Abstract
Free full text

Identification of protein phosphatase 1 as a regulator of the LRRK2 phosphorylation cycle
Associated Data
Abstract
A cluster of phosphorylation sites in LRRK2 (leucine-rich repeat kinase 2), including Ser910, Ser935, Ser955 and Ser973, is important for PD (Parkinson’s disease) pathogenesis as several PD-linked LRRK2 mutants are dephosphorylated at these sites. LRRK2 is also dephosphorylated in cells after pharmacological inhibition of its kinase activity, which is currently proposed as a strategy for disease-modifying PD therapy. Despite this importance of LRRK2 dephosphorylation in mutant LRRK2 pathological mechanism(s) and in LRRK2’s response to inhibition, the mechanism by which this occurs is unknown. Therefore we aimed to identify the phosphatase for LRRK2. Using a panel of recombinant phosphatases, we found that PP1 (protein phosphatase 1) efficiently dephosphorylates LRRK2 in vitro. PP1 activity on LRRK2 dephosphorylation was confirmed in cells using PP1 inhibition to reverse LRRK2 dephosphorylation induced by the potent LRRK2 kinase inhibitor LRRK2-IN1 as well as in R1441G mutant LRRK2. We also found that PP1 and LRRK2 can form a complex in cells. Furthermore, we observed that PP1 inhibition modulates LRRK2’s cellular phenotype by reducing skein-like LRRK2-positive structures associated with dephosphorylation. In conclusion, the present study reveals PP1 as the physiological LRRK2 phosphatase, responsible for LRRK2 dephosphorylation observed in PD mutant LRRK2 and after LRRK2 kinase inhibition.
INTRODUCTION
Mutations in the LRRK2 (leucine-rich repeat kinase 2) gene are a common cause of inherited forms of PD (Parkinson’s disease) [1,2]. Moreover, genome-wide association studies have identified LRRK2 as a risk factor for sporadic PD [3–6]. LRRK2 encodes a large multi-domain protein comprising, among others, domains with GTPase and kinase activity. The essential catalytic core encompasses the GTPase domain, termed ROC (Ras of complex proteins), which is linked to the kinase domain via the COR (C-terminal of ROC) domain. The catalytic segment of LRRK2 is flanked at its C-terminus by a WD40 domain and at the N-terminus by armadillo and ankyrin repeats and the namesake leucine-rich repeat domain [7] (Supplementary Figure S1 at http://www.biochemj.org/bj/456/bj4560119add.htm). Of the many LRRK2 substitutions described, the majority of those that are proven to be pathogenic are concentrated in the catalytic core: N1437H, R1441C/G/H and Y1699C in the ROC–COR bidomain and G2019S and I2020T in the kinase domain [8] (Supplementary Figure S1). The most common mutation in patients, G2019S, is located in the kinase domain and has consistently been shown to increase kinase activity 2–3-fold [9], suggesting that targeting kinase activity might be a therapeutic strategy [10]. The other pathogenic substitutions, R1441C/G/H, Y1699C and I2020T, have more modest effects on kinase activity [9,11–13].
The LRRK2 protein is phosphorylated at multiple sites, but the regulation of its phosphorylation is not fully understood [14–18]. LRRK2 displays two distinct clusters of phosphosites, one set of autophosphorylation sites with low or no basal phosphorylation levels in cells and a second set of serine residues with high basal phosphorylation levels located between the ankyrin and leucine-rich repeat domains (Supplementary Figure S1) [19]. Evidence suggests that these sites, including Ser910, Ser935, Ser955 and Ser973, are phosphorylated by kinases other than LRRK2 itself ([12,16,17,19,20] and Supplementary Figure S2 at http://www.biochemj.org/bj/456/bj4560119add.htm). Some nominated kinases are PKA (protein kinase A) or the IKK (inhibitor of κB kinase) family which phosphorylate LRRK2 when activated [16,21], although administration of the PKA inhibitor H89 does not significantly block total or Ser935 phosphorylation [22,23]. Antibodies against the Ser910/Ser935/Ser955/Ser973 sites label LRRK2 extracted from cells or from the brain, showing that these are physiologically relevant modifications of the protein in vivo [24].
Changes in the cellular LRRK2 phosphorylation status are linked to the pathogenesis of LRRK2-related PD as disease-associated mutations R1441G/C/H, Y1699C, I2020T and G2385R display reduced phosphorylation at these sites compared with WT (wild-type) LRRK2 [16,17,20]. Cellular LRRK2 phosphorylation governs a number of LRRK2-mediated processes, including regulation of the cellular LRRK2 complex and subcellular localization. Indeed, decreased cellular phosphorylation correlates with decrements in phospho-dependent 14-3-3 binding and consequent subcellular relocalization of LRRK2 [17]. PD mutations that induce cytoplasmic accumulation of LRRK2 can relocalize to both filamentous and large inclusions in different cell types [17,25–27].
Interestingly, many laboratories have observed that acute inhibition of LRRK2 kinase activity leads to dephosphorylation of LRRK2, loss of 14-3-3 binding [15,16,20,24] as well as subcellular relocalization to skein-like structures [14,15,24]. Typically, these structures are filamentous and co-localize with microtubules [15]. This is a crucial observation, as this potential therapeutic avenue for PD induces cellular phenotypes (decreased phosphorylation, loss of 14-3-3 binding and relocalization) that are similar to pathogenic PD mutations in LRRK2. The nexus of these events is the dephosphorylation of LRRK2.
Taken together, the available data show that phosphorylation of LRRK2 is a highly regulated physiological event that is relevant to the effects of PD-related mutations and represents a potential pharmacodynamic marker for the effects of kinase inhibitors [22]. However, little is known about the regulation of LRRK2 phosphorylation and the data indicate that dephosphorylation is a unifying theme of PD mutations and LRRK2 kinase inhibition. We therefore set out to identify the major phosphatase(s) responsible for regulating LRRK2 phosphorylation. Using a combination of in vitro, pharmacological and molecular approaches, we identified PP1 (protein phosphatase 1) as a key LRRK2 phosphatase in physiological and pathological conditions. These results provide a mechanistic explanation for the altered phosphorylation of pathogenic LRRK2 variants and following cellular treatment with the potent LRRK2 kinase inhibitor LRRK2-IN1 [24].
