Abstract
Free full text

ALK7, a Receptor for Nodal, Is Dispensable for Embryogenesis and Left-Right Patterning in the Mouse
Abstract
Mesendoderm formation and left-right patterning during vertebrate development depend upon selected members of the transforming growth factor β superfamily, particularly Nodal and Nodal-related ligands. Two type I serine/threonine kinase receptors have been identified for Nodal, ALK4 and ALK7. Mouse embryos lacking ALK4 fail to produce mesendoderm and die shortly after gastrulation, resembling the phenotype of Nodal knockout mice. Whether ALK4 contributes to left-right patterning is still unknown. Here we report the generation and initial characterization of mice lacking ALK7. Homozygous mutant mice were born at the expected frequency and remained viable and fertile. Viability at weaning was not different from that of the wild type in ALK7−/−; Nodal+/− and ALK7−/−; ALK4+/− compound mutants. ALK7 and ALK4 were highly expressed in interdigital regions of the developing limb bud. However, ALK7 mutant mice displayed no skeletal abnormalities or limb malformations. None of the left-right patterning abnormalities and organogenesis defects identified in mice carrying mutations in Nodal or in genes encoding ActRIIA and ActRIIB coreceptors, including heart malformations, pulmonary isomerism, right-sided gut, and spleen hypoplasia, were observed in mice lacking ALK7. Finally, the histological organization of the cerebellum, cortex, and hippocampus, all sites of significant ALK7 expression in the rodent brain, appeared normal in ALK7 mutant mice. We conclude that ALK7 is not an essential mediator of Nodal signaling during mesendoderm formation and left-right patterning in the mouse but may instead mediate other activities of Nodal and related ligands in the development or function of particular tissues and organs.
The formation of the mesodermal and endodermal germ layers is one of the earliest induction events during vertebrate embryogenesis. Genetic experiments with mouse and zebra fish have indicated that Nodal and related factors are required for mesendoderm formation (18). Mouse embryos lacking Nodal, the only family member identified in mammals, die shortly after gastrulation without forming a node, an organizing center similar to the dorsal lip of frog embryos that is responsible for the formation of axial mesoderm (5, 25). Although genetic experiments have indicated that the transforming growth factor β family member Activin is not required for mesendoderm formation, its ability to induce mesodermal markers upon overexpression in the early embryo has indicated that this protein may activate some component of the Nodal signaling pathway, including putative Nodal receptors. In agreement with this, the Activin type I receptor ALK4 has been shown to form a receptor complex for Nodal together with Activin type II receptors ActRIIA and ActRIIB and the extracellular protein Cripto, a member of the epidermal growth factor (EGF)-Cripto-FRL1-cryptic (CFC) family of glycosyl phosphatidylinositol-anchored proteins (16, 24). This notion is supported by loss- and gain-of-function studies indicating a role for Activin receptors and Cripto in Nodal signaling. Thus, a null mutation of Cripto abolishes mesendoderm formation (6). In addition, mice lacking ActRIIB have defects in left-right axis formation, and compound ActRIIA and ActRIIB mutants recapitulate all of the typical deficits of Nodal loss of function (2, 14, 19). Gain-of-function experiments with a constitutively activated form of ALK4 have demonstrated the ability of this receptor to induce mesendoderm and formation of a secondary axis in whole embryos (1, 3), and the phenotype of a null mutation in the mouse ALK4 gene resembles that of the Nodal knockout (9).
In addition to ALK4, the type I receptor ALK7 has also been found to mediate signaling by Nodal and related proteins (16). Unlike ALK4, however, ALK7 was capable of transmitting Nodal signals even in the absence of Cripto, albeit less efficiently (16), suggesting that ALK7 may have a higher affinity for Nodal ligands than ALK4. ALK7 mRNA has been localized to the ectodermal and organizer regions of Xenopus gastrula embryos, and low but detectable levels have also been found in the early mouse embryo (16). ALK7 is expressed predominantly in distinct subpopulations of cells in the rodent central nervous system during postnatal development, primarily cerebellum, hippocampus, cortex, and brain stem nuclei (12, 17, 20). ALK7 mRNA expression has also been detected to various degrees in a few peripheral organs and tissues, such as gut, kidney, ovary, pancreas, brown fat, and hair follicles (12, 17, 20). ALK7 is most similar in its serine/threonine kinase domain to ALK4 and ALK5 but is very divergent from all other ALK receptors in its extracellular domain (17). ALK7 signaling, as evaluated with a constitutively active mutant receptor, resembles that of Activin and transforming growth factor β in that it activates Smad2 and Smad3 but not Smad1 (10, 21).
