of other tumor characteristics, such as tumor percentage, immune cell infiltration, tumor size, proliferation or metastasis (P=4×10-13), and were significant in 7 of 8 tumor types (Supplementary tables 7-8). In line with an earlier report21, high proliferation was the only other variable significantly predicting HM (P=5.3×10-10), although only in 4 of 8 tumor types (Extended data figure 5g-h). Using multiple regression, we estimated contributions of tumor characteristics to HM variance. Based on partial correlation coefficients, proliferation predicted 12.1±4.1% and hypoxia 33.3±5.7% of HM events explained by the model (Extended data figure 5i).
Given the enrichment of HM events in hypoxic tumors, we next selected genes enriched for HM events in hypoxic versus normoxic tumors (5% FDR). This revealed 263±94 genes per tumor type, with 9.0±1.6% being shared between any 2 types (Supplementary table 9). Ontology analysis of hypermethylated genes revealed common biological processes, such as cell cycle arrest, DNA repair and apoptosis. In line with tumor hypoxia inducing glycolysis, angiogenesis and metastasis, HM was also observed in genes suppressing these processes (Extended data figure 6a-c).
Reduced TET activity underlies HM
Three strategies were used to confirm the role of TET activity in hypoxia-associated HM. First, we correlated TET expression with HM events, while correcting for hypoxia and proliferation. TET2 and TET3 expression correlated inversely with HM (P=0.046 and 0.0028, Extended data figure 7a), as did hypoxia and proliferation (P<1.2×10-13 for both). Similar to our in vitro observations, this implicates reduced TET activity in HM.
Secondly, we assessed the overlap of HM events induced by hypoxia and IDH1R132 mutations8 in 63 glioblastomas. Among IDH1-wildtype glioblastomas, the HM frequency was 3.4-fold higher in hypoxic tumors (Figure 4a, Extended data figure 7b). As expected, IDH1R132 tumors showed HM, albeit 3.9-fold more than hypoxic tumors (Figure 4a), indicating that TET enzymes, being fully inactivated in IDH-mutant tumors9, were only partially inactivated in hypoxia, similar to our in vitro observations. Of 228 genes frequently hypermethylated in glioblastomas, hypermethylated genes in the hypoxic and IDH-mutant subgroups displayed a 58% overlap (P<10-16; Figure 4b) and a reduced expression (Extended data figure 7c), indicating that loss of TET activity affects the same genes, regardless of the underlying trigger.

a, HM in 19 normoxic (blue), 21 intermediate (grey), 17 hypoxic (red) and 6 IDH1R132-mutated (yellow) glioblastomas. b, Overlap between genes hypermethylated in hypoxic versus IDH1R132-mutated glioblastomas. c-d, (c) 5hmC measured across 485,000 CpGs in 12 normoxic versus 12 hypoxic non-small-cell lung tumors, and (d) changes in 5(h)mC for unaltered CpGs (grey), and CpGs altered in both 5mC and 5hmC (25% FDR, blue; P<0.01, red). ***P<0.001 by Fisher’s exact (a), **P<0.01 by t-test (c).
Finally, to link hypoxia-associated HM to 5hmC loss, we profiled 24 non-small cell lung tumors for 5mC and 5hmC using 450K arrays (Extended data figure 7d). This revealed a generalized loss of 5hmC in hypoxic tumors (-7.1±1.1%; P=3.7×10-3; Figure 4c). Also individual probes mostly displayed 5hmC loss and 5mC gain in hypoxic tumors (respectively, 96.7% and 65.4% of probes altered at P<0.01; Supplementary table 10). Of all probes displaying 5mC gain, most (87%) also displayed 5hmC loss, and of probes altered both in 5hmC and 5mC (P<0.01), 92% showed 5hmC loss and 5mC gain (Figure 4d; P<10-16). This directly implicates hypoxia-induced loss of 5hmC in HM of hypoxic tumors.
Rescue and exacerbation of hypoxia-induced HM in murine breast tumors
To manipulate tumor oxygenation and confirm its impact on HM, we used mice expressing the polyoma middle T-antigen under the mouse mammary tumor virus promoter (MMTV-PyMT). These mice spontaneously develop breast tumors, with hypoxic areas emerging from 7 weeks onwards, encompassing ~20% of tumor at 16 weeks27. Hypoxic areas in these tumors were also depleted in 5hmC (Figure 5a-b).
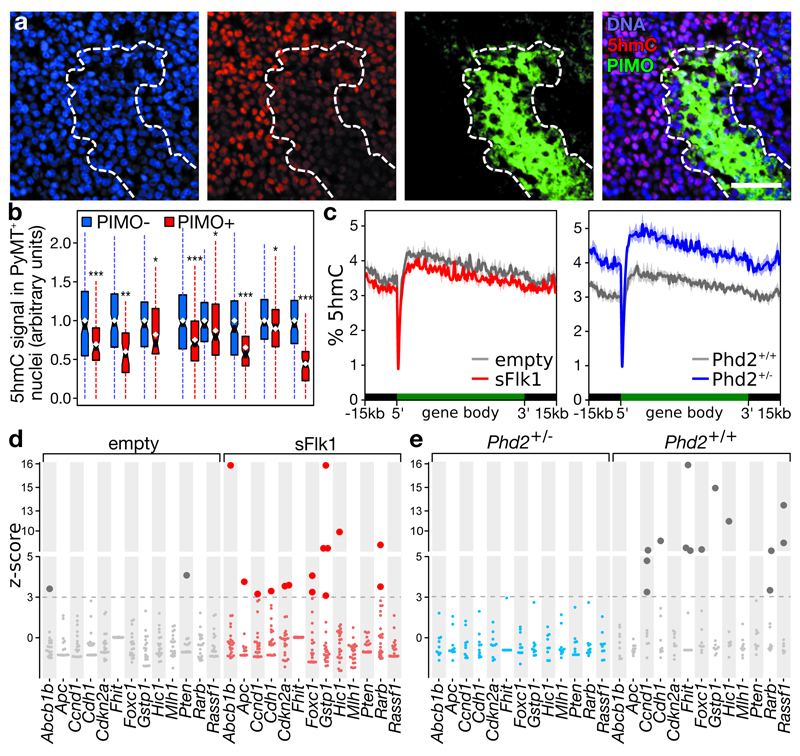
a-b, Immunofluorescence of breast tumors in tg(MMTV-PyMT) mice. (a) Representative image, scale: 50 μm. (b) Boxplot of 5hmC signal in >150 PyMT-positive nuclei from 8 tumors, stratified for pimonidazole (yes/no) and normalized to pimonidazole-negative nuclei. c, 5hmC levels ± s.e.m. across a metagene in tumors of 12-week-old mice receiving empty or sFlk1-overexpressing plasmid (left, n=3), or 16-week-old mice with the indicated genotype (right, n=3 for Phd2+/+; n=4 for Phd2+/-). d-e, HM in (d) tumors developing in 12-week-old mice receiving empty (n=19) or sFlk1-overexpressing plasmid (n=24) 3 weeks earlier, and in (e) tumors developing in 16-week-old Phd+/- (n=10) and Phd+/+(n=9) mice. Plotted are z-scores of HM, relative to normoxic tumors (empty and Phd2+/- for d and e). Dotted line: 5% FDR, darker dots: significant HM. Brca1 and Timp3: not shown (no HM event detected). Hypermethylated genes on average had 5.8% (d) and 4.7% (e) more methylation. *P<0.05, **P<0.01, ***P<0.001 by t-test.
We monitored HM changes by targeted BS-seq of TSG promoters commonly inactivated in cancer28. Hypoxic human breast tumors indeed display a specific increase in HM at these TSG promoters, whereas no effect was observed for oncogenes (Extended data figure 8a). In line with the age-associated increase in tumor hypoxia27, HM events increased dramatically with age or tumor size, but not in normal mammary glands (Extended data figure 8b-d). Importantly, >95% of cells in these tumors were PyMT-positive, whereas cell proliferation and immune cell infiltration were comparable between hypoxic and normoxic areas (Extended data figure 8e-g). HM changes are therefore unlikely secondary to changes in proliferation or cellular heterogeneity.
To test whether reduced tumor oxygenation increases HM, 9-week-old MMTV-PyMT mice were hydrodynamically injected with a soluble-Flk1 (sFlk1)-expressing plasmid. After 3 weeks, this caused tumor vessel pruning and hypoxia (Extended data figure 9a-d). Shallow whole-genome sequencing for 5hmC (TAB-seq) revealed a global loss of 5hmC upon sFlk1 overexpression (-12.4±3.5%, P=0.040), predominantly at gene-dense regions and affecting the entire gene (Figure 5c, Extended data figure 9e), consistent with previously described 5hmC distributions15. Moreover, targeted BS-seq revealed an exacerbated HM phenotype after sFlk1 overexpression at 12 weeks, and this in TSGs but not oncogenes (10 out of 15 TSGs contained ≥1 HM event; P=0.010, Figure 5d, Extended data figure 9f). Tumor growth and expression of proliferation markers, Tet paralogues and the immune cell marker CD45 were unaffected by sFlk1 overexpression, indicating that HM occurs independently (Extended data figure 9g-j).
To rescue this effect, we normalized the tumor vasculature by intercrossing a heterozygous Phd2 loss-of-function allele with the PyMT transgene. This significantly reduced tumor hypoxia at 16 weeks27 (Extended data figure 9k). TAB-seq revealed a 5hmC gain (+25.3±4.7%, P=0.0098), primarily at gene-dense regions and affecting the entire gene (Figure 5c, Extended data figure 9l). Interestingly, BS-seq revealed that, whereas 8 out of 15 TSGs displayed ≥1 HM event in Phd2+/+ tumors, no HM was observed in Phd2+/- tumors (P=2.6×10-7, Figure 5e). Again, oncogenes were unaffected (Extended data figure 9m). Importantly, effects were independent of Phd2 haplodeficiency in tumor cells, as similar effects were observed in PyMT mice having endothelial-cell-specific Phd2 haplodeficiency (Extended data figure 9n-o)27. Like the sFlk1 model, also increasing tumor oxygenation by Phd2 haplodeficiency did not affect tumor growth, expression of proliferation markers, Tets or CD45 (Extended data figure 9p-u).