MATERIALS AND METHODS
Cell culture and transfection
HEK (human embryonic kidney)-293T cells, NTH 3T3 cells and SH-SY5Y cells were maintained in DMEM (Dulbecco’s modified Eagle’s medium) (Gibco-Life Technologies) supplemented with 8% or 15% heat-inactivated FBS (Harlan Sera-Lab) and 50 μg/ml gentamycin at 37 °C in a humidified atmosphere containing 5% CO2. A549 cells were maintained in RPMI medium with L-glutamine supplemented with 10% FBS, and 0.1 mM MEM (modified essential medium) Non-Essential Amino Acids solution. Overexpression of LRRK2 in HEK-293T and SH-SY5Y cells was obtained by transduction with a lentiviral vector encoding 3×FLAG-LRRK2 or eGFP-LRRK2. Lentiviral vectors encoding human 3×FLAG-LRRK2 and eGFP-LRRK2 under control of the CMV (cytomegalovirus) promoter were prepared as described previously [28]. For transduction in cell culture, 50000 HEK-293T or SH-SY5Y cells were plated in a 24-well plate. The next day, vector was applied to the cells for 2 days. This procedure was repeated twice with the same cells to obtain triple transduced cells. These cells were then expanded for use in experiments. HEK-293 T-REx cell lines were cultured in DMEM supplemented with 10 % FBS and 2 mM glutamine, 1 × antimycotic/antibiotic, 15 μg/ml blasticidin and 100 μg/ml hygromycin. T-REx cultures were induced to express the indicated protein by inclusion of 1 μg/ml doxycycline in the culture medium for 24 h. Transfections were performed 24 h after seeding for 48 h with linear polyethylenimine (Polysciences Europe) according to the manufacturer’s protocol. For co-immunoprecipitation, T-REx HEK-293 cells were transfected using polyethylenimine [29]. All cultures were mycoplasma-free.
Pharmacological treatment of cells
Cells grown until 50–100% confluency were treated with the indicated compounds, dissolved in pre-warmed culture medium to the final treatment concentration, for the time indicated and with DMSO treatment as a negative control. Except where indicated, cells were rinsed twice in PBS and lysed in lysis buffer [20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 % Triton X-100, 10 % (v/v) glycerol, protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche)] or as described in [14]. Cell lysates were cleared by centrifugation at 15000 g for 10 min. Membranes were processed as described in the Supplementary Materials and methods section (at http://www.biochemj.org/bj/456/bj4560119add.htm).
Metabolic labelling
Metabolic labelling was performed essentially as described in [30]. Briefly, cells expressing LRRK2 were rinsed twice in DMEM without phosphates and then labelled with 5 μCi/cm2 [32P]Pi (PerkinElmer) at 37 °C. Following 4–8 h labelling, cells were lysed and LRRK2 was immunoprecipitated using FLAG-M2-agarose beads (Sigma). Immunoprecipitated protein was resolved on Nupage 3–8 % tris-acetate SDS/PAGE gels (Invitrogen) and immunoblotted. Membranes were stained with Ponceau S (Sigma) to visualize proteins. Membranes were processed as described in the Supplementary Materials and methods section.
Statistics
Unless otherwise indicated, the Figures shown are representative of at least three independent experiments. Phosphorylation levels were normalized for expression levels and experimental test conditions were normalized to control conditions. Statistical analysis was performed using one-way ANOVA or one-sample t test comparing test values with the hypothetical value of 1 with Bonferroni correction for multiple comparisons as appropriate. For counting of skein-like structures in cells, analysis was performed with Fisher’s exact test on the proportion of cells with skein-like inclusions out of all transfection-positive cells. Statistical significance was set at P < 0.05.
RESULTS
PP1α efficiently dephosphorylates LRRK2 in vitro
We first identified phosphatases able to dephosphorylate cellular LRRK2 phosphorylation sites in vitro from a panel of catalytic subunits of common serine/threonine phosphatases. To visualize total phosphorylation levels, LRRK2 was metabolically labelled with 32P in HEK-293T cells overexpressing 3×FLAG-LRRK2, affinity purified and incubated with phosphatases at a limiting concentration (12.5 units/ml, see the Supplementary Materials and methods section). Alkaline phosphatase (250 units/ml) was included as a positive control [17]. All phosphatases were active under these experimental conditions against the test substrates phosphorylated MBP (myelin basic protein) or pNPP (p-nitrophenyl phosphate) (Supplementary Figure S3 at http://www.biochemj.org/bj/4567bj4560119add.htm). We found that PPlα (HGNC symbol PPP1CA) was able to dephosphorylate LRRK2 to the same extent as the positive control. Using antibodies against pSer910, pSer935, pSer955 and pSer973 LRRK2, we found that all individual sites were efficiently dephosphorylated by PPlα. Partial dephosphorylation was observed after treatment with PP2A1 (HGNC symbol PPP2CA) as well as with PPλ (Figure 1), although this did not reach statistical significance. These results identify PPlα as a candidate phosphatase for the constitutive phosphorylation sites of LRRK2.
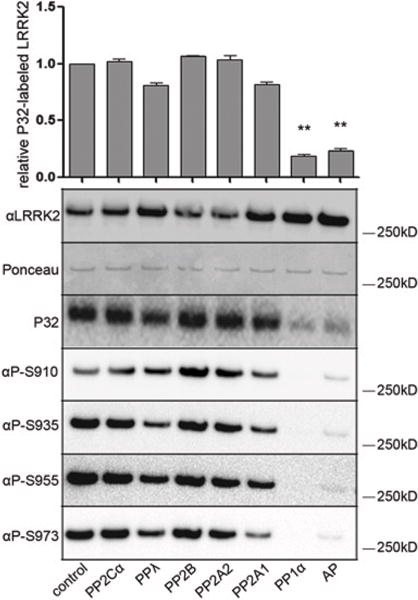
LRRK2, metabolically labelled with 32P, was immunopurified from HEK-293T cells overexpressing 3×FLAG–LRRK2, and incubated with the indicated phosphatases in vitro. Shown are the incorporated 32P levels as well as the levels of phosphorylation at Ser910, Ser935, Ser955 and Ser973 measured using phosphosite-specific anti-LRRK2 antibodies. Total LRRK2 levels were detected using both Ponceau S and immunoblotting. The graphs show the quantification of incorporated 32P in LRRK2 after in vitro dephosphorylation normalized to total LRRK2 levels using immunoblotting of three independent experiments, ±S.E.M. Blots show representative results. Statistical significance was assessed by single sample t tests combined with Bonferroni correction for multiple testing. **P < 0.01. Ap, alkaline phosphatase.