Here we report the generation and initial characterization of mutant mice lacking ALK7. In this study, we have focused our analysis on possible phenotypes resembling Nodal loss of function, as well as possible defects in brain structures where ALK7 mRNA expression is concentrated.
MATERIALS AND METHODS
Targeting of the ALK7 locus.
A murine 129/Sv lambda FIX II genomic phage library (Stratagene) was screened with a 110-bp probe from the ATG domain of rat ALK7 (corresponding to nucleotides −17 to +93). A 14.1-kb clone containing 220 bp with high similarity to the cDNA sequence of the rat ALK7 gene was obtained. A targeting vector was constructed by introducing the bacterial lacZ gene reporter in frame with the ALK7 start codon. A neomycin cassette was inserted in the reverse orientation after the lacZ gene. The targeting construct contained 11,177 bp of homology with the endogenous ALK7 gene, 4,132 bp at the 5′ end and 7,045 bp at the 3′ end (Fig. (Fig.1A).1A). The targeting construct was excised with NotI and SalI prior to electroporation into E14 embryonic stem (ES) cells. After neomycin selection, ES cell colonies were screened for homologous recombination events by Southern blotting with a 795-bp external probe (Fig. (Fig.1B).1B). Nine positive clones were obtained, of which six were injected into C57BL/6 blastocysts and implanted into the uteruses of pseudopregnant females. Three independent ES cell clones gave germ line transmission of the mutation, and a total of 15 chimeric mice were obtained. Chimeric males corresponding to two different ES cell clones were crossed to either Sv129 or C57BL/6 females. No phenotypic differences were found between mutant animals derived from either ES cell clone. Of note is that earlier attempts to target the kinase and GS domains of ALK7 failed to yield homologous recombination events after screening of more than 2,200 ES cell colonies.
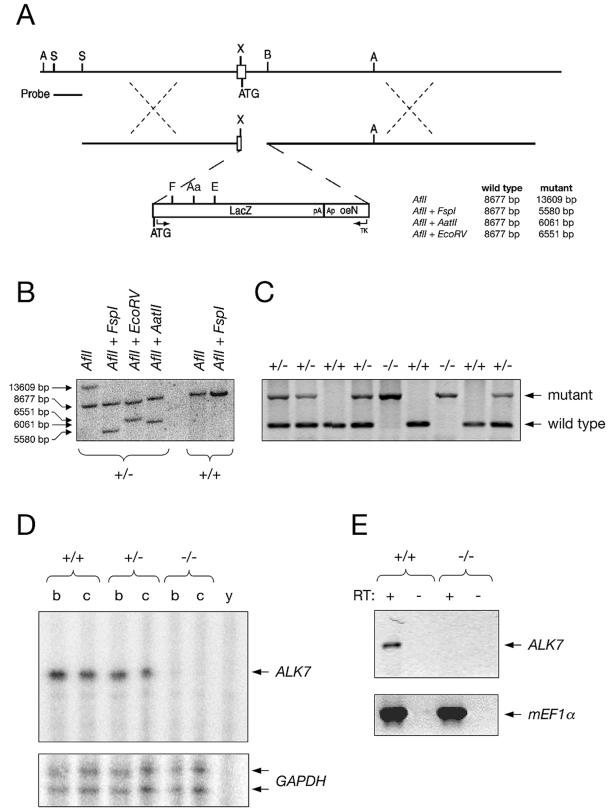
Generation of ALK7 mutant mice by homologous recombination in ES cells. (A) Diagrams of the targeted locus and construct used for the generation of ALK7 null mice. The external probe used for Southern blotting analysis is indicated. See Materials and Methods for details. A, AflII; Aa, AatII; E, EcoRV; F, FspI; S, StuI, X, XmaI; B, BspEI. Fragment sizes expected after the indicated restriction enzyme cleavages are noted. (B) Southern blotting analysis of two ES cell clones with the indicated restriction enzymes. One of the clones (+/−) carries a targeted allele. (C) PCR analysis of tail DNA from the progeny of an intercross between two heterozygous mutant mice. Bands corresponding to the mutant and wild-type alleles are indicated. (D) RNase protection assay of ALK7 mRNA expression in adult brain (b) and cerebellum (c) of wild-type (+/+), heterozygous (+/−), and homozygous (−/−) ALK7 mutant mice. y, yeast tRNA control. The signal obtained with the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe added to the reaction mixture (lower panel) was used as loading control. (E) RT-PCR analysis of ALK7 mRNA expression in E8.5 wild-type (+/+) and ALK7 mutant (−/−) embryos. RT, reverse transcriptase; c, control without input cDNA. Expression of the mouse elongation factor 1 alpha (mEF1α) (lower panel) was used as loading control.
