Abstract
Background
The gut microbiome in infancy influences immune system maturation, and may have an important impact on allergic disease risk.Objective
We sought to determine how prenatal and early life factors impact the gut microbiome in a relatively large, ethnically diverse study population of infants at age 3 to 6 months, who were enrolled in Vitamin D Antenatal Asthma Reduction Trial, a clinical trial of vitamin D supplementation in pregnancy to prevent asthma and allergies in offspring.Methods
We performed 16S rRNA gene sequencing on 333 infants' stool samples. Microbial diversity was computed using the Shannon index. Factor analysis applied to the top 25 most abundant taxa revealed 4 underlying bacterial coabundance groups; the first dominated by Firmicutes (Lachnospiraceae/Clostridiales), the second by Proteobacteria (Klebsiella/Enterobacter), the third by Bacteriodetes, and the fourth by Veillonella. Scores for coabundance groups were used as outcomes in regression models, with prenatal/birth and demographic characteristics as independent predictors. Multivariate analysis, using all microbial community members, was also conducted.Results
White race/ethnicity was associated with lower diversity but higher Bacteroidetes coabundance scores. C-section birth was associated with higher diversity, but decreased Bacteroidetes coabundance scores. Firmicutes scores were higher for infants born by C-section. Breast-fed infants had lower proportions of Clostridiales. Cord blood vitamin D was linked to increased Lachnobacterium, but decreased Lactococcus.Conclusions
The findings presented here suggest that race, mode of delivery, breast-feeding, and cord blood vitamin D levels are associated with infant gut microbiome composition, with possible long-term implications for immune system modulation and asthma/allergic disease incidence.Free full text

Factors Influencing the Infant Gut Microbiome at Age 3–6 months: Findings from the ethnically diverse Vitamin D Antenatal Asthma Reduction Trial (VDAART)
Abstract
Background
The gut microbiome in infancy influences immune system maturation, and may have an important impact allergic disease risk.
Objective
To determine how prenatal and early life factors impact the gut microbiome in a relatively large, ethnically diverse study population of infants at 3–6 months of age, who were enrolled in VDAART, a clinical trial of vitamin D supplementation in pregnancy to prevent asthma and allergies in offspring.
Methods
We performed 16S rRNA gene sequencing on 333 infants’ stool samples. Microbial diversity was computed using the Shannon Index. Factor analysis applied to the top 25 most abundant taxa revealed four underlying bacterial co-abundance groups; the first dominated by Firmicutes (Lachnospiraceae/ Clostridiales), the second by Proteobacteria (Klebsiella/Enterobacter), the third by Bacteriodetes, and the fourth by Veillonella. Scores for co-abundance groups were used as outcomes in regression models, with prenatal/birth and demographic characteristics as independent predictors. Multivariate analysis, using all microbial community members, was also conducted.
Results
Caucasian race/ethnicity was associated with lower diversity but higher Bacteroidetes co-abundance scores. Caucasian infants had lower Proteobacteria scores as compared to African Americans. C-section birth was associated with higher diversity, but with decreased Bacteroidetes co-abundance scores. Firmicutes and Proteobacteria scores were higher for infants born by C-section. Breastfed infants had lower proportions of Clostridiales. Cord blood vitamin D was linked to increased Lachnobacterium, but decreased Lactococcus.
Conclusions
The findings presented here suggest that race, mode of delivery, breastfeeding and cord blood vitamin D levels are associated with infant gut microbiome composition, with possible long-term implications for immune system modulation and asthma/allergic disease incidence.
Capsule Summary
The findings presented in this study suggest that race, mode of delivery, breastfeeding and cord blood vitamin D levels are associated with infant gut microbiome composition, with potential long-term implications for immune system modulation and asthma/allergic disease incidence.
Introduction
The gut microbiome plays several fundamental roles in maintaining physiological homeostasis. Resident microbes in the human gut discourage invasion by pathogens, process vital nutrients, and harvest energy otherwise inaccessible to human cells. (1, 2) In addition to producing beneficial effects locally within the human gastrointestinal tract, commensal microbiota in the human gut provide, quantitatively, the most important post-natal source of microbial stimulation of the immune system.(3, 4) Stimulation by commensal micro-organisms can influence both innate and adaptive immune system maturation, through tolerogenic dendritic cell activation, Th1 cell differentiation, and the generation and/or expansion of T regulatory cells in peripheral tissues.(5–8) Alterations in gut microbes have been linked to disorders of immune origin, including asthma and allergy. For instance, in a small cohort study, lower diversity of gut microbiota in infancy was linked with increased asthma risk by school age.(9) Lower prevalence of Bifidobacteria have been identified in the stool samples of atopic vs. non-atopic children.(10) Bacteroides antigens (specifically PSA (polysaccharide A)) from B. Fragilis), enhance T regulatory cell function, correct Th1/Th2 imbalance, and protect against autoimmune and inflammatory disease in animal models.(11–13) Lactobacillus strains suppress production of total IgE, and decreased relative abundance of Lactobacillus in the infant gut has been associated with allergic disease development.(14–16) While these studies suggest that specific bacterial taxa in the gut may be important for protection against immune-mediated disorders, the role of entire microbial communities in the proper functioning and maturation of the immune system must also be examined. The infant gut microbiome sets the stage for the composition of gut flora by 3 years of age and onward into adulthood, with potential implications for health both in early and later life. However, before we proceeding with trying to understand the effects of the early infant microbiome on later health, elucidation of the factors that impact on the establishment of this early life microbiome needs to be carried out.