LRRK2 interacts with PP1α
We next considered whether different PP1 isoforms were LRRK2 phosphatases in vivo. First, we examined localization in cells and found that LRRK2 strongly co-localized with PPlα and PPlβ in the cytoplasm (Figure 2A). PPlγ was primarily localized to the nucleus, although LRRK2 co-localized with the fraction of PPlγ present in the cytoplasm (Figure 2A). Secondly, we examined whether PP1 isoforms physically bound LRRK2. We transiently expressed FLAG-tagged PP1 isoforms or PP2Aα in HEK-293 T-REx cells expressing GFP or GFP-LRRK2. We found that PPlα formed a non-ionic detergent-stable complex with LRRK2, whereas interaction of LRRK2 with PP1β, PPlγ or PP2Aα was not detected under the same conditions (Figure 2B). We also observed that the LRRK2-PPlα interaction was enhanced after LRRK2-IN1 treatment and abolished after treatment with calyculin A (an inhibitor of PP1 and PP2A). The immunoblot for pSer935 and the 14-3-3 overlay assay were used to confirm the induced dephosphorylation of LRRK2 after inhibition [17,20] (Figure 2B).
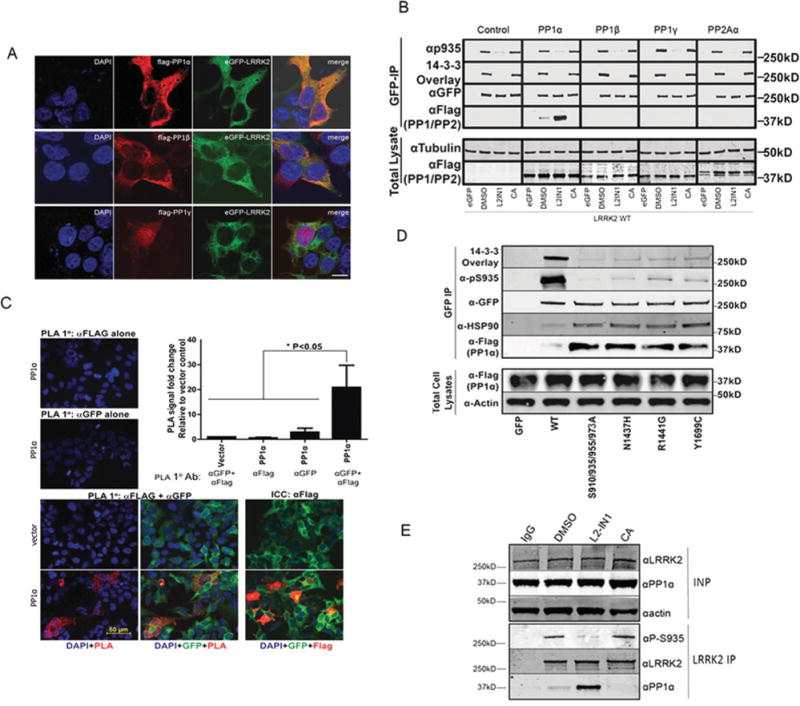
(A) HEK-293T cells with stable eGFP–LRRK2 expression were transfected with 3 × FLAG-tagged PP1α, PP1β or PP1γ. Cells were stained with DAPI (blue, left-hand panels), and proteins were detected by immunofluorescence using FLAG for PP1 isoforms (red, second panels) and eGFP for LRRK2 (green, third panels). Merged images are shown in the right-hand panels. The scale bar represents 10 μm and applies to all photomicrographs. (B) Co-immunopurification (IP) of PP1α with LRRK2, T-REx lines, stably expressing eGFP alone or WT eGFP–LRRK2 and transiently expressing FLAG-tagged PP1 isoforms or PP2Aα, were treated with 1 μM LRRK2-1N1 for 90 min or 20 nM calyculin A (CA) for 30 min. GFP complexes were isolated with GFP nano trap beads and analysed by immunoblotting with the indicated antibodies. Representative blots are shown for three independent experiments. p935, pSer935 (C) PLA. T-RExcells, stably expressing WT GFP–LRRK2, were transfected with FLAG vector or FLAG–PP1α as indicated. At 48 h after transfection, cells were subjected to PLA following the manufacturer’s protocol using the indicated primary antibodies. The far left-hand panels show the merged image of cells stained with DAPI (blue) and ligation amplifications (red). A merged image of GFP-LRRK2 with DAP/PLA is presented in the middle column. PLA signals per cell were quantitated by Duolink ImageTool and are presented as fold change from the number of vector signals per cell from n=3 experiments. Results are means+ S.D., analysis was one-way ANOVA against each control (*P < 0.05). The right-hand panels show immunocytochemistry with anti-FLAG antibody staining of transfected PP1α. Merge image shows DNA (blue), GFP–LRRK2 (green) and FLAG–PP1 (red). 1°, primary; ICC, immunocytochemistry, (D) Transfection of FLAG-tagged PP1α in T-REx HEK-293 cell lines overexpressing eGFP–LRRK2 WT or the mutants indicated. GFP-overexpressing cells were included as a negative control. Cells were lysed using Nonidet P40 lysis buffer and cell lysates were immunoprecipitated by GFP-Trap agarose. Immunopurified samples and total cell lysates were analysed by immunoblotting with the indicated antibodies. Representative blots are shown for three independent experiments. (E) Co-immunopurification of endogenous PP1α with endogenous LRRK2 in A549 cells after treatment with DMS0, 2 μM LRRK2-IN1 or 20 nM calyculin A. Input (INP) and co-immunoprecipitation (IP) samples were analysed with immunoblotting with indicated antibodies. Representative blots are shown for four independent experiments. Molecular masses in kDa are indicated to the side of Western blots.