Genotyping was done by PCR with tail DNA and the following oligonucleotide primers: ALK7wt_sense, TGGGGGACGAAATCATCAAG; ALK7wt_antisense, GCGCACCTGCACCCCTCCAA; ALK7mut_sense, CGCCCCGGGAACTTCAAAGC; and ALK7mut_antisense, TAACAACCCGTCGGATTCTC. Fifteen of the 51 agouti chimera offspring that were obtained could be genotyped as heterozygous for the ALK7 mutation. Genomic DNA from ES cells and mouse tails was prepared by treatment with lysis buffer (5 mM EDTA, 0.2% [wt/vol] sodium dodecyl sulfate [SDS], 200 mM NaCl, 100 mM Tris-HCl [pH 8.5]; 100 μg of proteinase K per ml), followed by isopropanol precipitation. For Southern blotting, the genomic DNA was cleaved and separated by electrophoresis on a 0.6% agarose gel. The gel was then blotted onto Hybond N+ nylon membranes (Amersham) by overnight alkali transfer according to the manufacturer's instructions. Hybridization was performed in Church buffer (7% [wt/vol] SDS, 1% [wt/vol] bovine serum albumin,10 mM EDTA, 0.5 M phosphate buffer [pH 7.4]) at 65°C with extended hybridization times (at least 1 day of prehybridization and 2 days of hybridization). After hybridization, the membranes were washed in 0.2% (wt/vol) SDS-0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C until they were clean. Bands were visualized and quantified by using a STORM840 PhosphorImager and ImageQuant software (Molecular Dynamics).
RNase protection assay and RT-PCR.
A 324-bp HindIII/BmrI fragment from a mouse ALK7 expressed sequence tag (mouse 1196242 vu65a06.r1 pT7T3D-Pac; American Type Culture Collection) corresponding to residues 277 to 381 from the kinase domain was subcloned into pBS, linearized with HindIII, and transcribed with T3 RNA polymerase to generate antisense RNA probes for RNase protection assay. [α-32P]CTP-labeled probes were hybridized with 10 μg of total RNA according to the instructions of the manufacturer (Ambion Inc.). Protected bands were visualized and quantified by using a STORM840 PhosphorImager and ImageQuant software (Molecular Dynamics).
Total RNA extraction was done with an RNeasy kit (Qiagen), followed by DNase I treatment (Invitrogen) and cDNA synthesis (ProStar first-strand reverse transcription-PCR [RT-PCR] kit; Stratagene). RT-PCR for ALK7 was done by using primer sequences located in different exons (ALK7 transmembrane, ACCGCCAGTGCACATACA; ALK7 kinase, TCGTGCTTCACAGCCA), generating a 623-bp fragment from the intracellular domain of the receptor.
Whole-mount in situ hybridization, skeletal preparations, and immunohistochemistry.
Whole-mount in situ hybridization was performed according to previously published procedures (22), but with proteinase K digestion omitted. The ALK7 probe was synthesized by using 563 bp of the mouse ALK7 gene amplified by PCR with primers TTCAAACTTCACCTGCCAAACG and CAGAGGCAGTAGCATCGTAG, cloned into pCRII, linearized with XbaI, and transcribed with SP6 RNA polymerase. The ALK4 probe has been described previously (9).