Colonization of the infant gut may begin prior, during or post delivery. Detection of microbes in cord blood and amniotic fluid suggests that the fetal gastrointestinal tract may harbor a limited prenatal microbiome,(17, 18) During birth and in the immediate post-natal period, the infant is exposed to microbes from maternal sources (originating from vaginal, stool and skin microbiota) as well as the environment,(19, 20) Many of these microbes have the potential to rapidly colonize the infant gut. In fact, bacteria are culturable from the stools of newborns within hours after birth.(21) Aspects of an infant’s prenatal, birth and post-natal experience may all contribute to the types of microbial flora that populate the infant gut.
Factors associated with microbial composition and diversity in the infant gut are understudied, and existing reports in this area rely mainly on small, restricted groups of infants (most less than 20 subjects) and diseased infants. As a result, these studies are likely underpowered, and findings from these reports may not be generalizable to a broader population. Our aim in this work was to determine how prenatal and early life factors (circumstances of labor and delivery, gestational age at birth, breastfeeding, and home exposures) impact the gut microbiome in a relatively large, ethnically diverse study population of infants at three to six months of age. While we studied the relationship of these predictors to overall microbial diversity, community level microbiome data (bacterial co-abundance groupings, multivariate analyses, and hierarchical taxonomy) are the main focus of this work. We assessed relative abundance of bacterial taxa by 16S rRNA gene sequencing in stool samples from over three hundred infants born to mothers in the VDAART (Vitamin D Antenatal Asthma Reduction Trial), a clinical trial of prenatal Vitamin D supplementation and outcomes in pregnant women and their offspring.(22)
Materials and Methods
VDAART Clinical Trial and Ancillary Study
For our analyses, we used characteristics and stool samples of infants born to mothers enrolled in the VDAART clinical trial, a two arm, double-blind, placebo controlled, randomized, clinical trial of Vitamin D, to determine whether higher vitamin D intake and levels in the pregnant mother will prevent asthma and allergy in childhood.(22) Pregnant women (who had or whose partner had allergies/asthma) were randomized (n=880) during the first trimester of pregnancy (10–18 weeks) to one of two treatment arms of a 4-year clinical trial: 4000 IU Vitamin D + prenatal vitamins or 400 IU Vitamin D + prenatal vitamins. Forty-one percent of infants in the VDAART clinical trial had a maternal history of asthma, while 23% had a paternal history of asthma (7% of subjects had both a maternal and paternal history of asthma).(22) Of the 810 infants, 333 infants donated stool samples.
Infant Characteristics
Characteristics of infants enrolled in the VDAART trial (including the subset enrolled in the gut microbiome ancillary study) were ascertained through medical records, questionnaires and data obtained at the time of delivery. Information on antibiotics administration during labor, mode of delivery, and gender was gathered at the time of delivery. Gestational age at birth was calculated based on estimated conception date (using data from first ultrasound) and date of delivery. Infant’s racial/ethnic group was derived from initial enrollment data on race/ethnicity of each biological parent. The ‘African American’ race/ethnicity category included infants of African American ancestry only, as well as those with mixed African and Hispanic ancestry. Infants of Native American or Asian descent were included in the ‘Other’ race category. Questionnaires administered to the mother at 6 months after birth were used to assess breastfeeding. We categorized infants into one of three groups: exclusively breastfed, breastfed with some formula supplementation, and exclusively formula fed. Infants in the exclusively breastfed category and those breastfed with some formula supplementation were each compared to exclusively formula fed infants (reference category)‥ Information about the specific timing of solid food introduction was gathered at the 18 month follow-up questionnaire (58% of infants). If these data were missing, we assessed whether solid food had been introduced at the 6 month questionnaire. Additional characteristics (gestational diabetes, infant day care attendance, cigarette smoking during pregnancy, maternal BMI at first obstetrician visit, vitamin D treatment group, cord blood vitamin D level) as well as home characteristics (dog ownership, cat ownership, mold/mildew on home walls or surfaces, and water damage in the home) were also examined as potential predictors of the infant gut microbiome.
Vitamin D Assessment in Cord Blood
Liquid chromatography-mass spectrometry (LC-MS/MS) was performed to measure 25(OH)Vitamin D on 261 infants’ umbilical cord blood samples. See Supplement for details.
Stool sample Collection and Sequencing of Bacterial 16S rRNA gene
Mothers were asked to collect ½ teaspoon of their infant’s stool from a dirty diaper using a tongue depressor, and to store the sample in the freezer, and bring it frozen to the next clinical visit. Stool samples were not collected if the infant had taken antibiotics within the past 7 days. Participants stored their infant’s stools sample in their home freezer for 1–2 days before bringing it to their clinical visit, whereupon the samples were immediately stored in −80° C. A total of 333 infant stool samples (age 3–6 months) were collected. DNA extractions were performed on stool samples, and the bacterial rRNA 16S gene (V3 to V5 hyper-variable regions) were amplified. Pyrosequencing data (Roche 454 Titanium) were produced by the Genome Center (TGI) at Washington University in St. Louis, MO. See supplement for quality filtering and bioinformatic processing of sequences.
Statistical Analyses of Microbiome Data
Diversity Analysis
Shannon diversity index and richness were computed using the VEGAN package in R statistical software. The Shannon diversity index accounts for both the number (richness) and distribution (evenness) of taxa.(23) The Shannon diversity index was normally distributed, and was analyzed as a continuous outcome in univariate linear regression models. A multiple regression model, using statistically significant (p<0.05) univariate characteristics, was then constructed.