Thirdly, we verified spatial proximity of these molecules in situ using the PLA (proximity ligation assay) [31], We performed in situ PLA on cells co-expressing FLAG-tagged PPlα and GFP-LRRK2 and quantified the number of individual ligation amplification signals per cell (Figure 2C). Control amplifications with only single antibody (anti-GFP or anti-FLAG alone) staining, where both antigens are present yielded no PLA signals. Additionally, no PLA signals were observed when both antibody probes were applied to vector-transfected cells, where tagged PP1 is not present but LRRK2 is expressed. We observed specific and significant PLA signals for PP1 and LRRK2 when antibodies against both epitope tags are used and both GFP-LRRK2 and FLAG-PPl are expressed, verifying in vivo interaction of these molecules.
The enhanced interaction of PPlα with LRRK2 after acute inhibition prompted us to test whether there is a similar recruitment of PP1 to mutants of LRRK2 that are dephosphorylated. We compared the interaction of FLAG-PPlα with GFP variants of LRRK2 expressed in inducible HEK-293 T-REx cells and found enhanced binding for mutants with reduced basal phosphorylation, such as N1437H, R1441G and Y1699C, as well as for the combined S910/935/955/973A (Figure 2D). As a positive control, we tested for HSP90 (heat-shock protein 90) co-immunoprecipitation and observed interaction with LRRK2 that was enhanced with LRRK2 aggregation-prone mutations as seen previously [17,20,32].
We confirmed the LRRK2–PP1 interaction for endogenous LRRK2 and PPlα in A549 cells (Figure 2E). Furthermore, we could show an LRRK2-IN1 -induced increase in the endogenous PPlα–LRRK2 interaction (Figure 2E).
Pharmacological inhibition of PP1 prevents LRRK2 dephosphorylation
We next tested whether inhibition of PP1 with small molecules would prevent dephosphorylation of LRRK2. We first treated HEK-293T cells, stably overexpressing LRRK2, with calyculin A or okadaic acid (a PP2A-selective inhibitor at the used concentration) [33] and found that neither compound affected overall LRRK2 phosphorylation under basal conditions (Figure 3A). Next, we tested phosphatase inhibition in conditions of induced dephosphorylation after inhibition of LRRK2 kinase activity and found that calyculin A prevents the dephosphorylation effect of LRRK2-IN1, whereas okadaic acid did not (Figure 3A). These results suggest that PP1 is an authentic phosphatase for LRRK2, and its effect is strongest under conditions where LRRK2 dephosphorylation is induced.
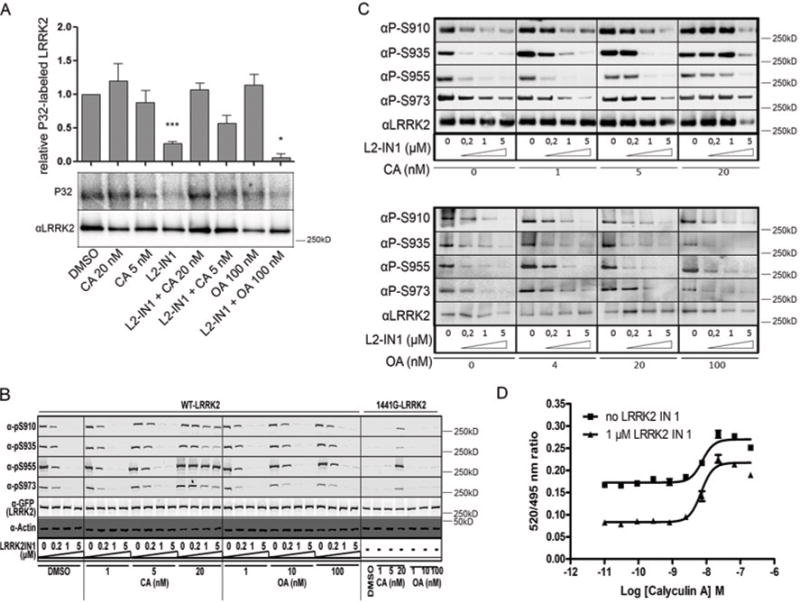
(A) Effect of phosphatase and kinase inhibitors on the overall LRRK2 phosphorylation level. HEK-293T cells stably overexpressing eGFP–LRRK2 were metabolically labelled and treated with phosphatase inhibitors and/or LRRK2-IN1 (1 μM) as indicated. Incorporated 32P was measured and normalized to total LRRK2 levels detected by immunoblotting, as described in the Materials and methods section. The graph shows quantification of three independent experiments as means ± S.E.M. Statistical significance was assessed by column statistics combined with Bonferroni correction for multiple testing. *P < 0.05, ***P < 0.001. (B) T-REx HEK-293 cells overexpressing eGFP–LRRK2 WT or the R1441G variant were treated with the indicated concentrations of okadaic acid or calyculin A alone, or combined with the indicated concentration of LRRK2-1N1. LRRK2 phosphorylation at Ser910, Ser935, Ser955 and Ser973 was determined by phosphorylation-site-specific anti-LRRK2 antibodies. Representative blots are shown for three independent experiments. (C) SH-SY5Y cells stably overexpressing eGFP–LRRK2 were treated with a concentration range of phosphatase inhibitors combined with LRRK2-IN1. Detection of phosphorylation level at the four cellular phosphorylation sites was performed as in (B). Blots show the representative results of three independent experiments (D) U20S cells expressing LRRK2–GFP were treated with 1 μM LRRK2-IN1 in the presence of the indicated dilution series of calyculin A. Cells were lysed in the presence of Terbium-labeIled anti-pSer935 antibody. TR-FRET (time-resolved FRET) was measured and the emission ratios of 520 nm/490 nm are plotted against calyculin A concentration. CA, calyculin A; OA, okadaic acid.