Ten-micrometer sections from fresh frozen brain tissue were prepared in a cryostat, mounted on Superfrost slides, and fixed in 4% paraformaldehyde prior to immunostaining. Primary antibodies (Calbindin [Chemicon] and Calretinin [Swant]) were applied overnight at 4°C at a 1:2,000 dilution in phosphate-buffered saline containing 10% goat serum, 0.1% Triton X-100, and 1% bovine serum albumin. Secondary biotin-conjugated anti-rabbit antibody (Vector Labs) was used at a 1:100 dilution in the same buffer for 2 h at room temperature, followed by the peroxidase substrate reaction with the Nove Red kit (Vector Labs). For hematoxylin and eosin stainings, the tissue was processed as described above, followed by staining with hematoxylin (Merck) and 1% eosin Y disodium salt (Sigma) according to standard protocols. Sections were embedded in Permount (Histolab). For radioactive in situ hybridization, sections were processed as described previously (8), using a 50-mer oligonucleotide probe derived from the ectodomain of mouse ALK7. lacZ expression was detected by β-galactosidase histochemistry in sections fixed in glutaraldehyde according to standard procedures. Tissue sections were examined in an Axioskop microscope (Zeiss) with Openlab image analysis software.
Skeletons from newborn mice were stained with Alcian Blue and Alizarin Red according to standard protocols, with the exception that Alcian Blue and Alizarin Red were added at the same time and incubated overnight.
RESULTS
Generation of ALK7 mutant mice by homologous recombination in ES cells.
A null mutation was introduced in the mouse ALK7 locus (Acvr1c) by homologous recombination in ES cells by standard techniques. The targeting construct contained over 11 kb of sequence homologous to the endogenous gene and was designed to introduce the lacZ reporter gene in frame with the ATG start codon of the ALK7 locus (Fig. (Fig.1A).1A). The mutation resulted in the deletion of 75 bp after the ATG start codon in the 3′ end of the first exon and of 664 bp from the 5′ end of the first intron. The correct targeting event was confirmed by restriction analysis and Southern blotting of genomic DNA from electroporated ES cell clones (Fig. (Fig.1B).1B). Three independent ES cell clones gave germ line transmission of the mutation, which was confirmed by PCR analysis of tail DNA (Fig. (Fig.1C).1C). Homozygous mutant mice were generated from two of these ES cell clones; no phenotypic differences were observed between mutant animals derived from either clone. No ALK7 mRNA could be detected in homozygous mutant mice by RNase protection assay (Fig. (Fig.1D)1D) or RT-PCR (Fig. (Fig.1E),1E), and ALK7 immunoreactivity was absent in pancreas from the mutants (not shown), confirming that we had generated a null mutation. In homozygous mutant mice, expression of the lacZ reporter gene could be detected in several organs and tissues that have previously been shown to express ALK7 mRNA, including gut, kidney, pancreas, and hair follicles. In the gut, strong lacZ expression could be detected at embryonic day 18.5 (E18.5) in the outer cell layer of the tips of the villi lining the internal wall of the intestine (Fig. (Fig.2A).2A). In the adult gut, lacZ staining was confined to the crypt, a proliferative region sandwiched between the smooth muscle layer and the intestine mucosa (Fig. (Fig.2B).2B). No lacZ staining was observed in the guts of wild-type animals (Fig. (Fig.2C),2C), and no gross histological abnormalities could be observed in the guts of ALK7 mutant mice compared to the wild type. In addition, glycemia and insulinemia in fasted young adult mutant mice were not significantly different from those in the wild type (data not shown). Intriguingly, no lacZ staining could be detected in the brains of homozygous mutant mice at any stage of development (not shown). Further RNase protection assay and RT-PCR analyses demonstrated a complete absence of lacZ mRNA expression in the brains of homozygous mutant mice (not shown), indicating that the mutation had inactivated the ALK7 locus in this tissue. Although the reason for the discrepancy between brain and other tissues is unknown, it is possible that the 739-bp segment deleted from the first exon and intron of the ALK7 gene contains regulatory sequences that are essential for the expression of this gene in brain.
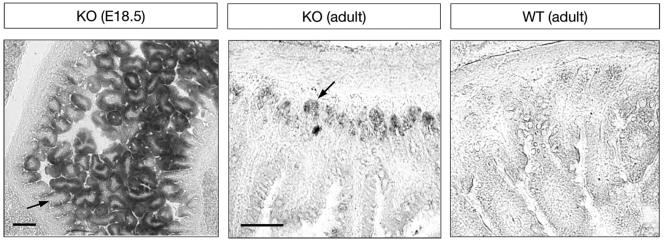
Expression of lacZ from the ALK7 locus recapitulates ALK7 expression in embryonic and adult gut. Shown are sections through E18.5 mutant gut (left) and adult mutant (knockout [KO]) and wild-type (WT) gut processed for β-galactosidase histochemistry. Arrows indicate lacZ-expressing cells in the outer layer of the tips of the villi lining the internal wall of the intestine (E18.5) and in the crypt, a proliferative region located between the smooth muscle layer and the intestine mucosa (adult). No lacZ staining was observed in the guts of wild-type animals. Bars, 100 (left panel) and 50 μm (right panel).