Co-abundance Groupings
To capture co-abundance groupings (or factor representations) of bacterial taxa within the infant gut, we computed spearman correlation coefficients for the top 25 taxa identified using the CCREPE (Compositionally Corrected by REnormalization and PErmutation) package in R biostatistical software, and used the correlation matrix for principal . factor analysis (with varimax rotation). Factor loadings were applied to16S sequencing counts, to compute an individual’s factor score for each bacterial co-abundance group. Scores for bacterial co-abundance groupings were used as the outcome variables in linear regression models.
Multivariate Associations
We used MaAsLin (Multivariate Associations with Linear Models) to further investigate predictors associated of the infant gut microbiome, as a way to capture additional taxa not identified as differentially abundant using the other statistical analysis techniques(24, 25) This analysis software performs boosted, additive, linear models to detect associations between one group of data (metadata/the predictors) and another group (microbial abundance of specific taxa. In this multivariate approach, all measured taxa are considered as outcomes simultaneously. Results of a MaAsLin analysis show the association between a specific predictor and abundance of one member of the microbial community.
Hierarchical (Tree) Structure of Microbiome
We computed a taxonomic tree structure for each infant’s gut microbiome sample (down to the genus level) with the HMPtrees (Human Microbiome Project Trees) package in R statistical Software.(26) Infant characteristics showing the strongest and most consistent associations with overall diversity (C-section, Breastfeeding) were used as a basis of comparison for taxonomic tree structure (See Supplementary Materials for methods, results and figures for Hierarchical Tree Comparisons).
Results
Infant Characteristics
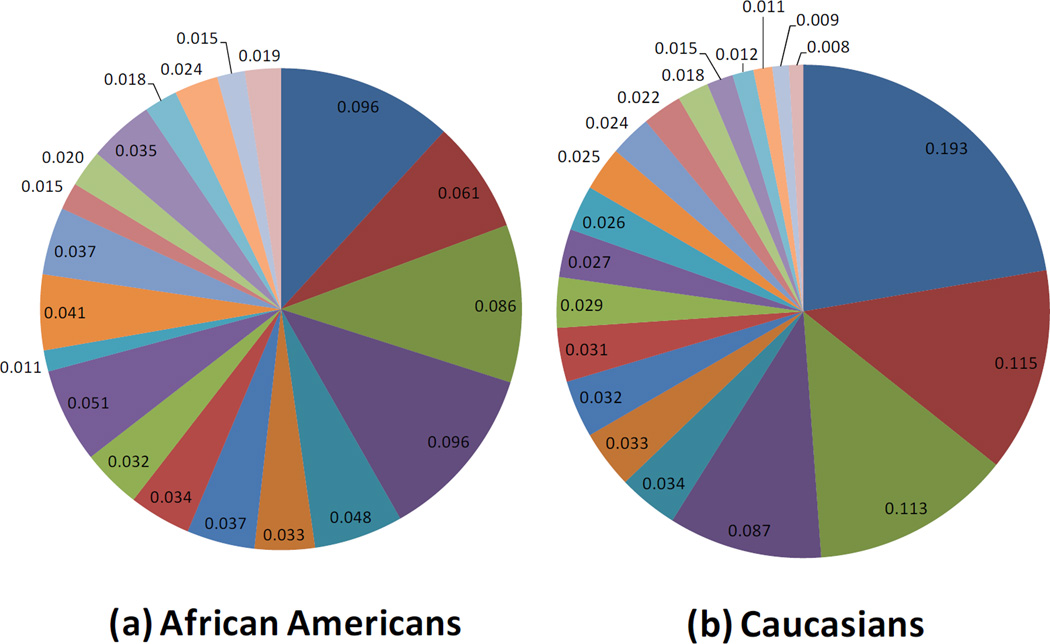
Infants with stool samples showed similar characteristics to infants in the whole VDAART cohort (Table 1). Approximately half of the infants were African American. Over half of all infants were breast fed (55%) in the first six months of life. Approximately one third (33%) of infants were breastfed with formula supplementation, 22% were exclusively breastfed, and 45% were exclusively formula fed at 6 months. One third of the infants were born by C-section. Solid foods had been introduced for 66% of the infants. The mean age at stool sampling was 5 months (20.1 weeks). Use of antibiotics during labor was relatively common (40%). Average relative abundance of the top 20 most abundant microbial taxa observed in the infant gut are shown by race/ethnic group (figure 1), mode of delivery (figure 2), and breast feeding (figure 3).