We confirmed these results using phospho-LRRK2-specific antibodies. First, T-REx HEK-293 cells with WT LRRK2 overexpression were treated with calyculin A or okadaic acid at different concentrations, in the presence or absence of LRRK2-IN1. Consistent with the metabolic labelling data, LRRK2-IN1-induced LRRK2 dephosphorylation could be prevented by calyculin A, but not by okadaic acid (Figure 3B). Interestingly, the low basal phosphorylation level of the LRRK2 R1441G mutant was restored to WT levels by treatment with calyculin A, but not with okadaic acid (Figure 3B). LRRK2-IN1-induced LRRK2 dephosphorylation was counteracted by calyculin A treatment, even at relatively low concentrations of 5 or 20 nM, but not by okadaic acid at concentrations up to 100 nM in a second cell type, stable SH-SY5Y cell lines (Figure 3C). We also employed a recently developed homogeneous and ratiometric assay to detect LRRK2 modification at Ser935 [22] in the absence or presence of LRRK2-IN1 (1 μM) across a dilution series of calyculin A. Calyculin A treatment in the low nanomolar range (EC50 = 6.98 nM) efficiently suppressed the induced dephosphorylation by 1 μM LRRK2-IN1 (Figure 3D) as well as by 93 nM or 10 μM LRRK2-IN1 (results not shown).
We next asked whether the regulation of LRRK2 phosphorylation is also similar for endogenous protein. Similar to our experiments on overexpressed LRRK2, treatment with calyculin A could also overcome endogenous LRRK2 dephosphorylation induced by LRRK2-IN1 in NIH 3T3 cells (Figure 4A) and in primary neurons (Figure 4B). In addition, calyculin A increased endogenous LRRK2 phosphorylation levels in primary neurons (Figure 4B). Okadaic acid treatment also showed a trend to increased pSer935 levels, suggesting that FP2A may also contribute in neurons, although this was to a much lower extent than calyculin A. These results show that, in all cell types tested, including in neurons, PP1 is the major phosphatase for LRRK2.
NIH 3T3 cells (A) or primary neurons from C57BL/6J mice (B) were treated with phosphatase inhibitor (5,20 or 100 nM), with LRRK2-IN1 (1 μM for 30 min) or with the combination of both. Monoclonal anti-pSer935 antibody was used to detect the phosphorylation level at Ser935. Histograms show the means ± S.E.M. of three independent experiments, *P < 0.05, ***P < 0.001. CA, calyculin A; OA, okadaic acid.
Molecular inhibition of PP1 counteracts LRRK2 dephosphorylation induced by kinase inhibition
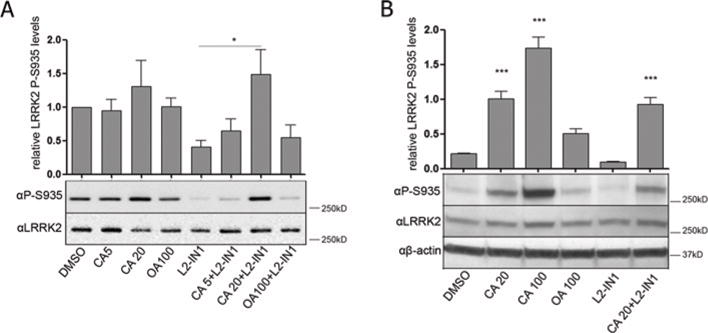
Since no specific pharmacological PP1 inhibitors are available, we wanted to confirm our data using highly specific and potent protein inhibitors of PP1, namely I2 (inhibitor-2, HGNC symbol PPP1R2) [34] or the central domain (residues 143–224) of NIPP1 (nuclear inhibitor of PP1, HGNC symbol PPP1R8) [35]. We overexpressed these protein inhibitors of PP1 in HEK-293T cells stably overexpressing LRRK2 (Supplementary Figure S4 at http://www.biochemj.org/bj/456/bj4560119add.htm) and investigated the effects on the phosphorylation level of LRRK2. Overexpression of I2 or NIPPl143–224 had no effect on basal LRRK2 phosphorylation level compared with an eGFP control. However, overexpression of NIPP1143–224 prevented LRRK2-IN1-induced LRRK2 dephosphorylation, consistent with the effects of pharmacological PP1 inhibition using calyculin A (Figure 5).
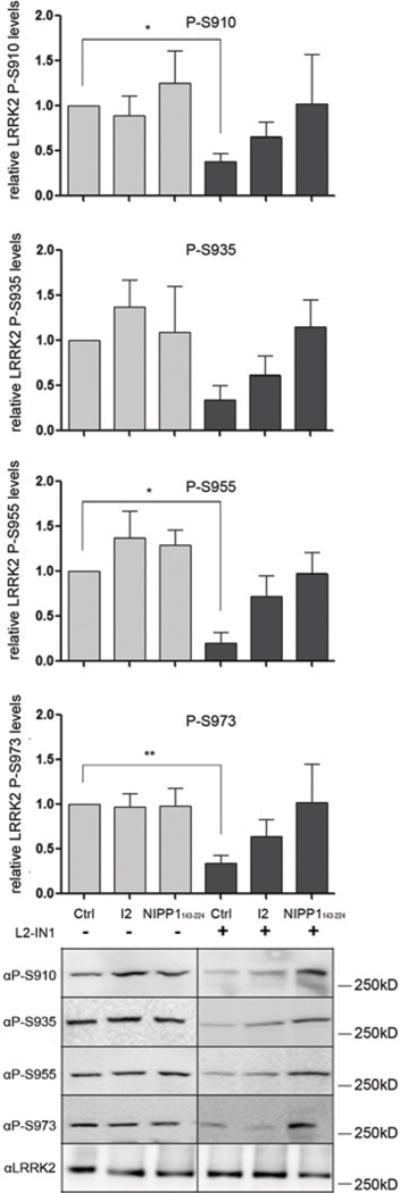
HEK-293T cells with stable eGFP–LRRK2 overexpression were transfected with expression constructs for the PP1 molecular inhibitors I2 or NIPP1143–224, or with a control construct encoding eGFP. Cells were treated with 1 μM LRRK2-IN1 or DMSO as a control for 30 min followed by cell lysis and immunoblotting with the different phosphorylation-site-specific anti-LRRK2 antibodies. Representative blots of three independent experiments are shown. Quantification of the blots are represented as a histogram. For quantification, the ratio of phosphorylation over total LRRK2 signal was normalized to control condition and are means ± S.E.M. The blots show representative results. Statistical significance was assessed by column statistics combined with Bonferroni correction for multiple testing. *P <0.05, **P <0.01.