ALK7 mutant mice are viable and fertile.
ALK7−/− mice developed to normal size and weight on a Chaw diet and were fertile in both inbred Sv129 and mixed Sv129 × C57BL/6 backgrounds. All data reported here are from the 129 inbred line unless otherwise indicated. Animals homozygous for the ALK7 mutation were observed at the expected frequency in the progeny of heterozygous intercrosses (Table (Table1),1), indicating that ALK7 function is dispensable for embryogenesis in the mouse, at least in the absence of additional mutations. We investigated possible genetic interactions between ALK7, Nodal, and ALK4 in mouse embryogenesis and viability by crossing homozygous ALK7 mutant mice with heterozygous Nodal and ALK4 mutants. Although both homozygous Nodal and ALK4 mutants die at early stages of embryonic development, heterozygous animals are normal (4, 9). However, both ALK7−/−; Nodal+/− and ALK7−/−; ALK4+/− compound mutants were observed at the expected frequencies at weaning (Tables (Tables22 and and33).
TABLE 1.
Normal frequency of ALK7 knockout mice at weaninga
| Genotype | No. of animals obtained | Frequency (%)
| |
|---|---|---|---|
| Observed | Expected | ||
| ALK7+/+ | 85 | 27.7 | 25 |
| ALK7+/− | 144 | 50.8 | 50 |
| ALK7−/− | 77 | 21.5 | 25 |
| Total | 306 | 100 | 100 |
TABLE 2.
Normal frequency of compound ALK7−/−; Nodal+/− mutant mice at weaninga
| Genotype | No. of animals obtained | Frequency (%)
| |
|---|---|---|---|
| Observed | Expected | ||
| ALK7+/−Nodal+/+ | 25 | 32 | 25 |
| ALK7+/−Nodal+/− | 12 | 16 | 25 |
| ALK7−/−Nodal+/+ | 22 | 28 | 25 |
| ALK7−/−Nodal+/− | 19 | 24 | 25 |
| Total | 78 | 100 | 100 |
TABLE 3.
Normal frequency of compound ALK7−/−; ALK4+/− mutant mice at weaninga
| Genotype | No. of animals obtained | Frequency (%)
| |
|---|---|---|---|
| Observed | Expected | ||
| ALK7+/−; ALK4+/+ | 26 | 28 | 25 |
| ALK7+/−; ALK4+/− | 19 | 20 | 25 |
| ALK7−/−; ALK4+/+ | 25 | 27 | 25 |
| ALK7−/−; ALK4+/− | 23 | 25 | 25 |
| Total | 93 | 100 | 100 |
Overlapping expression of ALK7 and ALK4 in the interdigital regions of developing limb bud.
Whole-mount in situ hybridization of E12 mouse embryos revealed distinct expression of ALK7 mRNA in interdigital regions in developing limb buds, in a pattern that overlapped with that of ALK4 mRNA (Fig. (Fig.3A).3A). Longitudinal sections of wild-type limb buds revealed a distinct patch of ALK7 mRNA expression, while no labeling could be observed in limb buds from ALK7 mutant mice (Fig. (Fig.3B).3B). Interestingly, mice carrying mutations in ActRIIA and ActRIIB have defects in axial vertebral patterning and limb skeletal malformations (14, 15). However, no skeletal abnormalities could be observed in mice lacking ALK7. The numbers of ribs and vertebrae were normal, and no limb or digit malformations, including webbing, could be detected in ALK7−/− mice (Fig. (Fig.3C3C and data not shown).
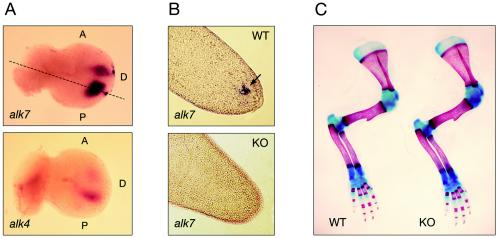
Overlapping expression of ALK7 and ALK4 in interdigital regions and lack of skeletal defects in limbs of ALK7 mutant mice. (A) Expression of ALK7 and ALK4 mRNAs in interdigital regions of limb buds from E12 mouse embryos analyzed by whole-mount in situ hybridization. A, anterior; D, dorsal; P, posterior. (B) Longitudinal sections through the tips of wild-type (WT) and knockout (KO) E12 limb buds (plane indicated by dashed line in panel A) hybridized for ALK7 mRNA. Note lack of ALK7 mRNA signal in mutant limb bud. (C) Alcian Blue and Alizarin Red staining of skeletal structures (bone in red and cartilage in blue) from forelimbs of newborn wild-type and ALK7 mutant (KO) mice.