Discussion
We here show that tumor hypoxia directly reduces TET activity, causing a 5hmC decrease predominantly at gene promoters and enhancers. Concomitantly, 5mC increases at these sites, and, similar to genetic mutations, becomes a substrate for oncogenic selection in vivo26. Since hypoxia prevails in tumors, 5mC changes in TSG promoters are enriched for, rendering hypoxic tumors hypermethylated at these sites. HM events in tumors have long been suspected to occur through selection of random DNA methylation variants29. However, the identification of genetically-encoded HM challenged this stochastic model2. By demonstrating that hypoxia drives HM, we show that genetically-encoded and tumor microenvironment-driven models of epimutagenesis co-exist. However, since hypoxia is pervasive, the mechanism described here is relevant for most solid tumors: up to 48% of HM events was hypoxia-related, and effects were replicated in all tumor types investigated, independently of mutation- and proliferation-induced HM. Importantly, modest hypoxia (2-5% O2) did not affect TET activity, indicating that TET enzymes are not physiological oxygen sensors like the PHDs, as reported30. TET activity only becomes limiting under pathophysiological oxygen concentrations found in tumors14, and analogous to somatic TET haploinsufficiency, this partial reduction in TET activity contributes to oncogenesis. Our findings also suggest intriguing avenues of investigation into other ischemia-related pathologies.
Our model provides an elegant mechanism for the association between hypoxia and (mal)adaptive oncogenic processes: genes affected by HM were involved in cell-cycle arrest, DNA repair and apoptosis, but also glycolysis, metastasis and angiogenesis. Interestingly, high levels of angiogenesis inhibitors stimulate metastatic spreading in murine cancer models, at least in specific settings31, and tumor hypoxia is considered a driver of this behavior. The mechanism described here, by which HM accumulates under hypoxia, may underlie these escape mechanisms. Contrastingly, low levels of VEGF inhibition can induce tumor vessel normalization and improve oxygenation32. Our observations in normalized PyMT tumors suggest that therapeutic benefits of vessel normalization, such as decreased metastatic burden27, might occur by inhibiting hypoxia-associated HM. Countering this HM, for instance through drugs inhibiting DNA methylation and/or by normalizing tumor blood supply, may thus prove therapeutically beneficial.
Methods
Materials
All materials were molecular biology grade. Unless noted otherwise, all were from Sigma (Diegem, Belgium).
Analysis of global 5mC and 5hmC levels in cultured cells
Cell lines
MCF7, MCF10A, A549, H1299, SHSY5Y, Hep G2, Hep 3B2, HT-1080, NCI-H358, LLC, Neuro-2a, 4T1 and SK-N-BE2c cells lines were obtained from the American Type Culture Collection and their identity was not further authenticated. These are not listed in the database of commonly misidentified cell lines maintained by ICLAC. LLC, Neuro-2a, 4T1, Hep G2, HT-1080, Hep 3B2, MCF7 and A549 cells were cultured at 37°C in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), 5ml of 100 U/ml Penicillin-Streptomycin (Pen Strep, Life Technologies) and 5ml of L-Glutamine 200mM. NCI-H358, H1299 and SK-N-BE2c cell lines were cultured at 37°C in Roswell Park Memorial Institute (RPMI) 1640 Medium (RPMI) 10% FBS 1% Pen Strep and 1% L-Glutamine. MCF10A cells were cultured at 37°C in DMEM/F-12 (Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12) supplemented with 5% horse serum (Life Technologies), 20 ng/ml human Epidermal Growth Factor (Prepotec), 0.5 μg/ml hydrocortisone, 100ng/ml cholera toxin, 10 μg/ml insulin, and 100 U/ml Pen Strep. The SHSY5Y cell line was cultured at 37°C in DMEM/F-12 supplemented with 10% FBS, 2% (PenStrep) and 1% Non Essential Amino Acids (MEM). Mouse J1 ES cells were cultured feeder-free in fibroblast-conditioned medium. Cell cultures were confirmed to be mycoplasma-free every month.
Cell line treatment conditions
Control cell cultures were grown at atmospheric oxygen concentrations (21%) with 5% CO2. To render cultures hypoxic, they were incubated in an atmosphere of 0.5% oxygen, 5% CO2 and 94.5% N2. Where indicated, IOX2 (50 µM), ascorbate (0.5 mM, a dose known to support TET activity19) or dimethyl α-ketoglutarate (0.5 mM) were added to fresh culture medium, using an equal volume of the carrier (DMSO) as a control for IOX2. Cells were plated at a density tailored to reach 80-95% confluence at the end of the treatment. Fresh medium was added to the cells just before hypoxia exposure. For glutamine-free culture experiments, dialysed FBS was added to glutamine-free DMEM, and supplemented with glutamine (4 mM) for the control. Mouse J1 ES cells and Tet1-gene-trap ES cells were cultured feeder-free in fibroblast-conditioned medium.
DNA extraction
After exposure to the aforementioned stimuli, cultured cells were washed on ice with ice-cold phosphate-buffer saline (PBS) with deferoxamin (PBS-DFO, 200 µM), detached using cell scrapers and collected by centrifugation (400 ×G, 4°C). Nucleic acids were subsequently extracted using the Wizard Genomic DNA Purification (Promega, Leiden, The Netherlands) kit according to instructions, with all buffers supplemented with DFO (200 µM), dissolved in 80 µL PBS-DFO with RNAse A (200 units, NEB, Ipswich, MA, USA), incubated for 10 minutes at 37°C. After proteinase K addition (200 units) and incubation for 30 minutes at 56°C, DNA was purified using the QIAQuick blood and tissue kit (all buffers supplemented with DFO), eluted in 100 µL of a 10 mM Tris, 1mM EDTA solution (pH 8) and stored at -80°C until further processing.
LC-ESI-MS/MS of DNA to measure 5mC, 5hmC and 8-oxoG levels
To measure the cytosine, 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and 8-oxo-7,8-dihydroguanine (8-oxo-G) content of DNA samples, three technical replicates were run for each sample. More specifically, 0.5 to 2 µg DNA in 25 µL H2O were digested as follows: an aqueous solution (7.5 µL) of 480 μM ZnSO4, containing 42 units Nuclease S1, 5 units antarctic phosphatase, and specific amounts of labeled internal standards were added and the mixture was incubated at 37 °C for 3 h in a Thermomixer comfort (Eppendorf). After addition of 7.5 μL of a 520 µM [Na]2-EDTA solution containing 0.2 units snake venom phosphodiesterase I, the sample was incubated for another 3 h at 37 °C. The total volume was 40 µL. The sample was then kept at -20 °C until the day of analysis. Samples were then filtered by using an AcroPrep Advance 96 filter plate 0.2 μm Supor (Pall Life Sciences) and then analyzed by LC-ESI-MS/MS, which are performed using an Agilent 1290 UHPLC system and an Agilent 6490 triple quadrupole mass spectrometer coupled with the stable isotope dilution technique. DNA samples were digested to give a nucleoside mixture and spiked with specific amounts of the corresponding isotopically labeled standards before LC-MS/MS analysis. The nucleosides were analyzed in the positive ion selected reaction monitoring mode (SRM). In the positive ion mode, [M+H]+ species were measured.
Determination and comparison of nucleoside concentrations
The resulting cytosine, 5mC, 5hmC and 8-oxo-G peak areas were normalized using the isotopically labeled standards, and expressed relative to the total cytosine content (i.e. C + 5mC + 5hmC). Concentrations were depicted as averages of independent replicates grown on different days, and compared between hypoxia and normoxia (21% O2), or between control and treated conditions, using a paired Student’s t-test. No statistical methods were used to predetermine sample size.
TET mRNA concentrations and hypoxia marker gene induction
RNA extraction, cDNA synthesis and qPCR
For RNA extractions, cell culture medium was removed, TRIzol (Life Technologies) added and processed according to manufacturers guidelines. Reverse transcription and qPCR were performed using 2× TaqMan® Fast Universal PCR Master Mix (Life Technologies), TaqMan probes and primers (IDT, Leuven, Belgium), whose sequence is available under Supplementary table 12. Thermal cycling and fluorescence detection were done using a LightCycler 480 Real-Time PCR System (Roche). Taqman assay amplification efficiencies were verified using serial cDNA dilutions, and estimated to be >95%.
mRNA concentration analysis and statistics
Ct values were determined for each sample and gene of interest in technical duplicates, and normalized according to the corresponding amplification efficiency. Per sample, TET expression was expressed relative to β-2-microglobulin (human) or Hypoxanthine Phosphoribosyltransferase 1 (mouse) levels by subtraction of their average Cts. Concentrations were expressed as averages of at least 5 replicates extracted on different days. For Figure 1a, copy number estimates for TET1, TET2 and TET3 were expressed for each cell line, relative to the summed copy number estimates of TET1, TET2 and TET3 under control conditions (21% O2). Concentrations were compared between hypoxia and normoxia, or between control and treatment conditions using a Student’s t-test. No statistical methods were used to predetermine sample size.
Hypoxia marker gene induction
To further verify induction of the hypoxia response program, hypoxia marker gene expression was verified. We analyzed mRNA levels of genes encoding the E1B 19K/Bcl-2-binding protein Nip3 (BNIP3) and fructose-bisphosphate aldolase (ALDOA), 2 established hypoxia marker genes33. RT-qPCR was performed as described for the TET mRNA concentration assays, and differential expression was calculated using the ΔΔCt method34. We moreover excluded that the increase in HIF1α protein concentrations was secondary to a transcriptional upregulation, by assessing HIF1A mRNA expression in parallel. mRNA concentrations were expressed relative to normoxic controls (21% O2). Differences in mRNA concentration were assessed using a Student’s t-test on 5 or more independent replicates grown on different days.
Validation of hypoxia induction and Tet protein expression
Western blotting for Hif1α, Tet1, Tet2 and Tet3
To assess HIF1α protein stabilization, proteins were extracted from cultured cells as follows: cells were placed on ice, and washed twice with ice-cold PBS. Proteins were extracted with extraction buffer (50 mM Tris HCl, 150 mM NaCl, 1% Triton X-100, 0.5% Na-deoxycholate and 0.1% SDS) with 1× protease inhibitor cocktail. Protein concentrations were determined using a bicinchoninic acid protein assay (BCA, Thermo Scientific) following the manufacture’s protocol, and an estimated 60 µg protein was loaded per well on a NuPAGE Novex 3-8% Tris-Acetate Protein gel (Life Technologies), separated by electrophoresis and blotted on polyvinylidene fluoride membranes. Membranes were activated with methanol and washed, and incubated with antibodies targeting β-actin (4967, Cell Signaling), Tet1 (09-872, Millipore) and Tet3 (61395, Active Motif), at 1:1000 dilution, targeting Tet2 (124297, Abcam) at 1:250 dilution, and targeting HIF-1α (C-Term) (Cayman Chemical Item 10006421) at 1:3000 dilution. Secondary antibodies and detection were according to routine laboratory practices. Western blotting was done on 6 independent replicates grown on different days.
Analysis of HIF1β target genes using ChIP-seq
To confirm that hypoxia-associated differential expression of TET genes is induced by the HIF pathway, we performed HIF1β ChIP-seq. Because HIF1β is the obligate binding partner of all 3 HIFα proteins stabilized and activated upon hypoxia35, HIF1β ChIP-seq reveals all direct HIF target genes.