Mode of Delivery and the Infant Gut Microbiome at Age 3–6 months: Composition of the Top 20 most abundant taxa
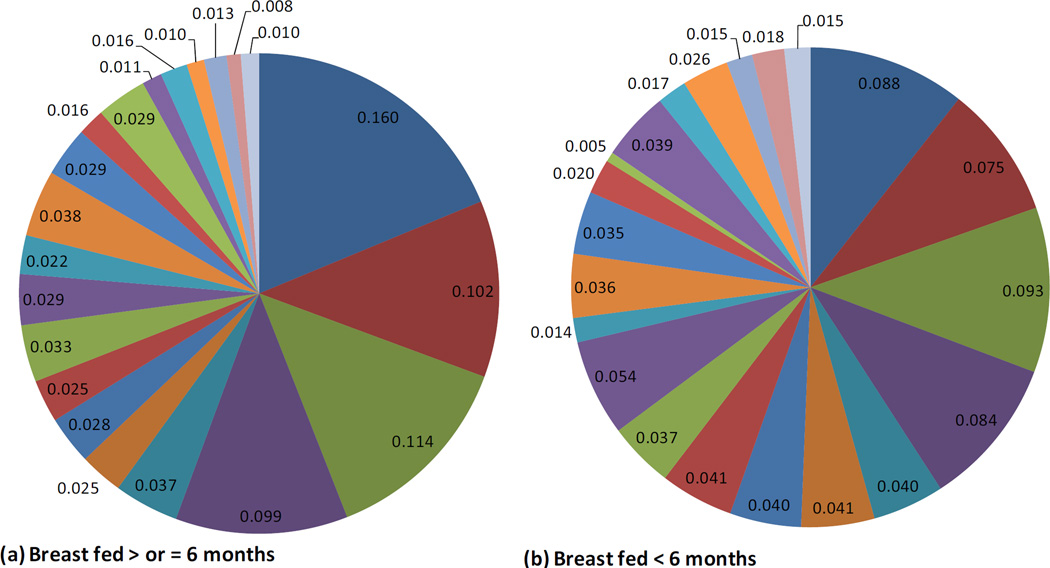

Breastfeeding and the Infant Gut Microbiome at Age 3–6 months: Composition of the Top 20 most abundant taxa
Table 1
Characteristics of Infants with age 3–6 months fecal flora sampling**
| Infants in VDAART Cohort (N=810) | Infants with Fecal Flora Sampling (N=333) | ||
|---|---|---|---|
| N (%) | N (%) | ||
| Gestational Age, weeks (mean, SD) | 39.0 (1.9) | 39.1 (1.9) | |
| Child’s Race | |||
| Black | 350 (43.1) | 149 (44.7) | |
| Hispanic | 112 (13.8) | 42 (12.6) | |
| Caucasian | 215 (26.5) | 92 (27.6) | |
| Other | 133 (16.4) | 50 (15.0) | |
| Male | 422 (52.1) | 180 ( 54.1) | |
| Breast Fed, Exclusively 1st 6 mths* | 126 ( 17.0) | 71 ( 21.7) | |
| Breast Fed and formula supplemented 1st 6 mths* | 287 (38.8) | 107 (32.9) | |
| Formula Fed, Exclusively 1st 6 mths* | 328 (44.2) | 147 (45.4) | |
| C-section birth | 240 (29.6) | 108 ( 32.4) | |
| Maternal antibiotics (during labor)* | 310 (38.5) | 133 ( 40.1) | |
| Infant antibiotics (1st days of life)* | 78 (9.7) | 26 (7.8) | |
| Age at Fecal Flora Sampling, weeks (mean, SD) | NA | 20.1 (4.6) | |
| Cord Blood Vitamin D, ng/ml (mean, SD) | 22.7 (11.9) | ||
Microbial Diversity
The mean Shannon diversity index was 2.2 (0.46 S. D, range = 0.5 to 3.15). Richness (total number of bacterial taxa detected in an infant’s stool sample) ranged from 9 to 55, with a median level of 32. A multiple regression model for Shannon diversity model (including all variables with p<0.05 in the univariate analysis) explained approximately 26% (adj. R2 = 26%) of the diversity level in infant gut (Table 2). Variables associated with the Shannon index had similar relationships to richness (data not shown). Caucasian and other race/ethnicity was associated with decreased diversity as compared to African American infants. C-section delivery was associated with higher bacterial diversity in the infant gut. Breastfeeding (both exclusive and breastfeeding with formula supplementation) predicted lower diversity, although the magnitude of the association was greater for exclusive breastfeeding. Age at fecal flora sampling had a small, but statistically significant effect on diversity. Solid food intake was associated with increased diversity in the infant gut. Vitamin D treatment group and antibiotic administration to the infant within days after birth were not associated with microbial diversity. Dog and cat ownership were associated with reduced diversity in univariate, but not adjusted models.
Table 2
Predictors of the infant gut microbiome at age 3–6 months (n=325)
| Overall Diversity (Shannon Diversity Index) | ||||||
|---|---|---|---|---|---|---|
| Univariate Associations | Multiple Regression Model | |||||
| Beta | (P value) | Beta | (P value) | Model adj. R2 | ||
| Child’s Race* | 26% | |||||
| Hispanic | −0.232 | 0.003 | −0.09 | 0.26 | ||
| Caucasian | −0.351 | <0.0001 | −0.15 | 0.02 | ||
| Other | −0.258 | 0.004 | −0.10 | 0.18 | ||
| Breast Fed exclusively** | −0.531 | <0.0001 | −0.45 | <0.001 | ||
| Breast Fed supplemented with formula** | −0.185 | 0.0005 | −0.13 | 0.02 | ||
| Solid Foods Introduced (Y/N) | 0.127 | 0.02 | 0.11 | 0.04 | ||
| C-section (Y/N) | 0.146 | 0.006 | 0.11 | 0.04 | ||
| Maternal Abx labor (Y/N) | 0.105 | 0.04 | 0.008 | 0.85 | ||
| Age at Fecal Sampling (weeks) | 0.013 | 0.02 | 0.009 | 0.11 | ||
| Maternal BMI at first Ob visit | 0.012 | 0.0009 | 0.006 | 0.10 | ||
| Pet dog (Y/N) | −0.140 | 0.01 | −0.03 | 0.50 | ||
| Pet cat (Y/N) | −0.153 | 0.04 | −0.05 | 0.54 | ||
| Child Abx in first days of life (Y/N) | 0.078 | 0.40 | ||||
| Treatment Group | 0.026 | 0.61 | ||||
| Smoking During Pregnancy (Y/N) | 0.162 | 0.39 | ||||
| Gestational Diabetes (Y/N) | 0.205 | 0.14 | ||||
| Gestational Age (weeks) | −0.02 | 0.08 | ||||
| Female (vs. Male) | −0.018 | 0.73 | ||||
| Day care attendance (Y/N) | 0.021 | 0.76 | ||||
| Mold/Mildew on walls (Y/N) | −0.087 | 0.39 | ||||
| Water damage in home (Y/N) | −0.086 | 0.39 | ||||
| Cord Blood Vitamin D level (ng/ml) | −0.004 | 0.06 | ||||
Because the infant microbiome in the first 6 months undergoes rapid changes, we examined the possibility that age at fecal flora sampling modifies the effects of mode or delivery or breastfeeding on infant gut diversity. Tests for interactions between C-section and age of fecal flora sampling and breastfeeding and age at fecal flora sampling were not significant, (p=0.72 and p=0.53, respectively) in models for infant gut microbial diversity. A stratified analysis (Supplemental table 8) by age at fecal flora sampling (categorized at age 12–16 weeks, >16weeks to 20 weeks and > 20 weeks) demonstrates that mode of delivery, breastfeeding and race all show similar magnitudes of association with infant gut diversity across this relatively wide age range. Lack of effect modification by fecal flora sampling age suggests that combining all age groups within the same analyses is acceptable for this study.