No left-right patterning defects in mice lacking ALK7.
Given the prominent role of Nodal signaling in left-right patterning, we investigated possible alterations in the positioning of several organs in ALK7 mutant mice. Fifty-nine ALK7−/− newborn mice (35 in inbred Sv129 and 24 in mixed Sv129 × C57BL/6 backgrounds, respectively) were examined for left-right patterning defects, including heart apex, gut looping, pulmonary isomerism, rostro-caudal position of the kidneys, and position of spleen and stomach. No left-right patterning defects were seen in any of the animals examined (Figs. (Figs.4A4A and B and data not shown). In addition, no signs of spleen hypoplasia, a phenotype found in compound ActRIIA ActRIIB mutant mice (11), could be detected in ALK7−/− mice (Fig. (Fig.4C4C).
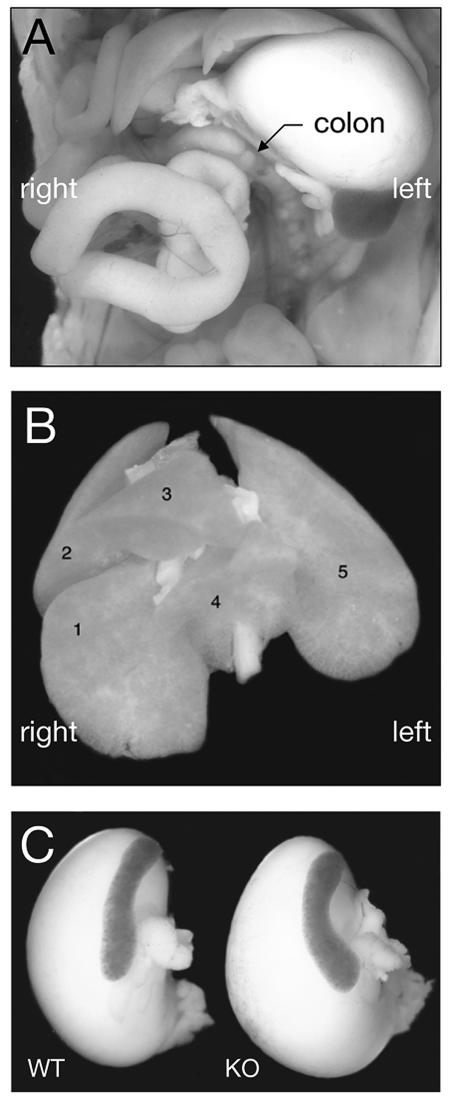
No left-right patterning defects in mice lacking ALK7. (A) Normal left-sided gut looping in newborn ALK7 mutant mice. The colon is indicated. (B) Normal lung lobation in newborn ALK7 mutant mice. Lung lobes are numbered as follows: 1, caudal lung lobe; 2, cranial lung lobe; 3, medial lung lobe; 4, accessory lung lobe; 5, left lung lobe. In right pulmonary isomerism, lobes 1, 2, 3, and 4 are duplicated on the left side. (C) Normal spleen size in newborn ALK7 mutant mice (knockout [KO]).
No histological abnormalities in cerebella, cortices, and hippocampi of ALK7 mutant mice.
Because of the predominant expression of ALK7 in the postnatal central nervous system, particularly in cerebellum, cortex, and hippocampus (12, 17, 20), we also investigated possible histological abnormalities in the brains of adult ALK7 mutant mice. Hematoxylin staining of cryostat sections of adult cerebellum revealed a normal lobation and lamination, with normal thicknesses of molecular and granule cell layers (Fig. (Fig.5A).5A). Purkinje cells, which express high levels of ALK7 mRNA (17), were found in normal numbers, and no defects could be observed in their arborization as assessed by Calbindin immunohistochemistry (Fig. (Fig.5A).5A). Cortical lamination appeared normal in cryosections of ALK7−/− brains stained with hematoxylin (Fig. (Fig.5B).5B). Moreover, no abnormalities in cell migration or positioning could be detected in postnatal day 6 cortex by using Calretinin immunohistochemistry (Fig. (Fig.5B),5B), which predominantly labels inhibitory interneurons in the cortex. Finally, the histological organization of the hippocampus also appeared to be normal in ALK7 mutant mice as assessed by hematoxylin staining (Fig. (Fig.5C5C).