Chromatin immunoprecipitation
25-30×106 cells were incubated in hypoxic conditions for 16 hours. Cultured cells were subsequently immediately fixed by adding 1% Formaldehyde (16% Formaldehyde (w/v), Methanol-free, Thermo Scientific) directly in the medium and incubating for 8 minutes. Fixed cells were incubated with 150 µM of glycine for 5 min to revert the cross-links, washed twice with ice-cold PBS 0.5% Triton-X100, scraped and collected by centrifugation (1000 ×G 5min at 4°C). The pellet was resuspended in 1400 µL of RIPA buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 2 mM EDTA pH 8, 1% Triton-X100, 0.5% Sodium deoxycholate, 1% SDS, 1% protease inhibitors) and transferred in a new eppendorf tube. The lysate was homogenized by passing through an insulin syringe, and incubated on ice for 10 min. The chromatin was sonicated for 3 min by using a Branson 250 Digital Sonifier with 0.7 s ‘On’ and 1.3 s ‘Off’ pulses at 40% power amplitude, yielding a size of 100 to 500 bp. The sample was kept ice-cold at all times during the sonication. The samples were centrifuged (10 min at 16000 ×G at 4°C) and the supernatant were transferred in a new eppendorf tube. The protein concentration was assessed using a BCA assay. Fifty µL of shared chromatin was used as “input” and 1.4 µg of primary ARNT/HIF-1β monoclonal antibody (NB100-124, Novus) per 1 mg of protein was added to the remainder of the chromatin, and incubated overnight at 4°C in a rotator. Pierce Protein A/G Magnetic Beads (Life Technologies) were added to the samples in a volume that is 4X the volume of the primary Ab and incubated at 4°C for at least 5 hours. A/G Magnetic Beads were collected and the samples were washed 5 times with the washing buffer (50 mM Tris-HCl, 200 mM LiCl, 2 mM EDTA, pH 8, 1% Triton, 0.5% Sodium deoxycholate, 0.1% SDS, 1% protease inhibitors), and twice with TE buffer. The A/G magnetic beads were resuspended in 50 µL of TE buffer, and 1.5 µL of RNAse A (200 units, NEB, Ipswich, MA, USA) were added to the A/G beads samples and to the input, incubated for 10 minutes at 37°C. After addition of 1.5 µL of proteinase K (200 units) and overnight incubation at 65°C, the DNA was purified using 1.8× volume of Agencourt AMPure XP (Beckman Coulter) according to the manufactory instructions, and then eluted in 15 µL of TE buffer. The input DNA was quantified on NanoDrop.
ChIP-seq, mapping and analysis
Five µg of input and all of the immunoprecipitated DNA were converted into sequencing libraries using the NEBNext DNA library prep master mix set. A single end of these libraries was sequenced for 50 bases on a HiSeq 2000, mapped using Bowtie and extended for the average insert size (250 bases). ChIP peaks were called by Model-based Analysis for ChIP-Seq36, with standard settings and using a sequenced input sample as baseline.
Patient-derived xenografted tumors
Patient-derived xenografts
To assess whether tumor-associated hypoxia reduces 5hmC levels in vivo, redundant material from 2 endometrial tumors and a breast tumor, removed during surgery, was grafted in the interscapular region of nude mice. Informed consent was obtained from the patient, following the ethical approval of the local ethical committee. All animal experiments were approved by the local ethical committee (P098/2014). Each tumor was allowed to grow until 1 cm3, after which it was harvested. 10% of this tumor was reimplanted in a nude mouse, and the tumor was thus propagated for 3 generations until it was used for this experiment. To mark hypoxic areas, mice were injected with pimonidazole (60 mg/kg, Hypoxyprobe, Massachusetts, USA) i.p. 1 hour before sacrifice.
Immunofluorescence staining and analysis
Tumors were harvested, fixed in formaldehyde and embedded in paraffin using standard procedures. Slides were deparafinated and rehydrated 2 xylene baths (5 minutes), followed by 5 times 3 minutes in EtOH baths at decreasing concentrations (100%, 96%, 70%, 50% and water) and a 3 minute Tris-buffered saline (TBS; 50 mM Tris, 150 mM NaCl, pH 7.6) bath.
The following antibodies were used for immunofluorescence staining: primary antibodies were FITC-conjugated mouse anti-pimonidazole (HP2-100, Hydroxyprobe), rabbit anti-5hmC (39791, Active Motif), rat anti-polyoma middle T (AB15085, Abcam), rat anti-CD31 (557355, BD Biosciences), rat anti-CD45 (553076, BD Biosciences), rabbit anti-Ki67 (AB15580, Abcam) and mouse anti-pan cytokeratin (C2562, Sigma). Secondary antibodies were Alexa fluor 405-conjugated goat anti-rabbit (A31556, Thermo Fisher), Alexa Fluor 647 conjugated goat anti-rat (A-21247, Life technologies), peroxidase-conjugated goat anti-FITC (PA1-26804, Pierce), biotinylated goat anti-rat (A10517, Thermo Fisher) and biotinylated goat anti-rabbit (E043201, Dako). Signal amplification was done using the TSA Fluorescein System (NEL701A001KT, Perkin Elmer) or the TSA Cyanine 5 System (NEL705A001KT, Perkin Elmer).
Different protocols were implemented depending on the epitopes of interest. Staining for the following epitopes was combined: CD45, 5hmC, pimonidazole and DNA; PyMT, 5hmC, pimonidazole and DNA; Ki67, pimonidazole and DNA; CD31 and pimonidazole; and pan-cytokeratin, 5hmC, pimonidazole and DNA.
Antigen retrieval for CD31, CD45 and pan-cytokeratin was done by a 7 min trypsin digestion, for pimonidazole and Ki67 using AgR at 100°C for 20 min, followed by cooling for 20 min. Slides were washes in TBS for 5 min, permeabilized in 0.5% Triton-X100 in PBS for 20 min. For 5hmC antigen retrieval, slides were next denatured in 2 N HCl for 10 min, with the HCl being neutralized for 2 min in borax, 1% in PBS pH 8.5, and washed twice for 5 min in PBS.
For all slides, endogenous peroxidase activity was quenched using H2O2 (0.3% in MeOH), followed by three 5 min washes in TBS. Slides were blocked using pre-immune goat serum (X0907, Dako; 20% in TNB; TSA Biotin System kit, Perkin Elmer, Waltham, MA). Binding of primary antibodies (anti-5hmC, anti-CD45, anti-CD31 and anti-pan cytokeratin or FITC-conjugated anti-pimonidazole; all 1/100 in TNB) was allowed to proceed overnight. Slides were washed 3× in TNT (0.5% Triton-X100 in TBS) for 5 min, after which secondary antibodies (all 1/100 in TNB with 10% pre-immune sheep serum) were allowed to bind for 45 min: sheep-anti-FITC-PO (for pimonidazole), goat anti-rabbit-Alexa Fluor 405 (for 5hmC), goat anti-rat-Alexa Fluor 647 (for CD45), and biotinylated goat anti-mouse (for pan-cytokeratin). Slides were washed 3× 5 min in TNT, after which signal amplification was done for 8 min using Fluorescein Tyramide (1/50 in amplification diluent).
Slides stained for pimonidazole that required co-staining slides for Ki67 or PyMT, or slides stained for pan-cytokeratin that required co-staining for pimonidazole were subjected to a second indirect staining for the latter epitopes: after 5 min of TNT and 5 min of TBS, slides were quenched again for peroxidase activity using H2O2 and blocked using pre-immune goat serum, prior a second overnight round of primary antibody binding (anti-Ki67, FITC-anti-pimonidazole or anti-PyMT, all 1/100). The next day, 3× 5 min washes with TNT were followed by a 1 h incubation with a biotinylated goat anti-rabbit antibody (for Ki67) or goat anti-rat (for PyMT), again 3× 5 min washes with TNT, a 30 min incubation with peroxidase conjugated to streptavidine (for Ki67 and PyMT) or to anti-FITC (for pimonidazole), again 3× 5 min washes with TNT and signal amplification for 8 min using, for pimonidazole, Fluorescein Tyramide and for others Cyanine 5 Tyramide (1/50 in amplification diluent).
Finally, slides were stained with propidium iodide + RNAse (550825; BD biosciences) for 15 min, washed for 5 min in PBS and mounted with Prolong Gold (Life Technologies).
Slides were imaged on a Nikon A1R Eclipse Ti confocal microscope. 3-5 sections per slide were imaged, and processed using Image J. More specifically, nuclei were identified using the propidium iodide signal, and nuclear signal intensities for Fluorescein and Cy3 (pimonidazole and 5hmC) measured. Analyses were exclusively performed on slide regions showing a regular density and shape of nuclei, in order to avoid inclusion of acellular or necrotic areas. The pimonidazole signal will also not stain necrotic/acellular areas 37, and was used to stratify viable cell nuclei into normoxic (pimonidazole negative) and hypoxic (pimonidazole positive) regions; and the 5hmC signal in both populations was compared using ANOVA. PyMT-negative and CD45-positive cells were counted directly. The fraction of pimonidazole and CD31-positive areas was directly quantified using ImageJ across 10 images per slide.
Metabolite levels
Metabolite and protein extraction
For metabolite extractions, 12-well cell culture dishes were placed on ice and washed twice with ice-cold 0.9% NaCl, after which 500 µL of ice-cold 80% methanol was added to each well. Cells were scraped and 500 µL was transferred to a vial on ice. Wells were washed with 500 µL 80% methanol, which was combined with the initial cell extracts. The insoluble fraction was pelleted at 4°C by a 10 minute 21,000×G centrifugation. The pellet (containing the proteins) was dried, dissolved in 0.2 N NaOH at 96°C for 10 minutes and quantified using a bicinchoninic acid protein assay (BCA, Pierce, Erenbodegem, Belgium), whereas the supernatant fraction was processed for metabolite profiling.
Derivation and measurement of metabolites
The supernatant fraction containing metabolites was transferred to a new vial and dried in a Speedvac. The dried supernatant fraction was dissolved in 45 µL of 2% methoxyamine hydrochloride in pyridine and held for 90 minutes at 37°C in a horizontal shaker, followed by derivatization through the addition of 60 µL of N-(tert-butyldimethylsilyl)-n-methyl-trifluoroacetamide with 1% tert-butyldimethylchlorosilane and a 60 minute incubation at 60°C. Samples were subsequently centrifuged for 5 minutes at 21,000 ×G, and 85 µL was transferred to a new vial and analysed using a gas-chromatography based mass spectrometer (triple quadrupole, Agilent) operated in Multiple Reaction Monitoring (MRM) mode.