Pharmacological Inhibition of PP1 prevents formation of LRRK2 skein-like structures
Dephosphorylation of LRRK2 induced by kinase inhibition [14,15,24] or by some pathogenic mutations [17,25–27] causes redistribution into skein-like structures. If PP1 dephosphorylates LRRK2 to induce these structures, then inhibition of PP1 should reverse the effects of mutations or acute kinase inhibition. We transiently expressed WT LRRK2 or a series of mutants (S910A/S935A, R1441C, R1441G, Y1699C, K1906M, G2019S and I2020T) in HEK-293-FT cells (Figures 6A and and6B)6B) and investigated the effect of PP1 inhibition in the presence or absence of LRRK2-IN1. Approximately 45% of cells expressing WT LRRK2 formed skein-like structures after LRRK2-IN1 treatment (1 μM for 30 min), which was significantly reduced in the presence of 5 nM calyculin A (only 22 % of cells had skein-like structures, Figure 6C). A similar decrease in cells with skein-like inclusions was observed for the pathogenic LRRK2 mutants R1441C, Y1699C and G2019S after treatment with both drugs. However, we did not observe a significant reduction in the number of cells with skein-like structures for S910A/S935A, R1441G and I2020T mutants, possibly related to a strong binding of these variants to PPlα (results not shown).
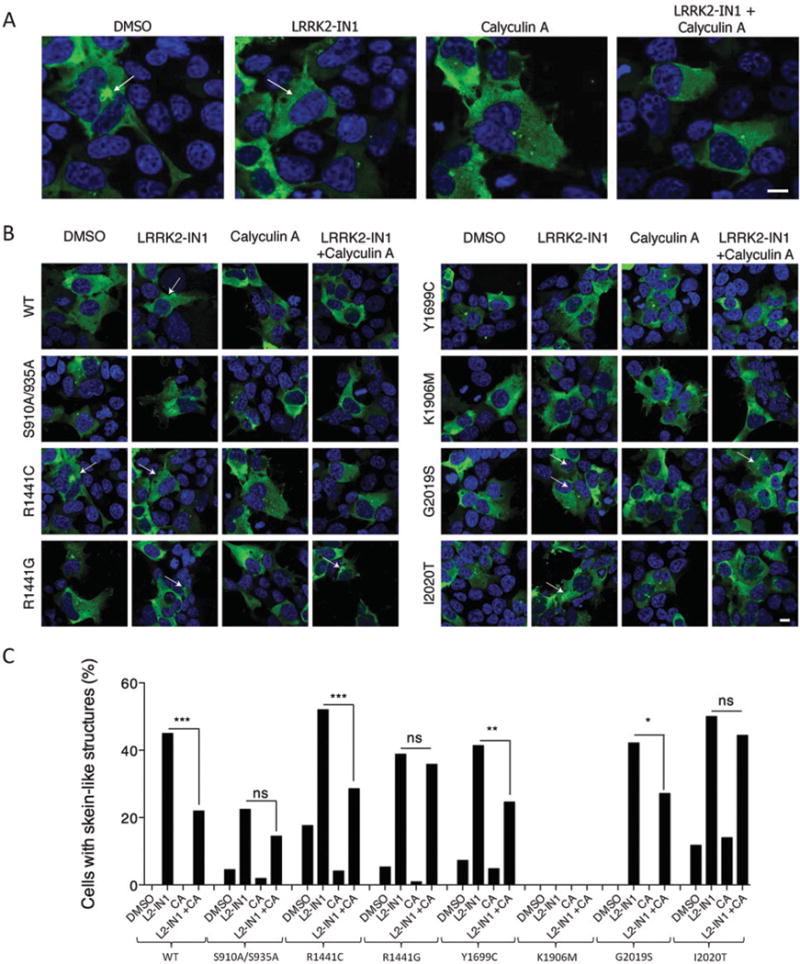
(A and B) Immunocytochemistry of HEK-293-FT cells transiently expressing 3×FLAG–LRRK2 WT and series of mutants (S910A/S935A, R1441C, R1441G, Y1699C, K1906M, G2019S and I2020T) were treated for 30 min with DMSO, 1 μM LRRK2-IN1, 5 nM calyculin A, or both 1 μM LRRK2-IN1 and 5 nM calyculin A. Arrows indicate skein-like inclusions. Enlarged images for R1441C are given in (A), whereas images for all tested LRRK2 isoforms are given in (B), Scale bar represents 10 μm and applies to all photomicrographs, (C) Quantification of cells with skein-like structures for samples described in (A) and (B), CA, calyculin A. Bars represent percentage of cells with inclusions out of all LRRK2-transfected cells counted per sample (n=100–150 cells). Statistical significances were tested using a Fisher’s exact test. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant.
The decreased percentage of cells with skein-like structures after treatment with only 5 nM of calyculin A, both in conditions of LRRK2-IN1 treatment and in mutations, confirms the link between LRRK2 dephosphorylation and the formation of skein-like inclusions and point to a role for PP1 in this process.
DISCUSSION
Similar to many kinases, LRRK2 is also a phosphoprotein and a particular cluster of phosphosites in the interdomain region between the ankyrin repeat and leucine-rich repeat domains, including sites Ser910/Ser935/Ser955/Ser973, is subject to regulation. The identification of regulators of LRRK2 cellular phosphorylation at these sites may provide an insight into the molecular basis of PD as well as into the consequences of kinase inhibition. Although studies researching kinases upstream of these sites have begun to be reported [16,21,23], there are no reports of phosphatases that can affect cellular LRRK2 phosphorylation. In the present study we employed a combination of in vitro, pharmacological and molecular cell biology approaches to identify phosphatases regulating LRRK2 phosphorylation at Ser910/Ser935/Ser955/Ser973.