C-section delivery was associated with a reduced odds of breastfeeding in the first 6 months of life (OR=0.57, 95% CI 0.36 to 0.91). We tested for an interaction between these two variables in our multiple regression model for infant gut diversity. We did not detect a significant interaction (p=0.84 for interaction term) suggesting that the impact of breastfeeding on infant gut diversity does not vary by mode of delivery.
Bacterial Co-Abundance Groupings
Factor analysis of the top 25 most abundant bacterial taxa in the infant gut (Supplemental Table 1) revealed four factors associated with bacterial co-abundance (a four factor solution was identified based on a scree plot). The first factor captured co-abundance of Lachnospiraceae genera (positive loadings 0.4 to 0.8), along with Unclassified Clostridiales (loading 0.65), both of which are in the Firmicutes phylum. Bacteroides was negatively associated with this Lachnospiraceae/U. Clostridiales co-abundance grouping (loading = −0.31). For the second factor positive loadings for Klebsiella, Enterobacter, and Unclassified Enterobacteriaceae were most substantial (0.46, 0.86, and 0.76, respectively). The third factor was defined mainly by Bacteroides abundance (0.70 for factor loading), with negative factor loadings for Escherichia/Shigella (−0.44) and Bifidobacterium (−0.47). The fourth factor showed substantial positive loadings for Veillonella and Clostridium. For ease of interpretability, these co-abundance groupings (factors) are labeled in the following way: Factor 1: Lachnospiraceae (+), U. Clostridiales (+), Factor 2: Klebsiella (+), Enterobacter (+), Factor 3: Bacteroides (+), Escherichia/Shigella (−), Bifidobacterium (−), and Factor 4: Veillonella (+) and Clostridium (+).
In Table 3, factor scores for these four bacterial co-abundance groups were used as outcomes in multiple regression models with infant characteristics as independent predictors. C-section delivery was associated with higher co-abundance scores for the Factor 1 and Factor 2, but with lower scores of Factor 3. Breastfeeding (both exclusive and formula supplemented breastfeeding) was linked to lower Factor 1 scores, but only exclusive breastfeeding was associated with reduced Factor 2 scores‥ Antibiotics administration during labor was associated with higher scores for Factor 1, but with lower scores for Factor 3. Race/ethnicity had an impact on three out of the four bacterial co-abundance scores. Caucasian infants demonstrated higher Factor 3 and lower Factor 2 scores as compared to African Americans. All racial groups (Caucasian, Hispanic, and Other) had higher Factor 4 scores as compared to African American infants. Cat ownership was associated with higher Factor 3 scores and lower Factor 4 scores. Solid food intake showed a borderline statistically significant association with increased Factor 3 scores. Cord blood vitamin D levels (ng/ml) were associated with higher scores for Factor 1. (Table 4).
Table 3
Infant Predictors of Bacterial Co-abundance Factor Scores (Linear Regression Results, n=325)
| Multiple Regression Models for Bacterial Co-Abundance Groupings | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Dominant Phyla for Grouping Dominant Genera | Firmicutes Lachnospiraceae (+) Clostridales U. (+) Factor 1 Score | Proteobacteria Klebsiella (+) Enterobacter (+) Factor 2 Score | Bacteroidetes Bacteroides (+) Shigella (−) Bifidobacterium (−) Factor 3 Score | Firmicutes Veillonella (+) Clostridium (+) Factor 4 Score | |||||
| Infant Characteristics: | Beta | (P value) | Beta | (P value) | Beta | (P value) | Beta | (P value) | |
| Gestational Age (weeks) | −0.03 | 0.43 | −0.02 | 0.37 | 0.001 | 0.97 | −0.02 | 0.37 | |
| Child’s Race | |||||||||
| Hispanic | 0.06 | 0.70 | −0.23 | 0.09 | 0.07 | 0.69 | 0.54 | 0.003 | |
| Caucasian | 0.08 | 0.56 | −0.14 | 0.20 | 0.29 | 0.05 | 0.42 | 0.002 | |
| Other | 0.10 | 0.47 | 0.05 | 0.64 | −0.05 | 0.78 | 0.31 | 0.04 | |
| Male (vs. Female) | −0.31 | 0.002 | 0.04 | 0.62 | 0.06 | 0.55 | 0.02 | 0.96 | |
| Breast Fed exclusively** | −0.85 | <0.001 | −0.27 | 0.02 | −0.23 | 0.17 | 0.22 | 0.07 | |
| Breast Fed supplemented with formula** | −0.28 | 0.02 | 0.01 | 0.90 | 0.01 | 0.83 | 0.08 | 0.50 | |
| Solid Foods Introduced (Y/N) | 0.08 | 0.47 | 0.15 | 0.13 | 0.23 | 0.07 | −0.003 | 0.90 | |
| C-section (Y/N) | 0.27 | 0.02 | 0.09 | 0.33 | −0.29 | 0.01 | 0.14 | 0.05 | |
| Maternal Abx labor (Y/N) | 0.19 | 0.07 | −0.04 | 0.64 | −0.22 | 0.04 | −0.006 | 0.71 | |
| Infant Abx, 1st days life (Y/N) | −0.02 | 0.95 | −0.05 | 0.80 | 0.18 | 0.42 | −0.35 | 0.06 | |