Analysis of metabolite concentrations
For each sample, metabolite measurements were normalized per sample to the corresponding protein concentration estimates, and expressed relative to control-treated samples. Four technical replicates were run for each sample, and the experiment was repeated 4 times using independent samples (n=16). Differences in metabolite concentration were assessed using a two-tailed paired Student’s t-test or using analysis of variance with post-hoc Tukey HSD when repeated measures were compared.
ROS measurement using 2',7'-dichlorodihydrofluorescein diacetate
MCF7 cells were cultured in 24 well plates and exposed to 21% (control) or 0.5% O2 (hypoxia) for 24 hours. DMEM used for staining was pre-equilibrated to the required O2 tension, and all steps performed at 21% (control) or 0.5% O2 (hypoxia) using a glove box. The cells were washed 2× with 500 µL DMEM, and incubated for 30 min in 2',7'-dichlorodihydrofluorescein diacetate (DCF-DA; 10 μM) in 500µL DMEM, keeping 2 wells unstained by DMEM without DCF-DA. Cells were treated with the indicated concentrations of H2O2 in DMEM for 30 min at 37 °C, and fixed by adding 33.3 µL of 16% methanol-free paraformaldehyde (Thermo Fisher) for 8 min at RT. The fixative was quenched using glycine (150 µM), cells were washed 2× in ice-cold PBS, scraped to detach them and transfer them to pre-cooled FACS tubes over cell strainers. Cells were kept on ice until they were analysed by flow cytometry using a FACSVerse (BD Biosciences).
Nuclear ROS measurement using Nuclear Peroxy Emerald 1
MCF7 cells were seeded on 12 well glass bottom plates and after 24 h exposed to 21% (control) or 0.5% O2 (hypoxia) for 24 h. PBS used for subsequent staining was pre-equilibrated to the required O2 tension, and all washing, treatment and staining steps were performed at the appropriate O2 tension (21% or 0.5%) using a glove box. Cells were loaded with Nuclear Peroxy Emerald 1 (NucPE1; 5 μM)38,39 and Hoechst 33342 (10 μg/mL) in PBS for 15 min at 37 °C. After washing 3× in PBS, control cells were incubated with H2O2 (0.5 mM in PBS) as a positive control, or with water (control and hypoxia cells) in PBS at 37 °C for 20 min. Cells were washed 3× in PBS, placed on ice and immediately imaged by confocal microscopy. The nuclear NucPE1 signal was measured, and averaged across >100 nuclei per replicate using ImageJ. This experiment was repeated 5 times on different days, and signals compared using a t-test.
Cell growth measurement using Sulforhodamine B
5,000 cells/well were seeded in three 96-well plates. After 48 h, one plate was fixed using trichloroacetic acid (3.3% wt/vol) for 1 h at 4 °C, one plate incubated for 24 h at 37 °C under hypoxic and one under control conditions (resp. 0.5% and 21% O2). The latter 2 plates were subsequently also fixed using trichloroacetic acid (3.3% wt/vol) for 1 h at 4 °C, and all 3 plates were next analyzed using the In Vitro Toxicology Assay Kit, Sulforhodamine B-based (Sigma) as per the manufacturers instructions. Growth inhibition was calculated as described40.
siRNA transfection
siRNA ON-TARGETplus SMART pools (Thermo) were diluted in Optimem I reduced serum medium using Lipofectamine RNAiMAX (Life technologies) to reverse transfect MCF7 cells in 10 cm dishes (for DNA) or 6 well plates (for RNA). Cells were transfected 72 h before RNA and DNA extraction as described.
Hydroxylation assay using nuclear extracts
MCF7 cells were cultured for 24 h under control or hypoxic conditions (resp. 21 and 0.5% O2), chilled on ice and processed for extraction of nuclear proteins using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific). The activity of control and hypoxic extracts was assessed in parallel using the Colorimetric Epigenase 5mC-Hydroxylase TET Activity/Inhibition Assay Kit (Epigentek, Farmingdale, USA) according to manufacturers instructions. Reactions were allowed to proceed for one hour, after which washing and detection of 5hmC were done according to manufacturers instructions. Differences between hypoxia and control were analyzed using ANOVA, for 5 independent experiments.
DNA hydroxymethylation assay using purified Tet enzyme
The genomic DNA used in this assay was extracted from Tet-triple-knockout ES cells (a gift from Prof. Guo-Liang Xu, State Key Laboratory of Molecular Biology, CAS, Shanghai, China), and it therefore was devoid of 5hmC41. To enable efficient denaturation, it was digested using MseI prior to the assay and purified using solid phase reversible immobilisation paramagnetic beads (Agencourt AMPure XP, Beckman Coulter, USA). The assays were performed in Whitley H35 Hypoxystations (don Whitley Scientific, UK) at 37° C, 5% CO2, N2, plus the following oxygen tensions: 0.1%, 0.25%, 0.5%, 1%, 2.5%, 5%, 10% and 21%. Hypoxystations were calibrated less than 1 month prior to all experiments. Optimized assay components were as follows: 1.0 µg/µL bovine serum albumin (New England Biolabs), 50 mM Tris (pH 7.8), 100 µM dithiothreitol (Life Technologies), 2ng/µL digested gDNA, 250 µM α-ketoglutarate, 830 µM ascorbate, 200 µM FeSO4 and 45 ng/µL Tet1 enzyme (Wisegene, USA). The major assay components (H20, BSA and Tris) used for all samples were allowed to pre-equilibrate at 0.1% O2 for 1 hour. These and the remaining assay buffer components (<100 µL) were then pre-equilibrated at the desired oxygen tension for 15 min, and mixed prior to addition of Tet1 enzyme in a total reaction volume of 25 µL. Reactions were allowed to proceed for 3 min, longer incubations showed a decrease in activity. Reactions were stopped with 80 mM EDTA and stored at -80° C. To measure the resulting 5hmC content of the DNA, reactions were diluted to 100 µL, denatured for 10 min at 98° C and analysed in duplicate using the Global 5-hmC Quantification Kit (Active Motif) following manufacturers instructions. Michaelis-Menten and Lineweaver-Burk plots and the resulting KM values were estimated using R.
Hypoxia-induced changes in genomic distribution of 5(h)mC in MCF7 cells
DIP-seq
To assess where in the genome the levels of 5mC and 5hmC were altered, we performed DNA immunoprecipitations coupled to high-throughput sequencing (DIP-seq). MCF7 cells were selected for these experiments as they were a cancer cell line with high levels of 5hmC and expression of TETs under control conditions, and a cell growth that is unaffected by hypoxia, thus enabling us to study effects of hypoxia on TET activity in a cell line that shows high endogenous activity, but that is isolated from hypoxia-induced changes in cell proliferation. MCF7 cell culture and DNA extractions were as described for LC/MS analyses. Library preparations and DNA immunoprecipitations were as described42, using established antibodies targeting 5mC (clone 33D3, Eurogentec, Liege, Belgium) and 5hmC (Active Motif catalogue number.39791, La Hulpe, Belgium). For 5hmC-DIP-seq, paired barcoded libraries prepared from DNA of hypoxic and control samples were mixed prior to capture, to enable a direct comparison of 5hmC-DIP-seq signal to the input. A single end of these libraries was sequenced for 50 bases on a HiSeq 2000, mapped using Bowtie and extended for the average insert size (150 bases). Mapping statistics are summarized in Supplementary table 11.
For analysis of sequencing data, MACS peak calling, read depth quantification and annotation with genomic features as annotated in EnsEMBL build 77 was done using using SeqMonk. Differential (hydroxy-)methylation was quantified by EdgeR43, using either 3 or 5 independent pairs of control and hypoxic samples (resp. for 5hmC-DIP-seq and 5mC-DIP-seq). These cells were cultured and exposed to hypoxia (0.5% O2) or control conditions (21% O2) on different days. Results were reported for 5hmC peak areas that exhibited a change significant at a P<0.05 and 5% FDR.
Target enrichment BS-seq using SeqCapEpi
To confirm enrichment of 5mC at gene promoters using an independent method, DNA libraries were prepared using methylated adapters and the NEBNext DNA library prep master mix set following manufacturer recommendations. Libraries were bisulfite-converted using the Imprint DNA modification kit (Sigma) as recommended, and PCR amplified for 12 cycles using barcoded primers (NEB) and the KAPA HiFi HS Uracil+ ready mix (Sopachem, Eke, Belgium) according to manufacturers instructions. Fragments were selected from these libraries using the SeqCapEpi CpGiant Enrichment Kit (Roche) following the manufacturers instructions, sequenced from both ends for 100 bases on a HiSeq 2000.
For analyzing these sequences, sequencing reads were trimmed for adapters using TrimGalore and mapped on a bisulfite-converted human genome (GRCh37) using BisMark. The number of methylated and unmethylated cytosines in captured regions were quantified using Seqmonk for each experiment. Differential methylation of regions of interest was assessed by Fisher’s exact test and for 5 independent replicates grown on different days. t-scores were averaged following Fisher’s method. Mapping statistics are summarized in Supplementary table 11.
RNA-seq
To assess the impact of the increased 5mC occupancy at gene promoters on their expression, RNA-seq was performed. Briefly, total RNA was extracted using TRIzol (Invitrogen), and remaining DNA contaminants in 17-20ug of RNA was removed using Turbo DNase (Ambion) according to the manufacturers instruction. RNA was repurified using RNeasy Mini Kit (Qiagen). Ribosomal RNA present was depleted from 5ug of total RNA using the RiboMinus Eukaryote System (Life technologies). cDNA synthesis was performed using SuperScript® III Reverse Transcriptase kit (Invitrogen). 3 µg of Random Primers (Invitrogen), 8 µL of 5× First-Strand Buffer and 10 µL of RNA mix was incubated at 94°C for 3 min and then at 4°C for 1 min. Then, 2 µL of 10 mM dNTP Mix (Invitrogen), 4 µL of 0.1 M DTT, 2 µL of SUPERase• In™ RNase Inhibitor 20U/ µL (Ambion), 2 µL of SuperScript™ III RT (200 units/µL) and 8 µL of Actinomycin D (1µg/µL) were added and the mix were incubated 5 min at 25°C, 60 min at 50°C and 15 min at 70°C to heat inactivating the reaction. The cDNA was purified by using 80 µL (2× volume) of Agencourt AMPure XP and eluted in 50 µL of the following mix: 5 µL of 10X NEBuffer 2, 1.5 µL of 10 mM dNTP mix (10mM dATP, dCTP, dGTP, dUTP, Sigma), 0.1µL of RNaseH (10 U/µL, Ambion), 2.5 µL of DNA Polymerase I Klenov (10U/µL, NEB) and water until 50 µL. The eluted cDNA was incubated for 30 min at 16°C, purified by Agencourt AMPure XP and eluted in 30 µL of dA-Tailing mix (2 µL of Klenow Fragment, 3 µL of 10X NEBNext dA-Tailing Reaction Buffer and 25 µL of water). After 30 min incubation at 37°C, the DNA was purified by Agencourt AMPure XP, eluted in TE buffer and quantified on NanoDrop. Subsequent library preparation was done using the DNA library prep master mix set and sequencing was performed as described for ChIP-seq. Expression levels (reads per million) of genes displaying significant increases in methylation at their gene promoter, as determined using SeqCapEpi, was compared between control and hypoxic samples using t-test. Mapping statistics are summarized in Supplementary table 11.