First, using a panel of recombinant serine-threonine phosphatases, we show that only PP1 efficiently dephosphorylated LRRK2 in vitro at the four different cellular phosphorylation sites tested. Secondly, pharmacological inhibition of PP1, but not of PP2A, prevents LRRK2-IN1-induced dephosphorylation in five different cell types, including in HEK-293T, SH-SY5Y and U20S cell lines as well as on endogenous LRRK2 in NIH 3T3 cells and mouse primary neuron cultures. It should be noted that a trend towards increased phosphorylation was observed after PP2A inhibition with okadaic acid in primary neurons, suggesting that PP2A may play an auxiliary role in dephosphorylation of LRRK2 under specific conditions or cell types. Inhibition of PP1 also restores the reduced phosphorylation level of the R1441G disease mutant to WT levels. Finally, the functional relevance of PP1-mediated LRRK2 phosphoregulation was confirmed by the observation that PP1 inhibition reduces the formation of skein-like structures, which have previously been shown to be associated with dephosphorylated LRRK2 [14,15,17,24–27], both for mutant forms of LRRK2 as well as after LRRK2 kinase inhibition. From these results, we conclude that PP1 is a regulator of the LRRK2 phosphorylation cycle (Figure 7).
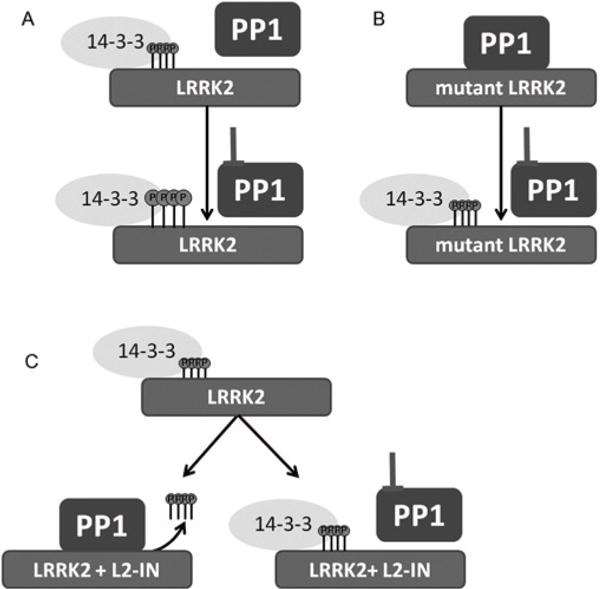
(A) WT LRRK2 is basally phosphorylated in cells at multiple sites clustering in the N-terminal half of the protein (see Supplementary Figure S1 at http://www.biochemj.org/bj/456/bj4560119add.htm for a schematic representation of the cellular phosphorylation sites relative to the functional domains). In its phosphorylated state, LRRK2 binds 14-3-3 proteins tightly and PP1 is loosely associated with the cellular LRRK2 complex Under conditions of PP1 inhibition (pharmacological inhibition with calyculin A or molecular PP1 inhibition with NIPP1143–224, see text), LRRK2 phosphorylation level is only modestly increased. (B) Several disease mutants are basally dephosphorylated and this coincides with the observation reported in the present study that PP1 binds tightly to dephosphorylated disease mutants. Mutant LRRK2 can be rephosphorylated by PP1 inhibition. (C) LRRK2 kinase inhibition (L2-IN) leads to dephosphorylation of LRRK2. The dephosphorylation induced by LRRK2-IN is blocked by PP1 inhibition.
The results of the present study suggest that LRRK2 dephosphorylation involves the enhanced access of PP1 to LRRK2 phosphosites. The PPlα interaction with LRRK2 is weak under basal conditions, consistent with rapid binding kinetics for phosphatases. However, we found that some pathogenic mutant forms of LRRK2, such as R1441G or Y1699C, had increased interaction with PPlα. These data are consistent with the previous observation that most LRRK2 PD mutations have decreased phosphorylation at Ser910/Ser935 [14,16,17]. Our data also show that transient inhibition of kinase activity of LRRK2 by small molecules leads to enhanced interaction between PPlα and LRRK2.
Several considerations point to conformational changes in LRRK2 to enable PP1 action as a regulatory mechanism of LRRK2 dephosphorylation. For example, pathogenic mutants with reduced phosphorylation levels such as R1441G and Y1699C display a weakened dimerization which could be explained by an enhanced intramolecular ROC–COR interaction [36,37]. Therefore enhanced PP1 recruitment/activity appears to correlate to altered properties of the LRRK2 catalytic core either via reduced ROC–COR dimerization or through structural or compound-induced changes in the ATP-binding pocket of the kinase domain. Resolving the crystal structure of the entire LRRK2 molecule may be required to further test these hypotheses.
Phosphorylation levels of cellular proteins are determined by the net effect of phosphorylation by kinases and dephosphorylation by phosphatases. To date, PKA [16,23] and IκB (inhibitor of nuclear factor κB) kinases [21] have been suggested to be responsible for the cellular phosphorylation of LRRK2. However, the rapid dephosphorylation induced by LRRK2-IN1 (>80% dephosphorylation after 90 min [21,24]) and the relatively long half-life of LRRK2 in cells [38,39], suggests that enhanced dephosphorylation rather than decreased phosphorylation of LRRK2 by kinases could be the primary mechanism of LRRK2 dephosphorylation. This conclusion is also in line with our finding that PP1 activity is necessary for LRRK2 to become dephosphorylated.
Cellular PP1 activity is provided by a holoenzyme, which consists of a catalytic subunit bound to one or two regulatory subunits that provide substrate specificity [40,41]. There are three different isoforms of the PP1 catalytic subunit, namely PPlα, PPlβ and PPlγ, which have identical catalytic properties and are all sensitive to the inhibitors used in the present study. As our results show that PPlα preferentially interacts with LRRK2, this suggests that PPlα is the primary PP1 isoform active on LRRK2. All three PP1 isoforms are widely expressed with high abundance in the brain where they have been shown to play a role in different neuronal processes such as synaptic plasticity and memory [42,43]. Intriguingly, both LRRK2 [44,45] and PPlα [46] are specifically enriched in the striatum.