| Age at fecal flora sampling (weeks) | 0.01 | 0.34 | −0.02 | 0.06 | −0.009 | 0.48 | 0.004 | 0.71 | |
| Dog Ownership (Y/N) | −0.01 | 0.91 | −0.14 | 0.16 | −0.03 | 0.80 | −0.09 | 0.40 | |
| Cat Ownership (Y/N) | 0.29 | 0.07 | −0.21 | 0.11 | 0.50 | 0.003 | −0.31 | 0.03 | |
| Daycare Attendance (Y/N) | −0.02 | 0.91 | −0.12 | 0.31 | 0.02 | 0.90 | 0.13 | 0.31 | |
| Maternal BMI | 0.003 | 0.69 | −0.006 | 0.26 | −0.002 | 0.74 | 0.004 | 0.56 | |
Beta and p values bolded if p<0.05
Table 4
Cord Blood Vitamin D Bacterial Co-abundance Factor Scores (Linear Regression Results)* (n=261)
| Bacterial Co-Abundance Groupings | ||||||||
|---|---|---|---|---|---|---|---|---|
| Dominant Phyla for Grouping Dominant Genera | Firmicutes Lachnospiraceae (+) Clostridales U. (+) Factor 1 Score | Proteobacteria Klebsiella (+) Enterobacter (+) Factor 2 Score | Bacteroidetes Bacteroides (+) Shigella (−) Bifidobacterium (−) Factor 3 Score | Firmicutes Veillonella (+) Clostridium (+) Factor 4 Score | ||||
| Beta | (P value) | Beta | (P value) | Beta | (P value) | Beta | (P value) | |
| Cord Blood Vitamin D (ng/ml) | 0.014 | 0.004 | −0.003 | 0.40 | 0.007 | 0.19 | −0.006 | 0.19 |
Some prenatal characteristics (smoking in pregnancy, gestational diabetes) and environmental variables (water damage, visible mold/mildew in the home), occurred at low frequency in our population (1–5%); therefore we did not have the power to examine the relationships between these variables and bacterial co-abundance groupings in the infant gut microbiome.
Multivariate Analysis (MaAsLin) Results
Multivariate analysis (adjusted for all covariates) showed similar findings, overall, to other statistical analysis methods used. For breast feeding as a predictor, MaAsLin (Supplemental Table 2a) helped to identify decreased abundance of taxa within the family Clostridales (Clostridium, Coprococcus, , Ruminococcus and Eubacterium, Lachnospiraceae Dorea, Lachnospiraceae Blautia), showing considerable overlap with the lower Lachnospiracaceae/ Clostridales bacterial co-abundance scores observed for breastfeeding. These relationships with specific taxa in our multivariate analysis were observed only for exclusive breastfeeding (Supplemental Table 2a), but not for breastfeeding with formula supplementation (Supplemental Table 2b). While breastfeeding was associated with increased lactobacillus (p<0.05) and a trend toward increased Bifidobacterium in univariate analyses, these associations were not observed in the multivariate models. For C-section (Supplemental Table 3), the strongest findings by multivariate analysis were for decreased Bacteroides and Parabacteroides abundance. With regard to racial comparisons, Megasphaera and Lactococcus were significantly higher in African Americans (Supplemental Table 4). Cord blood Vitamin D was associated with increased Lachnobacterium (re-iterating the finding of increased Lachnospiracacea/Clostridales co-abundance scores with higher Vitamin D). Additionally, decreased Lactococcus was observed for higher cord blood vitamin D (Supplemental Table 5). Although some associations were observed for cat ownership and bacterial co-abundance grouping analysis (Table 3), these findings were not reflected in the multivariate analysis results. None of the taxa showed significant associations with pet ownership after adjustment for multiple comparisons (Supplemental Table 6 and 7).
Discussion
In the present study, we identified predictors of the infant gut microbiome at three to six months of age, in one of the largest, most ethnically diverse study populations to date. In addition, compared with other studies investigating the microbiome in early infancy, this is a relatively healthy cohort of infants. In this early life window, the infant’s gut microbiota is plastic and undergoing rapid change, with potential implications for innate and adaptive immune system maturation, and for future overall health. Therefore, it is critical to understand the factors associated with microbial communities populating the infant gut during this key developmental time period. Through the use of complimentary analytical methodologies, we examined the relationships between infant characteristics and gut microbiota at age 3–6 months. As a first step, we identified predictors of microbial diversity. Next, we determined specific compositional differences in gut microbiota by infant characteristic, using bacterial co-abundance groupings (based on the top 25 most abundant taxa). Lastly, we employed multivariate analysis to assess the relationship between multiple independent predictors and the relative abundance of all detectable taxa simultaneously.