TCGA samples and data analysis
Sample description
From the TCGA pan-cancer analysis, we selected all solid tumor types for which >100 tumors were available with both gene expression data (RNA-seq) and DNA methylation data (Illumina Infinium HumanMethylation450 BeadChip). These were 408 bladder carcinomas, 691 breast carcinomas, 243 colorectal adenocarcinomas, 520 head and neck squamous cell carcinomas, 290 kidney renal cell carcinomas, 430 lung adenocarcinomas, 371 lung squamous cell carcinomas, and 188 uterine carcinomas, representing in total 3,141 unique patients. Corresponding RNA-seq read counts as well as DNA methylation data from Infinium HumanMethylation450 BeadChip arrays were downloaded from the TCGA server. Breast tumor subtype was annotated for 208 tumors, and for the remaining tumors imputed by unsupervised hierarchical clustering of genes in the PAM50 gene expression signature44. Other clinical and histological variables were available for >95% of tumors, and missing values were encoded as not available. Gene mutation data was available for 129 bladder carcinomas, 646 breast carcinomas, 200 colorectal adenocarcinomas, 306 head and neck squamous cell carcinomas, 241 kidney renal cell carcinomas, 182 lung adenocarcinomas, 74 lung squamous cell carcinomas, and 3 uterine carcinomas.
Stratification of tumors for hypoxia and proliferation
To identify which of these tumor samples were hypoxic or normoxic, we performed unsupervised hierarchical clustering based a modification (Ward.D of the clusth function in R`s stats package) of the Ward error sum of squares hierarchical clustering method45, on normalized log-transformed RNA-seq read counts for 14 genes that make up the hypoxia metagene signature (ALDOA, MIF, TUBB6, P4HA1, SLC2A1, PGAM1, ENO1, LDHA, CDKN3, TPI1, NDRG1, VEGFA, ACOT7 and ADM)25. In each case the top 3 subclusters identified were annotated as normoxic, intermediate and hypoxic. To identify which of these tumor samples were high- or low-proliferative, we performed unsupervised hierarchical clustering based a modification (Ward.D of the clusth function in R`s stats package) of the Ward error sum of squares hierarchical clustering method45, and this for all genes annotated to an established tumor proliferation signature (MKI67, NDC80, NUF2, PTTG1, RRM2, BIRC5, CCNB1, CEP55, UBE2C, CDC20 and TYMS)46. Tumors in the top 2 subclusters identified were labeled as high- or low-proliferative.
Analysis of the top 1000 CpGs most hypermethylated versus normal tissue
To identify tumor-associated HM events, we compared 450K methylation data from tumors and normal tissues. All available DNA methylation data from normal tissue (matched or unmatched to tumor samples, on average 59 per tumor type, representing 472 in total, range = 21-160) were downloaded. For each of the 8 tumor types investigated, we selected the top 1,000 CpGs that showed the highest average tumor-associated increases in DNA methylation. Per tumor type, unsupervised hierarchical clustering based on a modification of the Ward error sum of squares hierarchical clustering method (Ward.D of the clusth function in R`s stats package)45 annotated the first 3 clusters identified as having low, intermediate and high hypermethylation. Cluster co-membership for methylation and hypoxia metagene expression were analysed using the Cochran-Armitage test for trend. Analyses using the top 100, 500, 5,000 or 10,000 CpGs yielded near identical results (not shown).
Analysis of HM events
We next applied a method to identify those CpGs that exhibit exceptional increases in HM but that are hypermethylated only in a subset of all tumors. Such more rare events are typically found in cancer, where HM inactivates a gene in only a subset of tumors. HM of individual CpGs at gene promoters (i.e. on average 3.7 CpGs per promoter are represented on the 450K array) in individual tumors was assessed as follows: To achieve a normal distribution, all β-values were transformed to M-values47 using M = log2(β/(1-β)). For each tumor type, the mean μ and standard deviation σ of the M value across all control (normoxic) tumors was next calculated, irrespective of mutational status, for each CpG, and used to assign Z-values to each CpG in each tumor using Z = (M - μ)/σ. These Z-values describe the deviation in normal methylation variation for that probe. To identify CpGs that display an extreme deviation, we selected those for which the Z-value exceeded 5.6 (i.e. the mean plus 5.6 times the standard deviation, corresponding to a Bonferroni-adjusted P-value of 0.01); they were considered as hypermethylation events in that particular tumor. This analysis was preferred over Wilcoxon-based models that assess differences in the average methylation level between subgroups, as the latter do not enable the identification or quantification of the more rare HM events in individual promoters or CpGs.
To identify genes with frequently hypermethylated CpGs in their promoter, the number of HM events in that promoter was counted in all tumors, and contrasted to the expected number of HM events in that promoter (i.e. the general HM frequency × the number of CpGs assessed in that promoter × the number of tumors) using Fisher’s exact test. Genes with an associated Bonferroni-adjusted P-value below 0.01 were retained and considered as frequently hypermethylated in that tumor type.
To assess what fraction of these HM events are hypoxia-related, we assumed that the fraction of events detected under normoxia was hypoxia–unrelated, and that all excess events detected in intermediate and hypoxic tumors were hypoxia-related. For example, in 691 breast carcinomas, 0.25% of CpGs were hypermethylated in 251 normoxic tumors, 0.81% in 350 intermediate and 1.40% in 90 hypoxic tumors. So, 0.56% and 1.15% of HM events in respectively intermediate and hypoxic tumors were hypoxia-related. Taking into account the number of tumors, 0.25% of HM events (i.e. (0.25% × 251 + 0.25% × 350 + 0.25% × 90) ÷ 691) are not hypoxia-related, and 0.43% are hypoxia related (i.e. (0% × 251 + 0.56% × 350 + 1.15% × 90) ÷ 691). Hence, 63% of all HM events (i.e. 0.43÷(0.43+0.25)). To assess the contribution of hypoxia to HM relative to other covariates, partial R2 values were calculated for the contribution of each covariate in a linear model, and compared to the total R2 achieved by the model.
To identify genes with CpGs in their promoter that are more frequently hypermethylated in hypoxic than normoxic tumors, the number of HM events in that promoter was counted in all hypoxic tumors, and contrasted to the number found in normoxic tumors. Differences in frequencies were assessed using Fisher’s exact test, and genes with a Bonferroni-adjusted P<0.01 were retained and considered hypermethylated upon hypoxia. Enrichment of ontologies associated with these genes was assessed using Fisher’s exact test as implemented in R`s topGO package.
Analysis of the impact of HM events on the expression of associated genes
To enable a direct comparison between the expression of different genes, we transformed gene expression values (reads per million) to their respective z-scores. To assess the impact of HM events on the expression of associated genes, we compared the expression z-scores of all frequently HM genes that contain one or more HM events in their promoter (i.e., on average each promoter contains 3.7 CpGs; if one of these is hypermethylated the associated gene is considered hypermethylated in that particular tumor), to the expression of all frequently HM genes that do not contain a HM event. The effect of HM on gene expression was plotted for the 8 main tumor types stratified into normoxic, intermediately hypoxic and hypoxic tumors, and for glioblastomas stratified into normoxic, intermediately hypoxic, hypoxic and IDH-mutant tumors (n=4). The difference in expression z-scores between genes not carrying and carrying a HM event in their promoter was assessed using a t-test.
Analysis of the impact of frequent mutations on the occurrence of HM events
To assess the impact of somatic mutations on hypoxia-associated HM frequencies, we analyzed the top 20 genes described to be most frequently mutated in the pan-cancer analysis,24 and supplemented this list with genes known to cause HM upon mutation (i.e. IDH1, IDH2, SDHA, FH, TET1, TET2 and TET3). Mutations in IDH1 and IDH2 were retained if they respectively affected amino acid R132, and amino acids R140 or R172. Mutations in other genes were scored using Polyphen, and only mutations classified as probably damaging were retained. 7 mutations were found in lung tumors, 3 mutations in colorectal tumors, 8 mutations in breast tumors and 6 mutations (all IDH1R132) in glioblastomas. None of these mutations were enriched in hypoxic subsets. In multivariate analyses of variance, in each of the tumor types analyzed, mutations in these genes were significantly associated with increased HM frequencies, but also hypoxia was independently and significantly associated with the HM frequency.
Correlation between HM and expression of TET or DNMT enzymes
Gene expression values (reads per million) of DNMT and TET enzymes were determined for each tumor using available RNA-seq data. The number of HM events at significantly hypermethylated genes in each tumor was determined as described above. Hypermethylation in each tumor was subsequently correlated to TET or DNMT gene expression in that tumor, correcting for hypoxia and proliferation status, using ANOVA.
5mC and 5hmC profiling using 450K arrays for 24 lung tumors
Tumor samples
Newly diagnosed and untreated non-small-cell lung cancer patients scheduled for curative-intent surgery were prospectively recruited. Included subjects had a smoking history of at least 15 pack-years. The study protocol was approved by the Ethics Committee of the University Hospital Gasthuisberg (Leuven, Belgium). All participants provided written informed consent. In the framework of a different project48, RNA-seq was performed on 39 tumors from these patients. Gene expression for these samples was clustered for their hypoxia metagene signature25. This yielded 2 clear clusters, containing respectively 24 and 15 normoxic and hypoxic tumors. Twelve samples were randomly selected from each cluster, in order to perform 5hmC and 5mC profiling.
Illumina Infinium HumanMethylation450 BeadChips
For Tet-assisted bisulfite (TAB)-chip, DNA was glycosylated and oxidized as described49, using the 5hmC TAB-Seq Kit (WiseGene, Chicago, USA). Subsequently, bisulfite conversion, DNA amplification and array hybridization were done following manufacturers instructions.
Analysis of TAB-chip and BS-chip
Data processing was largely as described50. In brief, intensity data files were read directly into R. Each sample was normalized using Subset-quantile within array normalization (SWAN) for Illumina Infinium HumanMethylation450 BeadChips49. Batch effects between chips and experiments were corrected using the runComBat function from the ChAMP bioconductor package51. For obtaining 5mC-specific beta values, TAB-chip generated normalized beta values were substracted from the standard 450K generated normalized beta values, exactly as described50.
Murine cancer models
All the experimental procedures were approved by the Institutional Animal Care and Research Advisory Committee of the KU Leuven.