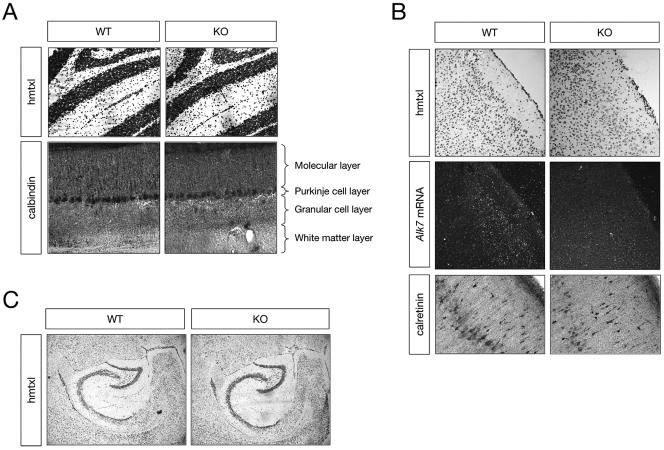
No histological abnormalities in cerebella, cortices, and hippocampi of ALK7 mutant mice. (A) Histological analysis of sections through the adult cerebellum of wild-type (WT) and ALK7 mutant (knockout [KO]) mice. Hematoxylin (hmtxl) and Calbindin stainings are shown. Cerebellar layers are indicated. (B) Histological analysis of sections through the adult cerebral cortex of wild-type and ALK7 mutant mice. Hematoxylin staining, ALK7 mRNA in situ hybridization, and Calretinin immunohistochemistry are shown. (C) Histological analysis of sagittal sections through the adult hippocampus of wild-type and ALK7 mutant mice. Hematoxylin staining is shown.
DISCUSSION
The secreted factor Nodal has been shown to play a crucial role in at least four developmental processes: mesendoderm formation (5, 25), elongation of the primitive streak (13, 19), formation of the anterior head (19), and left-right patterning (13, 15, 23). Nodal has been shown to interact with two type I and two type II receptor serine/threonine kinases, namely, ALK4, ALK7, ActRIIA, and ActRIIB. ALK4 is known to contribute to mesendoderm formation in the mouse (9), but due to the early lethality of ALK4 null mutants, the role of ALK4 in anterior head formation and left-right patterning remains to be demonstrated. On the other hand, all four Nodal-related phenotypes can be phenocopied in ActRIIA ActRIIB compound mutant mice (14, 15, 19). In addition, the Nodal EGF-CFC coreceptors Cripto and Cryptic have also been implicated in left-right patterning by genetic experiments (7, 23).
In this study, we report the generation and initial characterization of mutant mice lacking ALK7. None of the known Nodal-related phenotypes could be found in these mice, suggesting that ALK7 is not an obligatory component of the Nodal signaling pathway, at least during early development. It should be noted, however, that due to possible compensatory effects by ALK4 and other genes in this pathway, the participation of ALK7 in Nodal signaling cannot be completely ruled out at present. The gene network responsible for Nodal function includes several genes in addition to Nodal itself, such as those encoding type I and type II receptors; EGF-CFC coreceptors; Smad2, -3, and -4; and others. Previous genetic studies have indicated that this gene network is robust to different types of perturbations, suggesting that the true contribution of individual network components may emerge only through the generation and analysis of compound mutations. In an effort to address this problem, we have initiated the analysis of intercrosses between ALK7 mutants and mouse lines carrying mutations in other components of this gene network. In the present study, we generated ALK7−/−; Nodal+/− and ALK7−/−; ALK4+/− compound mutants but found no reduction in viability at weaning compared to ALK7−/− or wild-type mice. However, as our analysis was necessarily limited to the use of heterozygous mutants for Nodal and ALK4, it is possible that both of these genes are expressed in sufficient excess during development to preclude haploinsufficiency effects in the absence of ALK7. Only after analysis of further crosses between ALK7 mutants and other mouse lines, such as those carrying mutations in Cripto, ActRIIA, and ActRIIB, will it be possible to fully establish the role of ALK7 in Nodal signaling during mouse development.
