
| PMC full text: |
|
FIG 4
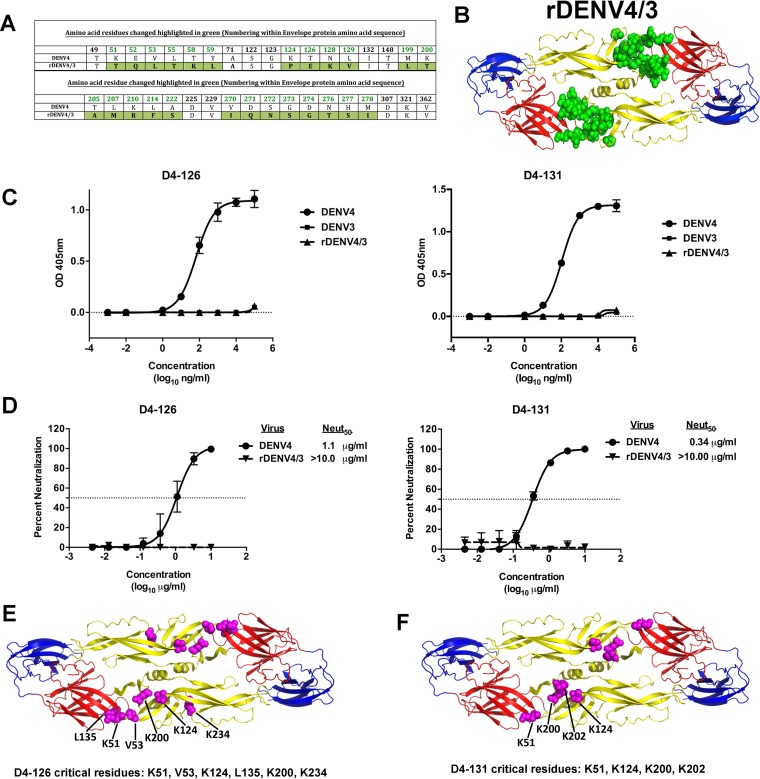
The EDI/II hinge region is important for the binding and neutralization of DENV4 by hMAbs D4-126 and D4-131. (A) Amino acid alignment of DENV4 and rDENV4/3 E protein sequence in the EDI/II hinge; EDII regions with mutated residues are colored in green. (B) Recombinant DENV4 containing EDI/II hinge; EDII residues from DENV3 (rDENV4/3) used to map the epitopes of hMAbs D4-126 and D4-131 are shown. The figure depicts a model of the DENV4 E protein dimer, with the mutated residues colored in green. (C) Binding of hMAbs D4-126 and D4-131 to DENV4, DENV3, or rDENV4/3. (D) DENV4 and rDENV4/3 neutralization by hMAbs D4-126 and D4-131. Results are from 2 technical replicates in an experiment. Each point represents the mean from the two replicates, and the error bars depict the standard deviations. Mapping of the critical residues for D4-126 and D4-131 was performed by shotgun mutagenesis by screening for loss of MAb binding to a DENV4 alanine scan mutation array. Critical residues are visualized as magenta spheres on the DENV E dimer structure (PDB identifier [ID] 1OAN). (E) Six critical residues were identified for D4-126 in the E protein DII and DI/II hinge region. (F) Four critical residues were identified for D4-131 in the E protein DII and DI/II hinge region.