Hypoxia induction using sFlk1-overexpression
For sFlk1-overexpression studies, male Tg(MMTV-PyMT) FVB mice were intercrossed with WT FVB female mice. Female pups of the Tg(MMTV-PyMT) genotype were retained, and tumors allowed to develop for 9 weeks. Subsequently, 2.5 µg of plasmid (Flk1-overexpressing or empty vector; randomly assigned within litter mates) per gram of mouse body weight was introduced in the blood stream using hydrodynamic tail vein injections52. Flk1 overexpression was monitored at 4 days after injection and at the day of sacrifice (18 days after the injection), by eye bleeds followed by an enzyme-linked immunosorbent assay for sFlk1 (R&D Systems, Abingdon, UK) in blood plasma. At 12 weeks of age, mice were sacrificed and mammary tumors harvested blinded for treatment.
Hypoxia reduction using Phd2 haplodeficiency
For the Phd2+/- experiments, male Tg(MMTV-PyMT) FVB mice were intercrossed with female Phd2-/+ mice, yielding litters of which half have either a Tg(MMTV-PyMT) genotype or a Tg(MMTV-PyMT);Phd2-/+ genotype. For the Phd2wt/fl experiments, male Tg(MMTV-PyMT) FVB mice were intercrossed with female Tie2-cre;Phd2wt/fl mice as described27, yielding litters of which half have either a Tie2-cre;Tg(MMTV-PyMT);Phd2wt/wt genotype or a Tie2-cre;Tg(MMTV-PyMT);Phd2-/+ genotype. At 16 weeks of age, female mice were sacrificed and mammary tumors harvested.
qPCR analysis of expression of Tets and marker genes
RNA was extracted from fresh-frozen tumors (stored at -80°C) using TRIzol (Life Technologies), and converted to cDNA and quantified as described for the cell lines. TaqMan probes and primers (IDT, Leuven, Belgium or Life technologies) are described under Supplementary table 12.
TAB-sequencing (TAB-seq) of PyMT tumors
TAB-seq libraries were prepared as described, using the 5hmC TAB-Seq Kit (WiseGene). DNA was bisulfite-converted using the EZ DNA Methylation-Lightning Kit (Zymo Research) and sequenced as described for SeqCapEpi experiments. Reads were mapped to the mouse genome (build Mm9) and further data processing was as for SeqCapEpi experiments. DNA from 3 independent tumors were selected per condition. TET oxidation efficiency was required to exceed 99.5% as estimated using a fully CG-methylated plasmid spike-in, 5hmC protection by glycosylation was 65% as estimated using a fully hydroxymethylated plasmid spike-in, bisulfite conversion efficiencies were estimated to exceed 99.8% based on nonCG methylation (=hmCpH %). Mapping statistics are summarized in Supplementary table 11.
Targeted deep BS-seq
As no murine capture kit was available for targeted BS-seq, a specific ampliconBS was developed for a set of 15 tumor suppressor gene promoters and 5 oncogene promoters. More specifically, DNA was bisulfite-converted using the Imprint DNA modification kit and amplified using the MegaMix Gold 2× mastermix and validated primer pairs. Per sample, PCR products were mixed to equimolar concentrations, converted into sequencing libraries using the NEBNext DNA library prep master mix set and sequenced to a depth of ~500×. Mapping and quantification were done as described for SeqCapEpi. The average and variance of methylation level M values in normal mammary glands were used as baseline, and amplicons displaying over 3 standard deviations more methylation (FDR-adjusted P<0.05) than this baseline were called as hypermethylated. At least 9 different tumors, each from different animals, were profiled per genotype or treatment, and differences in HM frequencies between sets of tumors were assessed using Mann-Whitney’s U-test.
Statistics
Data entry and analysis was performed in a blinded fashion. Statistical significance was calculated by two-tailed unpaired t-test (Excel) or analysis of variance (R) when repeated measures were compared. Data were tested for normality using the D’Agostino–Pearson omnibus test (for n > 8) or the Kolmogorov–Smirnov test (for n ≤ 8) and variation within each experimental group was assessed. Data are presented as means ± standard error of mean. DNA methylation and RNA-seq gene expression data distributions were transformed to a normal distribution by conversion to M values and log2 transformation respectively. Sample sizes were chosen based on prior experience for in vitro and murine experiments, or on sample and data availability for human tumor analyses. Other statistical methods (mostly related to specific sequencing experiments) are described together with the experimental details in other sections of the methods.
Extended Data
Extended data figure 1

Global 5hmC/C and 5mC/C content of DNA, TET1, TET2 and TET3 mRNA expression and hypoxia marker gene expression in 15 cell lines grown for 24 h under control (21% O2, white) or hypoxic (0.5% O2, red) conditions. TET mRNA copy number is expressed relative to B2M for human cell lines (HepG2, HT-1080, MCF10A, H358, MCF7, Hep3B, A549, H1299, SK-N-Be2c and SHSY5Y), and to Hprt for murine cell lines (LLC, N2A, 4T1, ESC-wt and ESC-Tet1-KO). Shown are cell lines derived from liver cancer (HepG2 and Hep3B), lung cancer (H358, A549, H1299 and LLC), breast cancer (MCF7 and 4T1), fibrosarcoma (HT1080), neuroblastoma (SK-N-Be2c and SHSY5Y), normal breast epithelium (MCF10A) and the inner cell mass of blastocyst-stage mouse embryos (ESC-wt and ESC-Tet1-KO). ALDOA and BNIP3 are expected to be increased, and HIF1A to be decreased upon hypoxia. The global 5fC content of ES cells is depicted, but was undetectable in cancer cell lines. Bars represent the mean ± s.e.m. of 5 different replicate samples. DNA and RNA from these replicates was extracted from cells derived from the same stock vial but grown on different days. * P<0.05, ** P<0.01, *** P<0.001 by paired t-tests.
Extended data figure 2

a, Changes in Tet1, Tet2 and Tet3 expression in mouse cell lines, at the protein level (top row, n=6) and the mRNA level (bottom row, n=5). middle row: representative immunoblot images of Hif1a, Tet1, Tet2 and Tet3. α-Tubulin serves as loading control, and expression of the corresponding coding gene (Tuba) was used to normalize mRNA expression, enabling a direct comparison of relative protein and relative mRNA expression changes. For the same reason, mRNA expression was depicted relative to control conditions, in contrast to the absolute levels shown in Extended data figure 1. Changes in Tet mRNA and protein expression correlate strongly (Pearson’s R: 0.855, P=4 × 10-4). For example, both 4T1 and N2A cells displayed increased Tet2 expression at the protein and mRNA level. Likewise, ES cells showed no pronounced changes at the protein nor at mRNA level. The overall expression of Tet enzymes was moreover not altered in any of these cell lines. For gel source data, see Supplementary figure 1. b, HIF1β ChIP-seq at the promoters of TET1, TET2 and TET3 and at hypoxia markers genes (BNIP3 and ALDOA), with peaks or promoter regions highlighted using colored boxes. Green and red boxes correspond to respectively overexpression and no overexpression (specified in the figure panel) of the corresponding gene, as determined using TaqMan in Extended data figure 1. Scale: reads per million reads and per basepair. c, (left) TET2 expression in MCF7 cells transfected with control (white) or TET2-targeting (purple) siRNAs, and (right) corresponding 5hmC levels as determined using LC/MS. d, 5hmC levels as determined using LC/MS, in wild-type (white) and Tet1-knockout (purple) ES cells grown under 21% (left) and 0.5% (right) oxygen tensions. Bars in c and d represent the mean ± s.e.m. of 5 replicate samples from cells derived from the same stock vial but grown on different days. * P<0.05, ** P<0.01, *** P<0.001 by paired t-tests (a, c, d)
Extended data figure 3

a-e, ROS production and redox state of MCF7 cells cultured for 24 h under control (21% O2, white) or hypoxic (0.5% O2, red) conditions. Shown are (a) GC/MS quantifications of changes in the cellular energy state as represented by the adenylate energy charge (AEC) (calculated as [ATP + 0.5 × ADP]/[ATP + ADP + AMP]); the cell’s reducing equivalents as represented by the relative NADH and NADPH levels (calculated as NADH/[NAD+ + NADH] and NADPH/[NADP+ + NADPH]) and the cell’s reductive capacity as represented by the levels of glutathione (calculated as GSH/[GSH + GSSG × 2]). b-c, Quantication (b) and representative FACS intensity traces (c) of total ROS levels in MCF7 cells exposed to hypoxia or H2O2, as assessed using 2',7'-dichlorodihydro-fluorescein diacetate (DCF-DA). d, Nuclear ROS in MCF7 cells as assessed using the Nuclear Peroxy Emerald 1 probe (NucPE1)39. MCF7 cells were exposed to 21% (control) or 0.5% O2 (hypoxia) for 24 h, after which live cells were loaded with NucPE1 (5 µM) and Hoechst 33342 (10 µg/mL) in O2 pre-equilibrated PBS for 15 minutes. After washing, control cells were incubated with H2O2 (0.5 mM in PBS) as a positive control, or with water (control and hypoxia cells) in PBS for 20 minutes. Cells were washed again and immediately imaged by confocal microscopy. Representative images are shown; scale: 50 µm. e, The nuclear NucPE1 signal, averaged across >100 nuclei and expressed relative to control conditions. f, LC/MS quantification of 8-oxoguanine concentrations in DNA of cells lines cultured for 24 h under control (21% O2, white) and hypoxic (0.5% O2, red) conditions. 8-oxoguanine serves as a marker of nuclear ROS53. g-i, GC/MS quantification of changes in the indicated metabolite levels in mouse embryonic stem cells (g), MCF10A cells (h) and MCF7 cells (i) grown for 24 h under control (21% O2, white), hypoxic (0.5% O2, red) or glutamine-free conditions (21% O2, black). j, Quantities of α-ketoglutarate and 2-hydroxyglutarate in MCF7 cells, expressed relative to α-ketoglutarate levels in MCF7 cells grown under control conditions (21% O2). k, LC/MS quantification of 5hmC levels in response to hypoxia (0.5% O2) and glutamine-free culture conditions. l, Growth of cell lines cultured for 24 h under control (21% O2, white) and hypoxic (0.5% O2, red) conditions, as assessed using a sulforhodamine B colorimetric assay. Changes in cell density after 24 h are depicted relative to control conditions (21% O2). m, IOX2-induced changes in the global 5hmC content of DNA, in TET mRNA expression and in hypoxia marker gene expression of 5 cell lines treated for 24 h with DMSO (carrier, white) or IOX2 (50 µM, blue). n, 5mC hydroxylation activity of nuclear lysates from MCF7 cells grown for 24 h under 21% or 0.5% O2 (white or red). Bars represent the mean ± s.e.m. of 5 (b, k, m), 6 (a, e), 16 (g-j) or 24 (l) samples prepared on different days. * P<0.05, ** P<0.01, *** P<0.001 by t-test (b, e, h-m).