Overall mean diversity within the infant gut microbiome was consistent with other studies of fecal flora within this age range.(27, 28) On average, the infant gut microbiota were dominated by gram negative anaerobes (Bacteroides, Veillonella), with significant representations of gram positive anaerobes (Bifidobacterium) and facultative microorganisms (Eshcerichia/Shigella). Extraction of four factors from 16S sequence data across all infant’s stool samples revealed a latent phylum-specific substructure of microbiota in the infant gut that demonstrated the following relationships among taxa: (1) Lachnospiracaceae co-occur with Clostridiales (both Firmicutes), (2) Klebsiella and Enterobacteriaceae (both Proteobacteria) show co-abundance, (3) Bacteroides (Bacteroidetes) is associated with decreased abundance Bifidobacterium (Actinobacteria) and Escherichia/Shigella (Proteobacteria), and lastly (4) Veillonella and Clostridium (both Firmicutes) show co-abundance. This latent substructure of the infant gut microbiome, identified using factor analysis, was associated with four predictors: mode of delivery, breastfeeding, race and Vitamin D. Furthermore, these groupings capture taxa with a prior associations to disease status.
Mode of Delivery
Mode of delivery was consistently one of the most important factors associated with the infant gut microbome in our study. The relationship between delivery mode and the microbial composition of the infant gut likely originates from different source contributions of microbiota (infants born vaginally are exposed to both maternal fecal and vaginal flora, while infants born by C-section may have increased colonization from maternal skin or environmental microbes).(29) Surprisingly, infants born by C-section had higher Shannon indices than infants born vaginally. On the surface, this finding is counter-intuitive, and conflicts with other reports of decreased gut diversity with C-section.(30) (31) It may be that the increased evenness of the overall taxonomic profiles in C-section born infants (as a result of lower predominance of Bacteroides) may partially explain the relationship between C-section and increased Shannon diversity.
The relationship between C-section delivery and lower abundance of Bacteroides was consistent across all of our statistical models (bacterial co-abundance, multivariate analyses, and hierarchical tree comparison (Supplementary Materials)), and has been reported previously in other studies.(29, 29, 30, 32) However, in addition to this association, our bacterial co-abundance analysis also suggests that with decreased Bacteroides abundance, Proteobacteria (Escherichia, Enterobacteriaceae, and Klebsiella) thrive more in the gut microbiota of infants born by C-section, replacing Bacteroidetes as the dominant gram negative phylum. This alternative gram negative profile (dominant Proteobacteria with minimal Bacteroides) may have important consequences for immune stimulation by PAMPs (i.e. heterogeneity of endotoxin TLR4 stimulation), or even potential implications for microbial function in the gut. Jakobsson et al have demonstrated that lower Bacteroidetes abundance in C-section born infants is associated with lower levels of Th1-associated chemokines (CXCL10 and CXCL11) during infancy, as well as reduced Th1 responses, which may ultimately increase risk of asthma or allergic disease.(30)
Breastfeeding
Breastfeeding was another independent predictor linked to altered gut microbiota across different types of microbiome analyses Breastfeeding has the potential to shape the gut microbiome as a source of prebiotics, or by directly interacting with microbes via its IgA content.(33) Breastfeeding was associated with reduced diversity (potentially driven by reduced evenness of taxa), a finding that is consistent with other reports.(32, 34) We also observed a trend for increased Bifidobacterium with breastfeeding, which may have resulted from increased availability of prebiotics in breast milk (oligosaccharides)(35) that favor proliferation of this taxa. Interestingly, breastfeeding was associated with a lower score for the main Firmicutes co-abundance grouping (representing Lachnospiraceae and Clostridiales), and in multivariate analysis, lower abundance of specific genera within Clostridiales (Clostridium, Ruminococcus, Coprococcus, Eubacterium). Lower abundance of Clostridia in breastfed infants has also been reported in much smaller, more homogenous study populations and is replicated here in a much larger, ethnically diverse cohort(31, 32) In emerging studies, Clostridia (C. Difficle) abundance in the infant gut at age 1 month has been linked to wheeze and sensitization in early life, and total Clostridia abundance is associated with atopic dermatitis risk. (36, 37) If the protective effects of breast feeding on allergic disease risk are indeed mediated by the gut microbiome, Clostridia may have an important role. Exclusive, rather than formula supplemented, breastfeeding may also be crucial, as our bacterial co-abundance grouping analysis showed the greatest impact on Clostridales reduction for exclusive breastfeeding, and our multivariate findings showed reductions in Clostridiales for only those infants who were exclusively breast fed.
Race/Ethnicity
Race/ethnicity was the third main factor associated with alterations in the infant gut microbiome in our work and, as a predictor of gut microbiota, is probably the least understood. Gut microbiota may vary by race due to differences in dietary patterns, personal care product use, or host genetics. Caucasian infants showed reduced overall diversity of the gut microbiome but also demonstrated higher abundance of Bacteroides, In keeping with the overall negative correlations we have seen for Bacteroides and Proteobacteria, Caucasian infants demonstrated lower scores for the Proteobacteria bacterial co-abundance group, represented mainly by Enterobacteriaceae (Klebsiella and Enterobacter). Lower abundance of taxa with potential protective benefits (i.e. Bacteroides) in the microbiome of minority children could, at least partially, explain the increased asthma incidence and morbidity in these populations. Further investigation is needed to uncover why infant gut microbial communites vary by race. While these relationships were observed after adjusting for potential confounding due to factors such as breastfeeding or mode of delivery, there is almost certainly residual confounding by factors tightly associated with race (i.e. factors linked to socioeconomic status), that could not be accounted for in these analyses. For example, race was associated with breast-feeding type (Hispanics and Caucasians were more likely to exclusively breastfeed than African American subjects), suggesting that race could also be linked to other unmeasured infant dietary trends.