The expression pattern of ALK7 during postnatal and adult stages indicates possible functional roles in several tissues and organs. Because some of the most prominent sites of ALK7 expression have been localized to the postnatal and adult central nervous system, we have investigated possible abnormalities in several brain regions where expression of this receptor is particularly concentrated, including cerebellum, cortex, and hippocampus. Our analyses, however, have not revealed major histological defects in any of those structures in ALK7 mutant mice. More detailed anatomical and functional studies, including perhaps stress paradigms, will be required to elucidate the precise function of ALK7 in the mammalian brain. Because of the significant functional overlap with ALK4, future studies on the role of ALK7 in the brain may also require examination of compound mutant animals with conditional mutations in the ALK4 locus. Finally, ALK7 expression has also been detected in a few peripheral organs, including kidney, ovary, and pancreas (12, 17, 20). The mutant animals described in this study open the possibility to investigate possible functions of this receptor and the Nodal signaling pathway in several other tissues and organs.
ADDENDUM IN PROOF
The role of ALK4 in left-right axis determination has been documented in a recent study (Y. Chen, E. Mironova, L. L. Whitaker, L. Edwards, H. J. Yost, and A. F. Ramsdell, Dev. Biol. 268:280-294, 2004).
Acknowledgments
We thank Patrik Ernfors for advice regarding construction of targeting vectors, Elizabeth Robertson for Nodal mutant mice, En Li for ALK4 mutant mice, Dagmar Galter for assistance with radioactive in situ hybridization, and Xiaoli Li for secretarial help.
This work was funded by grants from the Swedish Foundation for Strategic Research, the Swedish Cancer Society (3474-B97-05XBC), and the Swedish Medical Research Council (K99-33X-10908-06C). E.R. was supported during part of this project by an Individual Marie Curie Fellowship.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.24.21.9383-9389.2004
Read article for free, from open access legal sources, via Unpaywall:
https://mcb.asm.org/content/24/21/9383.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.24.21.9383-9389.2004
Article citations
Targeting the activin receptor 1C on CD4+ T cells for cancer immunotherapy.
Oncoimmunology, 13(1):2297503, 08 Jan 2024
Cited by: 0 articles | PMID: 38235319 | PMCID: PMC10793694
Heparan sulfate inhibits transforming growth factor β signaling and functions in cis and in trans to regulate prostate stem/progenitor cell activities.
Glycobiology, 30(6):381-395, 01 May 2020
Cited by: 6 articles | PMID: 31829419 | PMCID: PMC7416765
Control of brown adipose tissue adaptation to nutrient stress by the activin receptor ALK7.
Elife, 9:e54721, 05 May 2020
Cited by: 6 articles | PMID: 32366358 | PMCID: PMC7200161
Downregulation of Nodal inhibits metastatic progression in retinoblastoma.
Acta Neuropathol Commun, 7(1):137, 26 Aug 2019
Cited by: 4 articles | PMID: 31451106 | PMCID: PMC6709548
TGF-β Family Signaling in Early Vertebrate Development.
Cold Spring Harb Perspect Biol, 10(6):a033274, 01 Jun 2018
Cited by: 73 articles | PMID: 28600394 | PMCID: PMC5983195
Review Free full text in Europe PMC
Go to all (28) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Synergistic interaction between Gdf1 and Nodal during anterior axis development.
Dev Biol, 293(2):370-381, 27 Mar 2006
Cited by: 65 articles | PMID: 16564040
Gene-dosage-sensitive genetic interactions between inversus viscerum (iv), nodal, and activin type IIB receptor (ActRIIB) genes in asymmetrical patterning of the visceral organs along the left-right axis.
Dev Dyn, 224(3):279-290, 01 Jul 2002
Cited by: 12 articles | PMID: 12112458
Smad2 role in mesoderm formation, left-right patterning and craniofacial development.
Nature, 393(6687):786-790, 01 Jun 1998
Cited by: 372 articles | PMID: 9655392
A conserved role for the nodal signaling pathway in the establishment of dorso-ventral and left-right axes in deuterostomes.
J Exp Zool B Mol Dev Evol, 310(1):41-53, 01 Jan 2008
Cited by: 33 articles | PMID: 16838294
Review