Extended data figure 4

Shown are results from DIP-seq of DNA from MCF7 cells cultured for 24 h under 21% or 0.5% O2 (control and hypoxia), with examples of 5hmC-DIP-seq (top) and 5mC-DIP-seq (bottom) read depths (FPM, fragments per basepair per million fragments) at regions surrounding the transcription start site of NSD1, FOXA1 and CDKN2A. These show 5hmC loss (FDR<5%) and a 5mC gain that is more subtle, perhaps because the resolution of 5mC-DIP-seq is limiting: regions rich in 5hmC tend to be poorer in 5mC54, and thus have less substrate available for pull-down. 5mC-DIP-seq moreover captures all methylated sites, so most of the 5mC-DIP-seq signal does not derive from sites that are actively turning over 5hmC.
Extended data figure 5

a, Immunofluorescence analysis of patient-derived tumor xenografts, stained for pimonidazole (PIMO, white), 5hmC (red), DNA (propidium iodide, blue) and pan-cytokeratin (green). Shown are representative images of a breast and 2 endometrial tumor xenografts. The inset on the right shows boxplots illustrating the signal in normoxic pimonidazole-negative nuclei (blue), and in hypoxic pimonidazole-positive nuclei (red). b, Hypoxia marker gene expression clusters, with the first 3 clusters used to define normoxic, intermediate and hypoxic tumors. c, Unsupervised clustering of 1,000 CpGs showing the highest average methylation increase in tumor versus corresponding normal tissues. The first 3 clusters were used to define tumors of low, intermediate and high HM. The color bar above the clusters annotates each tumor as normoxic, intermediate or hypoxic, as determined in Extended data figure 5b. d, Boxplots showing the relative expression (z-score) of genes in tumors wherein they have either 0 or ≥1 hypermethylation (HM) event in their promoter, stratified into normoxic, intermediate and hypoxic tumors (resp. blue, grey and red). Diamonds indicate means, boxplot wedges indicate 2 times the standard error of the median. Genes having ≥1 HM events in their promoters have a lower average expression level (P<0.01 for each tumor type). e, Fraction of genes having a promoter that is rich, intermediate or poor in CpGs, out of all gene promoters that are assessed on the 450K array (450K), and out of all gene promoters that are frequently hypermethylated in the indicated tumor types. f, Fraction of 1,742 TET-wildtype tumors and 39 TET-mutant that are normoxic, intermediate and hypoxic. P>0.2 for all comparisons. g, Cell proliferation marker gene46 expression clusters, with the first 2 clusters used to define high-proliferative and low-proliferative tumors. h, HM frequencies in low and high-proliferative tumors, with asterisks representing P-values from linear models correcting for variables specified in Supplementary table 8. i, Partial correlation coefficient (partial R2) estimates of the relative contribution of tumor characteristics (annotated in TCGA) to the variance in HM observed in these tumors. Partial R2 values were obtained from linear model estimation using ordinary least squares, and expressed as a fraction of the total variance (i.e. total R2) explained by the model when taking into account all indicated variables, as indicated between brackets under each tumor type. * P<0.05, ** P<0.01, *** P<0.001 by t-test (a) or by generalized linear model (h); BLCA bladder carcinoma; BRCA breast carcinoma, COAD colorectal adenocarcinoma, HNSC head and neck squamous cell carcinoma, KIRC kidney renal clear cell carcinoma, LUAD lung adenocarcinoma, LUSC lung squamous cell carcinoma, UCEC uterine corpus endometrial carcinoma.
Extended data figure 6
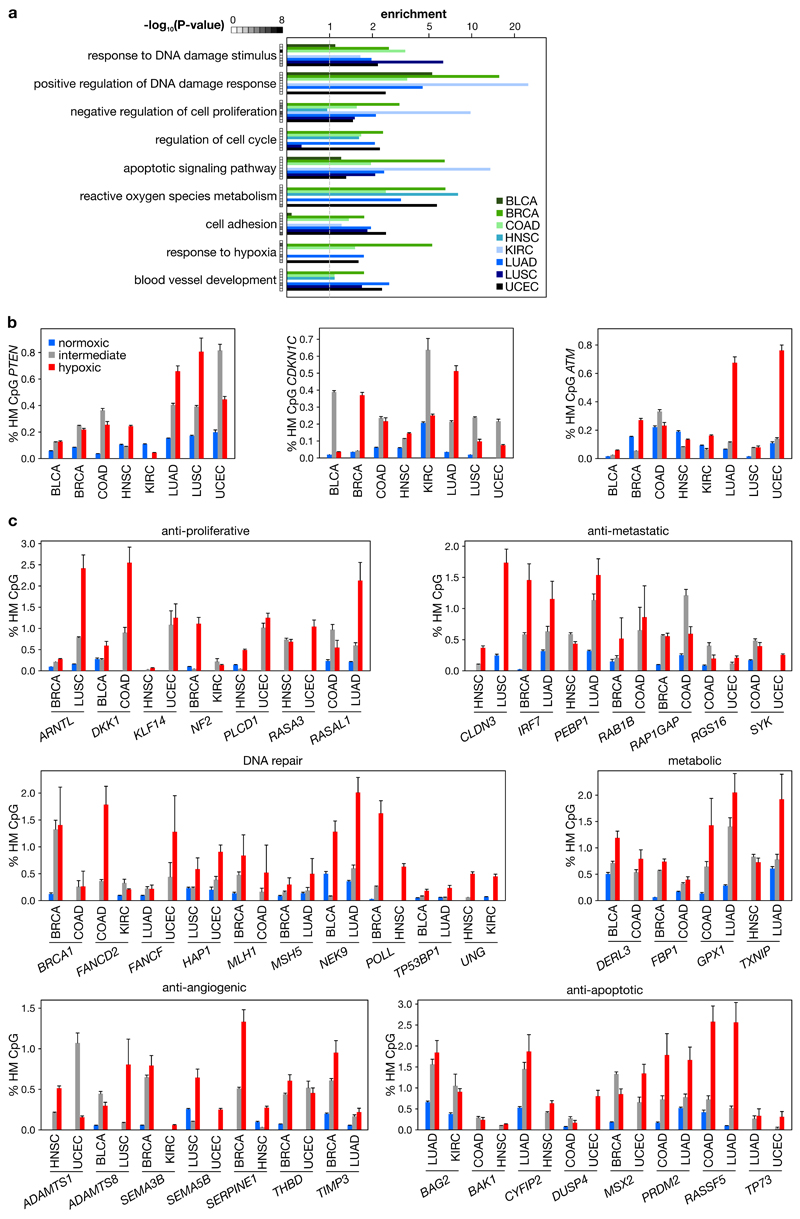
a, Ontology terms enrichment analysis of genes that are more frequently hypermethylated at their gene promoters in hypoxic than normoxic tumors, for 8 tumor types characterized in the TCGA Pan-Cancer effort. A representative set of terms is displayed, selected from terms enriched in most tumor types. P-values as defined by the grey-scale insert. Enrichment calculated using topGO. b, Selected examples of HM frequencies in the promoter of key tumor suppressor genes (PTEN, CDKN1C, ATM) more frequently hypermethylated in normoxic than hypoxic tumors. c, HM frequency in the promoter of selected genes involved in the processes indicated. P<0.05 for all genes (asterisks are not displayed). Bars in b and c represent the HM frequency ± s.e.m. P-values in (a) by Fisher’s exact test.
Extended data figure 7

a, The t-value of correlation between HM and expression of TET or DNMT genes across 3,141 tumors of 8 tumor types (bladder, breast, colorectal, head and neck, kidney, lung adeno, lung squamous, and uterine carcinoma) profiled in TCGA for gene expression and DNA methylation, while correcting for tumor type, hypoxia and proliferation. The dotted line represents P<0.05, negative t-values represent inverse correlations. b, Hypoxia metagene signature applied to 63 glioblastoma multiforme tumors from TCGA. c, Boxplots showing the relative expression (z-score) of genes in tumors wherein they have either 0 or ≥1 hypermethylation (HM) event in their promoter, stratified into IHD1WT tumors that are normoxic (n=19), intermediate (n=21) and hypoxic (n=17) (resp. blue, grey and red), and IDH1R138-mutated tumors (n=4, yellow). Diamonds indicate means, boxplot wedges indicate 2 times the standard error of the median. Genes having ≥1 HM events in their promoters have a lower average expression level. No HM events were detected in IHD1WT normoxic tumors. d Hypoxia metagene signature applied to 12 normoxic and 12 hypoxic non-small-cell lung tumors. * P<0.05, *** P<0.001 by t-test (c).
Extended data figure 8

a, Frequency of HM events in the promoters of all genes, all oncogenes and all tumor suppressor genes (TS genes) as annotated28, in 695 human breast tumors available through TCGA and stratified into normoxic, intermediate and hypoxic subsets. b-c, DNA was extracted from 53 tumors developing in MMTV-PyMT mice of the indicated ages (c) or weights (d) and sequenced to a depth of ~500x. Plotted are z-scores of HM (y axis, exponential) for 15 TSGs, relative to the tumors from 11-week-old mice. The dotted line represents the threshold for a Bonferroni-adjusted P<0.05, and bold darker dots are used for tumors displaying significantly increased HM events. d, DNA extracted from 20 normal mammary glands from 14-week-old mice, PCR-amplified for 15 TSGs and sequenced to a depth of ~500x. Plotted are z-scores of HM relative to 11-week-old tumors. e, Staining of PyMT tumors for 5hmC (red), DNA (propidium iodide, blue), pimonidazole (white) and PyMT (green), and fraction of PyMT-positive cells in normoxic and hypoxic areas. The area outlined corresponds to the hypoxic, pimonidazole-positive section, arrowheads point to PyMT-negative cells, scale: 25µm. The bar chart inset illustrates the relative number of PyMT-positive cells in normoxic and hypoxic areas (resp. grey and red; n=19). f, Ki67-positive cells in PyMT tumors: representative image of staining for DNA (propidium iodide, blue), Ki67 (red) and pimonidazole (green); scale: 50 µm. The bar chart inset illustrates the quantification of Ki67-positive cells in normoxic and hypoxic areas (resp. grey and red) across 6 tumors, analysing 3 field of view with over 150 cells per field of view. g, CD45-positive cells in PyMT tumors: representative image of staining for DNA (propidium iodide, blue), 5hmC (red), pimonidazole (green) and CD45 (white); scale: 100 µm. The bar chart inset illustrates the quantification of CD45-positive cells in normoxic and hypoxic areas (resp. white and red) of 11 tumors, capturing on average ~2,500 nuclei per analysis. *** P<0.001 in (a) by Fisher’s exact test, significance relative to all genes.