The present study points to new possibilities in targeting LRRK2 function. PP1 can be targeted in a highly specific manner to its substrate through its interaction with regulatory subunits [41]. This unique complex, including the PP1 substrate, PP1 and PP1 regulatory subunits could be disrupted with specific protein–protein interaction inhibitors of the interaction between the PP1 catalytic subunit (PPlα) and its regulatory subunits or its substrate [47]. Further research is needed to identify the specific regulatory components of the LRRK2-specific PP1 holoenzyme in order to test this approach.
In summary, we provide the first report confirming a phosphatase, PP1, as a physiological regulator of LRRK2 cellular phosphorylation in cell lines and in neurons. Mechanistically, we find that dephosphorylation is perhaps mediated by enhanced accessibility of PP1 to the LRRK2 complex, thereby explaining why LRRK2 is dephosphorylated following kinase inhibition or in disease mutants. By revealing a major regulator of LRRK2 phosphorylation in cells, the present study provides new insight into the regulation of LRRK2 physiological and pathological functions and points to the molecular consequences of a much discussed potential therapy for PD, namely LRRK2 kinase inhibition.
Acknowledgments
We thank Professor Johan Hofkens and Charlotte David (Molecular Imaging and Photonics, KU Leuven) for the use of the confocal laser-scanning microscope.
FUNDING
This study was supported by the Michael J. Fox Foundation (to J.-M.T., V.B. and R.J.N.). We thank the FWO (Research Foundation – Flanders)-Vlaanderen FWO project [number G.0666.09 (to V.B.)] and fellowships to E.L and J.M.T., the IWT (Agency for Innovation by Science and Technology) SBO/80020 project Neuro-TARGET to VB, and the KU Leuven [grant numbers OT/08/052A and IOF-KP/07/001 (to V.B.)] for their support. This research was also supported in part by the Intramural Research Program of the National Institutes of Health, National Institute on Aging (to M.R.C.). This research was also supported in part by the benevolence of the Brin/Wojcicki foundation (to R.J.N.) and the Fund Druwé-Eerdekens managed by the King Baudouin Foundation (to J.-M.T).
Abbreviations used
| COR | C-terminal of ROC |
| DMEM | Dulbecco’s modified Eagle’s medium |
| HEK | human embryonic kidney |
| I2 | inhibitor-2 |
| LRRK2 | leucine-rich repeat kinase 2 |
| NIPP-1 | nuclear inhibitor of protein phosphatase 1 |
| PD | Parkinson’s disease |
| PKA | protein kinase A |
| PLA | proximity ligation assay |
| PP | protein phosphatase |
| ROC | Ras of complex proteins |
| WT | wild-type |
Footnotes
AUTHOR CONTRIBUTION
Evy Lobbestael, Jing Zhao, Veerle Baekelandt, R. Jeremy Nichols and Jean-Marc Taymans designed the research. Evy Lobbestael, Jing Zhao, lakov Rudenko, Aleksandra Beylina and Fangye Gao performed the research. Monique Beullens, Mathieu Bollen, Mark Cookson, Veerle Baekelandt, R. Jeremy Nichols and Jean-Marc Taymans contributed new reagents. Evy Lobbestael, Jing Zhao, lakov Rudenko, Aleksandra Beylina, Mark Cookson, R. Jeremy Nichols and Jean-Marc Taymans analysed the data. Evy Lobbestael, Jing Zhao, Mark Cookson, Veerle Baekelandt, R. Jeremy Nichols and Jean-Marc Taymans wrote the paper. All authors read and approved the paper.References
Full text links
Read article at publisher's site: https://doi.org/10.1042/bj20121772
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc5141581?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1042/bj20121772
Article citations
G2019S selective LRRK2 kinase inhibitor abrogates mitochondrial DNA damage.
NPJ Parkinsons Dis, 10(1):49, 01 Mar 2024
Cited by: 4 articles | PMID: 38429321 | PMCID: PMC10907374
PAK6-mediated phosphorylation of PPP2R2C regulates LRRK2-PP2A complex formation.
Front Mol Neurosci, 16:1269387, 18 Dec 2023
Cited by: 1 article | PMID: 38169846 | PMCID: PMC10759229
Post-translational modification and mitochondrial function in Parkinson's disease.
Front Mol Neurosci, 16:1329554, 11 Jan 2024
Cited by: 1 article | PMID: 38273938 | PMCID: PMC10808367
Review Free full text in Europe PMC
LRRK2 is involved in the chemotaxis of neutrophils and differentiated HL-60 cells, and the inhibition of LRRK2 kinase activity increases fMLP-induced chemotactic activity.
Cell Commun Signal, 21(1):300, 30 Oct 2023
Cited by: 0 articles | PMID: 37904222 | PMCID: PMC10614378
Leucine-rich repeat kinase 2 at a glance.
J Cell Sci, 136(17):jcs259724, 12 Sep 2023
Cited by: 2 articles | PMID: 37698513 | PMCID: PMC10508695
Review Free full text in Europe PMC
Go to all (64) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Characterization of a selective inhibitor of the Parkinson's disease kinase LRRK2.
Nat Chem Biol, 7(4):203-205, 06 Mar 2011
Cited by: 256 articles | PMID: 21378983 | PMCID: PMC3287420
Protein phosphatase 2A holoenzymes regulate leucine-rich repeat kinase 2 phosphorylation and accumulation.
Neurobiol Dis, 157:105426, 16 Jun 2021
Cited by: 5 articles | PMID: 34144124
Phosphorylation of LRRK2 serines 955 and 973 is disrupted by Parkinson's disease mutations and LRRK2 pharmacological inhibition.
J Neurochem, 120(1):37-45, 11 Nov 2011
Cited by: 64 articles | PMID: 22004453
LRRK2 GTPase dysfunction in the pathogenesis of Parkinson's disease.
Biochem Soc Trans, 40(5):1074-1079, 01 Oct 2012
Cited by: 17 articles | PMID: 22988868 | PMCID: PMC3701022
Review Free full text in Europe PMC
Funding
Funders who supported this work.
Intramural NIH HHS (1)
Grant ID: ZIA AG000948-09