Cord Blood Vitamin D
Our findings suggest that vitamin D levels in utero (and shortly after birth) could influence the microbial community that colonizes the infant gut. We observed higher levels of Lachnospiraceae/U. Clostridales in infants with higher cord blood vitamin D, and multivariate analysis also revealed an increased abundance of Lachnobacterium, but a decreased abundance of Lactococcus. While very few studies have examined the impact of vitamin D on the developing gut microbiome, data from animal models suggests that vitamin D may influence microbial community composition. In vitamin D receptor (VDR) knockout mice, Lactobacillus was depleted, whereas Clostridium and Bacteroides were enriched.(38) In an animal model of ulcerative colitis (where mice lacked the enzyme to produce the active form of vitamin D (1,25 (OH) 2D3)), supplementation with either the active form of vitamin D (1,25 (OH) 2D3) or antibiotic treatment ameliorated the disease, suggesting that microbial dysbosis may arise when vitamin D is insufficient, thereby promoting severe inflammation in the gut.(39) Future work is needed to understand whether vitamin D plays a key role in dictating which microbes colonize the gut, or if it potentially modifies the effects of the microbiome, without directly determining community composition.
Conclusion
Until further translational studies are conducted, the precise role of altered gut communities on immune system function and risk for allergic disease will remain unknown. In our study, infant characteristics that may potentially increase risk of asthma and allergic disease development (African American Race/ethnicity, C-section birth, and formula feeding were associated with alterations in the infant gut microbiome profile. Both African American race/ethnicity and C-section were independent predictors of lower levels of Bacteroides, and higher levels of Proteobacteria (Klebsiella, Enterobacteriaceae) in the infant gut. Formula feeding was linked to higher levels of Clostridia in the infant gut, which may be associated with increased risk of allergic disease. Future work is required to uncover how alterations of gut microbial communities influence diseases of immune origin, and will necessitate functional genetic and possibly metabolomic approaches to complete our understanding of how the gut microbiome mediates asthma and allergic disease risk. Greater taxonomic resolution (down to species and/or strain level if possible) will also be critical. Lastly, it is important to note that this work is a cross sectional study of the infant gut microbiome conducted at 3–6 months of age, when the microbiome is still highly unstable. Longitudinal analysis to assess the impact of early life factors on the developing microbiome over time is needed, and, with respect to health outcomes analyses, repeated measures in infancy/early childhood will likely give a more definitive test of the microbiome’s relationship to asthma and allergy.
Acknowledgments
The VDAART investigators would like to acknowledge the enormous contributions to the study by Dr. Robert Strunk, an esteemed colleague and friend. Dr. Strunk was instrumental in the design, implementation, and success of the VDAART clinical trial and continuation study. We are forever grateful for his guidance, input, and collegiality.
Funding Sources: U01HL091528, R01HL108818
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Full text links
Read article at publisher's site: https://doi.org/10.1016/j.jaci.2016.08.045
Read article for free, from open access legal sources, via Unpaywall:
http://www.jacionline.org/article/S0091674916311344/pdf
Citations & impact
Impact metrics
Article citations
CTPAD: an interactive web application for comprehensive transcriptomic profiling in allergic diseases.
J Transl Med, 22(1):935, 14 Oct 2024
Cited by: 0 articles | PMID: 39402558 | PMCID: PMC11475762
Randomized control trial of moderate dose vitamin D alters microbiota stability and metabolite networks in healthy adults.
Microbiol Spectr, 12(10):e0008324, 27 Aug 2024
Cited by: 0 articles | PMID: 39189761 | PMCID: PMC11448053
Development of systemic and mucosal immune responses against gut microbiota in early life and implications for the onset of allergies.
Front Allergy, 5:1439303, 17 Jul 2024
Cited by: 1 article | PMID: 39086886 | PMCID: PMC11288972
Review Free full text in Europe PMC
Vitamin D supplementation for women during pregnancy.
Cochrane Database Syst Rev, 7:CD008873, 30 Jul 2024
Cited by: 1 article | PMID: 39077939
Review
Maternal vitamin D status during pregnancy and infant's gut microbiota: a prospective cohort study.
Front Nutr, 11:1428356, 29 Jul 2024
Cited by: 0 articles | PMID: 39135559 | PMCID: PMC11317374
Go to all (91) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Association of the Infant Gut Microbiome With Early Childhood Neurodevelopmental Outcomes: An Ancillary Study to the VDAART Randomized Clinical Trial.
JAMA Netw Open, 2(3):e190905, 01 Mar 2019
Cited by: 53 articles | PMID: 30901046 | PMCID: PMC6583279
Development of the Microbiota and Associations With Birth Mode, Diet, and Atopic Disorders in a Longitudinal Analysis of Stool Samples, Collected From Infancy Through Early Childhood.
Gastroenterology, 158(6):1584-1596, 18 Jan 2020
Cited by: 123 articles | PMID: 31958431
Distinguishable Influence of the Delivery Mode, Feeding Pattern, and Infant Sex on Dynamic Alterations in the Intestinal Microbiota in the First Year of Life.
Microb Ecol, 86(3):1799-1813, 03 Mar 2023
Cited by: 3 articles | PMID: 36864279
Influence of mode of delivery on infant gut microbiota composition: a pilot study.
J Obstet Gynaecol, 44(1):2368829, 24 Jun 2024
Cited by: 1 article | PMID: 38913773
Funding
Funders who supported this work.
NHLBI NIH HHS (4)
Grant ID: R01 HL091528
Grant ID: R01 HL108818
Grant ID: R01 HL091075
Grant ID: U01 HL091528