Extended data figure 9
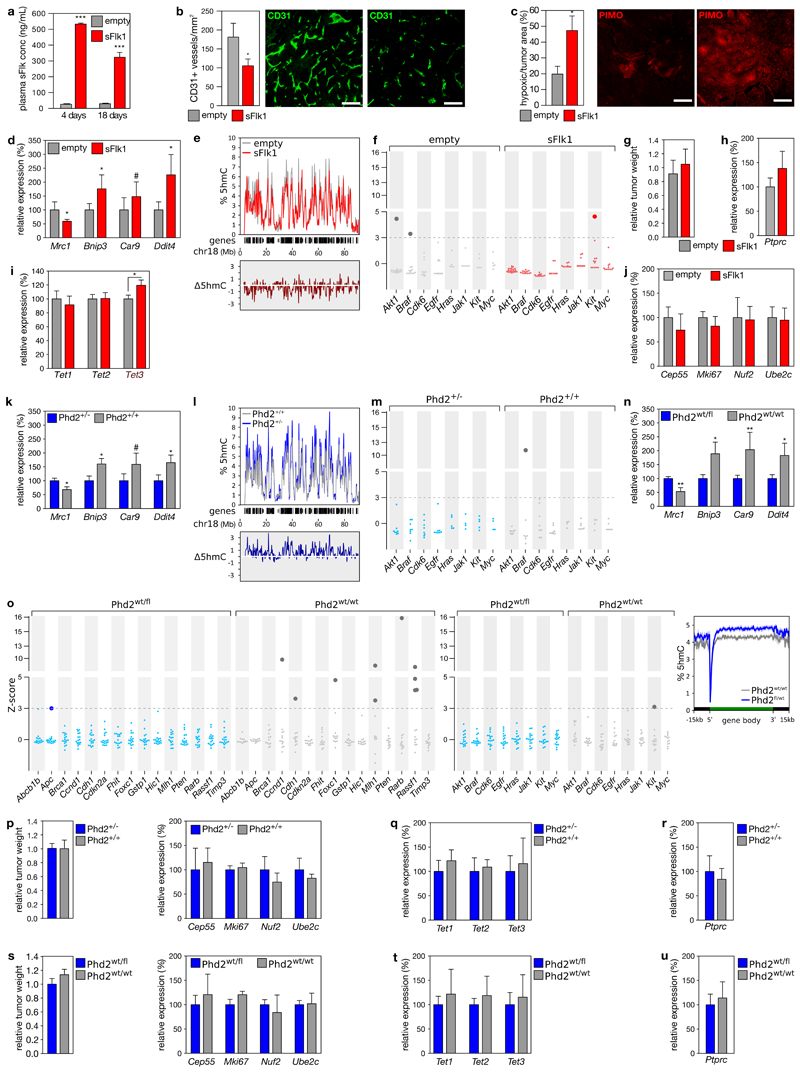
a, Plasma sFlk1 concentrations at the indicated times after hydrodynamic injection with an empty (n=7) or sFlk1-overexpression plasmid (n=5) (resp. grey and red). b-cQuantification of tumor vessel number (b) and hypoxic areas (c) of tumors from tg(MMTV-PyMT) mice, hydrodynamically injected with an empty or sFlk1-overexpression plasmid, with representative images of blood vessels stained for CD31 (b) and hypoxic areas stained for pimonidazole adducts (c); scale: 100µm. d Changes in RNA expression of hypoxia marker genes that are known to be downregulated (Mrc1) or upregulated (Bnip3, Car9, Ddit4) in hypoxic conditions. e, 5hmC levels (y axis) across mouse chromosome 18 (x axis) in 400kb bins, with the location of RefSeq genes (middle), and differences in 5hmC levels (lower). 5hmC levels were determined using shallow TAB-seq, and chromosome 18 was selected because it has large stretches of gene deserts that illustrate the 5hmC depletion in these areas (n=3). 5hmC levels decrease by 12.4±3.5% after sFlk1 overexpression, although technical limitations of TAB-seq (incomplete 5hmC protection or bisulfite conversion) may partially obscure the magnitude of effects. f, HM in tumors developing in 12-week-old mice receiving hydrodynamic injection with an empty (n=19) or sFlk1-overexpressing plasmid (n=24) 3 weeks earlier. DNA was bisulfite converted, PCR-amplified for the indicated oncogenes, and sequenced to a depth of ~500x. Plotted are z-scores of HM (y axis, exponential), relative to the more normoxic tumors (i.e. empty). The dotted line represents the threshold at 5% FDR, and bold darker dots the tumors displaying significantly increased HM events. g-j,(g) Relative weights of tumors from tg(MMTV-PyMT) mice, hydrodynamically injected with an empty (grey, n=19) or sFlk1-overexpression plasmid (red, n=24), and corresponding RNA expression of Ptprc (the gene encoding CD45, n=5) (h), of Tet enzymes (i, n=15 for empty plasmid, n=12 for sFlk1-overexpressing plasmid) and of cell proliferation markers (j, n=5 for each). k-m, As in (d-f), but for 16-week old tg(MMTV-PyMT) mice of the indicated genotype. n=5 (k), n=3 for Phd2+/+; n=4 for Phd2+/- (l) and n=9 (m) n, as in (d), but for 16-week old Tie2-Cre;tg(MMTV-PyMT) mice of the indicated genotypes (n=5). o, DNA was extracted from 17 breast tumors developing in Tie2-Cre;Phd2fl/wt;tg(MMTV-PyMT) mice (blue) and 13 breast tumors developing in Tie2-Cre;Phd2wt/wt;tg(MMTV-PyMT) mice (grey), all 16 weeks old. DNA was bisulfite converted, PCR-amplified for the indicated TSGs (left) and oncogenes (middle) and sequenced to a depth of ~500x. Plotted are z-scores of HM (y axis, exponential), relative to the more normoxic, Phd2wt/fl, tumors. The dotted line represents the threshold for a Bonferroni-adjusted P<0.05, and bold darker dots the tumors displaying significantly increased HM events. (right) 5hmC levels ± s.e.m. across a metagene in tumors of 16-week-old mice with the indicated genotype (n=3 for Phd2fl/fl; n=4 for Phd2wt/fl). p-u, Relative weights of tumors from Phd2+/-;tg(MMTV-PyMT) mice and Phd2+/+;tg(MMTV-PyMT) mice (n=10 and 9 resp.) (p-r) and from Tie2-Cre;Phd2fl/wt;tg(MMTV-PyMT) and Tie2-Cre;Phd2wt/wt;tg(MMTV-PyMT) mice (n=17 and 13 resp.) (s-u), and the corresponding RNA expression of cell proliferation markers (n=5) (p, s), of Tet enzymes (n=5) (q, t) and of Ptprc (n=5), the gene encoding CD45 (r, u). # P<0.10, * P<0.05, ** P<0.01, *** P<0.001 by t-test.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
supp_tables
Acknowledgements
We thank Gilian Peuteman, Thomas Van Brussel, Jens Serneels and Kerstin Kurz for assistance, Christopher Chang for NucPE1, Guo-Liang Xu for Tet-TKO ESCs. HZ and BT hold FWO-F postdoctoral fellowships. This work was supported by a ERC consolidator grant (CHAMELEON- 617595) to D.L.
Footnotes
Contributions B.T. and D.L. conceived and supervised the project, designed experiments, wrote the manuscript. B.T. and F.D.A. performed in vitro experiments and analysed data, helped by L.V.D.; M.C. and A.P. analysed Tet Michaelis-Menten kinetics; animal models provided by E.H., F.A. (xenografts), M.M. (sFlk1), A.K. and P.C. (Phd2+/-); V.N.K. contributed ideas, L.S. and K.P.K. reagents; J.S. quantified nucleotides by LC/MS, supervised by T.C.; B.G. quantified metabolites. H.Z. analysed TCGA; B.T., H.Z. and B.B. performed bioinformatics and statistics.
Author Information Microarray and sequencing data are available at GEO under accession GSE71403. Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper.
The authors declare no competing financial interests.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nature19081
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5133388
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1038/nature19081
Article citations
Epigenomic heterogeneity as a source of tumour evolution.
Nat Rev Cancer, 16 Oct 2024
Cited by: 0 articles | PMID: 39414948
Review
ALKBH4 functions as a hypoxia-responsive tumor suppressor and inhibits metastasis and tumorigenesis.
Cell Oncol (Dordr), 14 Oct 2024
Cited by: 0 articles | PMID: 39400679
Hypoxic Memory Mediates Prolonged Tumor-Intrinsic Type I Interferon Suppression to Promote Breast Cancer Progression.
Cancer Res, 84(19):3141-3157, 01 Oct 2024
Cited by: 0 articles | PMID: 38990731
Epithelial-Mesenchymal Plasticity and Epigenetic Heterogeneity in Cancer.
Cancers (Basel), 16(19):3289, 27 Sep 2024
Cited by: 0 articles | PMID: 39409910 | PMCID: PMC11475326
Review Free full text in Europe PMC
NF-κB and TET2 promote macrophage reprogramming in hypoxia that overrides the immunosuppressive effects of the tumor microenvironment.
Sci Adv, 10(38):eadq5226, 18 Sep 2024
Cited by: 0 articles | PMID: 39292770 | PMCID: PMC11409945
Go to all (320) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
GEO - Gene Expression Omnibus
- (1 citation) GEO - GSE71403
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
TET proteins and 5-methylcytosine oxidation in hematological cancers.
Immunol Rev, 263(1):6-21, 01 Jan 2015
Cited by: 137 articles | PMID: 25510268 | PMCID: PMC4617313
Review Free full text in Europe PMC
TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling.
Nature, 538(7626):528-532, 19 Oct 2016
Cited by: 117 articles | PMID: 27760115
Structure and Function of TET Enzymes.
Adv Exp Med Biol, 945:275-302, 01 Jan 2016
Cited by: 19 articles | PMID: 27826843
Review
TET enzymes as oxygen-dependent tumor suppressors: exciting new avenues for cancer management.
Epigenomics, 8(11):1445-1448, 13 Oct 2016
Cited by: 8 articles | PMID: 27733058
Funding
Funders who supported this work.
Biotechnology and Biological Sciences Research Council (1)
Grant ID: 1734215
European Research Council (1)
Cellular Hypoxia Alters DNA MEthylation through Loss of Epigenome OxidatioN (CHAMELEON)
Prof Diether Lambrechts, Flanders Institute for Biotechnology (VIB)
Grant ID: 617595
Medical Research Council (1)
Defining a protein hydroxylase tumour suppressor pathway
Dr Mathew Coleman, University of Birmingham
Grant ID: MR/N021053/1





